Abstract
Background
Alzheimer disease (AD) is a genetically complex disorder. Mutations in 3 genes, presenilin 1, amyloid precursor protein, and presenilin 2, lead to early-onset familial AD in rare families with onset of disease occurring prior to age 65. Specific polymorphisms in apolipoprotein E are associated with the more common, late-onset AD occurring after age 65. In this review, we discuss current advances in AD genetics, the implications of the known AD genes, presenilin 1, presenilin 2, amyloid precursor protein, and apolipoprotein E, and other possible genes on the clinical diagnosis, treatment, and genetic counseling of patients and families with early- and late-onset AD.
Review Summary
In addition to the mutations in 4 known genes associated with AD, mutations in other genes may be implicated in the pathogenesis of the disease. Most recently, 2 different research groups have reported genetic association between 2 genes, sortilin-related receptor and GAB2, and AD. These associations have not changed the diagnostic and medical management of AD.
Conclusions
New research in the genetics of AD have implicated novel genes as having a role in the disease, but these findings have not been replicated nor have specific disease causing mutations been identified. To date, clinical genetic testing is limited to familial early-onset disease for symptomatic individuals and asymptomatic relatives and, although not recommended, amyloid precursor protein apolipoprotein E testing as an adjunct to diagnosis of symptomatic individuals.
Keywords: alzheimer disease, genetics, genetic testing, genetic counseling
Alzheimer disease (AD) is the most common form of age-related Adementia with more than 5 million Americans currently affected.1 Although the majority of cases occur in individuals age 65 or older, the 2007 AD Facts and Figures published by the Alzheimer’s Association estimates 500,000 people have early-onset AD (EOAD) or another dementia with symptoms appearing before the age of 65 years. One out of 8 people age 65 and older and nearly 1 out of every 2 over age 85 have AD. AD is the seventh leading cause of death in the United States.
A major focus of AD research has been to understand the genetic etiology of AD and its relationship to AD neuropathology. The key neuropathological features of AD are abundant neurofibrillary tangles composed of tau protein and senile plaques made of β-amyloid (Aβ). Over 2 decades of genetic research has resulted in a better understanding of the pathway leading to the accumulation of insoluble protein, Aβ42. The accumulation of Aβ42 is considered a central component in the pathogenesis of AD and has been associated with the 3 autosomal dominant, deterministic genes known to be involved in EOAD, presenilin 1 (PSEN1), presenilin 2 (PSEN2), and amyloid precursor protein (APP).2–8 A fourth gene, apolipoprotein E (APOE), has been confirmed as a susceptibility factor/risk factor for late onset AD (LOAD).9 The APOE gene has 3 common variants, ε2, ε3, and ε4. The ε2 allele is associated with the lowest LOAD risk, whereas the ε4 allele is associated with an increased risk for LOAD and a younger age of onset of dementia in a dose-dependent manner.10,11 The increased risk for LOAD has also been demonstrated in different ethnic populations including African-Americans and Caribbean Hispanics.12,13 Since the discovery of APOE in 1993, there have been hundreds of other possible AD genes described in the literature.14 Due to sampling and diagnostic issues, population stratification (the differences in case and control allele frequencies due to differences in ethnicity), and sample size, identifying other genes involved in this complex disease is challenging.15 Despite these challenges, twin studies, which demonstrate a 60% concordance rate in monozygotic twins, and other investigations indicate that there may be several susceptibility genes along with APOEε4 that play a role in late-onset AD.16,17
As our understanding of the genes involved in the disease evolves, so may the ability to identify at- risk individuals who would be eligible for early treatment and prevention. In this article we will review the genetics of AD including several genes for which clinical genetic testing is currently unavailable for the diagnosis and prediction of the disease. However, with new advances in the genetics of AD, neurologists should be able to determine whether genetic testing for an AD patient is appropriate, understand recent genetic advances in AD and the current limitations of these findings in clinical practice, and recognize the importance of genetic counseling using the information we do know.
GENES OTHER THAN APP, PSEN1, PSEN2, AND APOE?
New technologies using extensive coverage of genetic markers through genome-wide association studies, advanced statistical methods and collaborative projects to increase the number of cases available for study may help overcome some of the obstacles to finding other genes associated with the disease. In addition, several resources have been established to assist in the discovery of AD related genes. The National Cell Repository for Alzheimer disease stores and maintains clinical data and biologic samples and makes them available to genetic researchers. (www.ncrad.org) The National Institute on Aging (NIA) has organized a multicenter study collecting large families with multiple members affected with late-onset AD through the NIA-Late-Onset Alzheimer disease Genetics Initiative. (www.niageneticsinitiative.org). Clinical data and biologic samples are made available to researchers to include in their genetic analyses. In addition, the NIA has established several mechanisms to store genetic data generated through NIH funded studies. These data can be reanalyzed and combined with other datasets.
Another resource, the AlzGene database, an online database (www.alzgene.org) that compiles up-to-date information on genetic association studies performed on AD and AD phenotypes, has organized and reviewed over 900 AD association studies and approximately 400 genes.14 The AlzGene database organizes and ranks these findings in an attempt to sift through the hundreds of positive and negative association studies with possible AD susceptibility genes. Using the AlzGene database as a guide, we will discuss several examples of other genes reported to be associated with AD including the most recent associations of sortilin-related receptor (SORL1) and GAB2 (both on chromosome 11) and other possible genes/loci on chromosomes 9 and 10.
CHROMOSOME 9
Nineteen candidate genes on chromosome 9 have been reported as associated with LOAD, including DAPK1 (death-associated protein kinase 1)18 and ubiquilin 1 (UBQLN1).19 AlzGene currently ranks DAPK1 as a top candidate gene for LOAD. DAPK1 is an enzyme involved in apoptosis, including neuronal apoptosis. Animal studies demonstrate the expression of the enzyme in the hippocampus and cortex.20 UBQLN1 has been shown to be involved in protein degradation and may interact with PSEN1 and PSEN2,21 however, 2 independent investigators were not able to confirm the association between the risk of AD and UBQLN1, but did see a significant genetic association between single-nucleotide polymorphisms or SNPs in the UBQLN1 gene and the age at onset of AD.22
CHROMOSOME 10
A broad region of chromosome 10 has demonstrated associations with dozens of potential candidate genes for AD. The IDE gene, which produces the insulin-degrading enzyme, is a possible candidate enzyme responsible for the degradation and clearance of Aβ. A recent longitudinal study of a multiethnic community of elders has further indicated that hyperinsulinemia may increase the risk of AD.23 Genetic studies have also shown that IDE gene variants are associated with the clinical symptoms of AD as well as the risk of type 2 diabetes. IDE is located on chromosome 10q24, close to a region for which previous linkage and association with late-onset AD has been reported.24 Additional case-control studies have reported both positive and negative results. Recently, a study demonstrated an association between IDE and late-onset AD in a Finnish population.25
Mitochondrial dysfunction has been implicated in AD, and the mitochondrial transcription factor A (TFAM) gene, on chromosome 10q21, is located in a region that showed linkage to late-onset AD in several study.26 This gene along with CH25H and SORCS1 are also considered as top candidate AD genes by Alzgene standards.
CHROMOSOME 11
Another genome scan published by Reiman et al27 used clinically and neuropathologically characterized cases and controls and discovered associations between a GAB2 and LOAD in APOE ε4 carriers only. Studies including immunohistochemistry and microarray indicated GAB2 is biologically relevant to AD neuropathology. GAB2 was overexpressed in pathologically vulnerable neurons; the Gab2 protein was detected in neurons, tangle-bearing neurons, and dystrophic neurites; and interference with GAB2 gene expression increased tau phosphorylation. The findings suggest that GAB2 modifies LOAD risk in APOE ε4 carriers and may influence Alzheimer neuropathology.27
Most recently, the neuronal (SORL1) on chromosome 11 was reported associated with AD.28 This study used an independent series of family studies and case-control studies (6 independent data sets) in several ethnic groups (Hispanic, European, Israeli Arab, White, and African-American) to demonstrate the association of SORL1 and AD. In addition, functional data showed reduced expression of SORL1 in cases with the SORL1 risk haplotype (a set of alleles on a chromosome that are so close together that they are inherited together). However, a specific mutation or variant has not been identified in SORL1. In order for SORL1 to be useful for clinical diagnostic testing, the specific variant associated with the disease needs to be identified. However, there may be different SORL1 variants in different populations. In addition, similar to APOE, the variants in SORL1 may not be sufficient to predict disease but still may play a role in APP processing and thereby affect the risk for AD. Therefore, SORL1 may be a susceptibility factor rather than a deterministic gene.28 Several groups have attempted to replicate these findings. One reports a “partial replication” using 2000 samples.29 A second study reports a replication of the original finding using an independent dataset including of 1408 individuals from the TGEN database (http://www.tgen.org/neurogenomics/data Translational Genomics Research Institute; TGEN) which included 1044 autopsied individuals (641 cases, 403 controls) and 364 clinically examined individuals from the Mayo Clinic (218 cases and 146 controls).30 However, another study, using this same dataset, was not able to replicate the association.31 The original SORL1 publication was a well-designed study with a large sample size, multiple populations and functional studies. The difficulty consistently replicating the original study emphasizes the challenges of studying these complex, common genetic diseases.
The search for genes causing late-onset AD through linkage and association studies has suggested associations of broad regions of chromosomes 9, 10, 11, and other chromosomes. In addition, case-control studies in sporadic AD have also implicated a large number of individual genes on other chromosomes, whose function can also be potentially linked to the known pathophysiology of AD. Although there are several interesting candidate genes, the evidence that any of these candidate genes are truly the site of genetic variants that modulate risk for AD is uncertain. Without sufficient confirmation in independent samples and a known variant or mutation (similar to APOE), clinical genetic testing is not available. Additionally, genes such as GAB2 and SORL1 with small individual effect size are likely to have limited predictive utility; however, perhaps when grouped together, such as in a panel in combination with APOE, they eventually may become a useful predictor of LOAD risk. Furthermore, they provide insight into the molecular mechanism of LOAD.
GENETIC COUNSELING IN THE ABSENCE OF A GENETIC TEST
Genetic testing is not recommended for every person with a family history or diagnosis of AD. In fact, association with a gene (or SNPs or a haplotype within a gene) and disease does not imply causation or confirm that the gene is a definite risk factor involved in the disease. Recent publications have thoroughly explained the steps of mapping genes in common, complex disorders or specifically in AD.19,32 In order for an association to be confirmed, the positive association must be replicated by independent studies and in different populations. Then the specific variant or mutation within the gene needs to be identified. Depending on the gene, there may be several variants found in different populations. Functional studies of the gene and gene product need to confirm that the variant/mutation affects the function of the gene such as the extensive functional studies in SORL1 and GAB2. However, it is possible to demonstrate a new disease gene based on the genetic evidence alone. For example, the risk effect APOE-ε4 in late-onset AD has been consistently replicated in a many studies and ethnic groups.24 Finally, the effect on risk and/or penetrance (chance that a gene carrier will develop the disease) needs to be assessed. (www.cdc.gov/genomics/gtesting/ACCE.htm).
With every new genetic discovery in AD, patients and their family members may ask questions about whether genetic testing for “the new AD gene” is available. While a test may not yet be available, genetic counseling is valuable to every person who has concerns about the genetic risk and the role of genetics in the disease. Family history of the disease remains the most important risk factor (other than age) for this disease. Individuals with a first-degree relative with AD have a risk about 2.5 times that of the background population risk.33 The counseling should educate families on several key issues including: 1) information on the current understanding of genetics of the disease in the family; 2) risk and risk factors; 3) testing options and opportunities to participate in genetic research; 4) DNA banking for future clinical genetic testing, and 5) the option of autopsy to confirm the diagnosis. Although educating individuals on the facts of genetics is important, the counseling should also focus on the issues of being at risk for a disease, the experience of being a caregiver, and the psychologic burdens of these diseases.34
ALZHEIMER DISEASE GENES AND CLINICAL PRACTICE
Clinical genetic testing is available for individuals with AD symptoms or at-risk children or siblings of patients with early-onset disease and a known mutation (Table 1). In the United States, PSEN1, PSEN2, and APP are available for symptomatic and presymptomatic testing in a clinical or federally approved laboratory under the Clinical Laboratory Improvement Amendments (www.genetests.org). According to the AD and Frontotemporal Dementia Mutation Database (http://www.molgen.ua.ac.be/ADMutations), 28 mutations have been identified within APP from 76 early-onset families. To date, 165 pathogenic PSEN1 mutations have been identified in 354 early-onset families (http://www.molgen.ua.ac.be/ADMutations), making PSEN1 mutations the most common known genetic cause of familial EOAD. The Gly206Ala mutation in PSEN1 was found in unrelated Caribbean Hispanic families with early-onset familial AD. This genetic change may be a prevalent cause of familial EOAD in the Caribbean Hispanic population.35 Unlike PSEN1 mutations, PSEN2 mutations are a rare cause of familial EOAD. With 11 mutations identified from 19 pedigrees, most PSEN2 mutations are thought to be private to mainly Volga-German families, although they have been found in several other populations. (http://www.molgen.ua.ac.be/ADMutations) EOAD accounts for less than 5% of all AD and familial EOAD meeting criteria for autosomal dominant inheritance accounts for about 13% of EOAD. Mutations in APP, PSEN1, and PSEN2 may explain up to 71% of the familial EOAD cases, but other unknown EOAD genes may be identified.36
TABLE 1.
Alzheimer Disease Genes Discussed in This Review
| Gene/Protein/Chromosomal Location | Mode of Inheritance | Penetrance | Age of Onset | No. Mutations/No. Families* | Availability of Clinical Testing |
|---|---|---|---|---|---|
| APP/amyloid precursor protein/21q21.3 | Autosomal Dominant | Complete | 40–60 | 28 in 76 families | Yes (limited) |
| PSEN1/presenilin1/14q24.3 | Autosomal Dominant | Complete | 30–60 (generally before age 65) | 165 in 354 families | Yes |
| PSEN2/presenilin2/1q31–42 | Autosomal Dominant | Incomplete | 40–75 | 10 in 18 families | Yes (limited) |
| APOE/apolipoproteinE/19q13.32 | Complex (susceptibility) | n/a | Late onset | Not a mutation e4 polymorphism associated with increased risk | Yes (recommended for symptomatic only) |
| SORL1/neuronal sortilin-related receptor/11q23 | Complex/susceptibility? | n/a | Late onset | n/a | No |
| GAB2/11q14 | Complex/susceptibility? | n/a | Late onset | n/a | No |
As a susceptibility factor rather than causal gene, APOE testing is not currently recommended for asymptomatic individuals with a family history of late-onset disease except through research studies. The American College of Medical Geneticists and the American Society of Human Genetics do not recommend APOE genotyping for presymptomatic identification of AD37,38 The rationale for this guideline is the low specificity and predictability of the test. Individuals can carry APOE ε4 and never develop the disease during their lifetime. Likewise, individuals without the ε4 variant do develop AD. Although APOE genotyping may be readily available in the management of other conditions, currently, APOE genotyping for presymptomatic individuals with a family history of late-onset AD is only available through research studies such as the REVEAL study (Risk Evaluation and Education for Alzheimer disease). Additionally, there is much debate about the clinical utility of APOE testing for diagnosis given APOE genotyping does not provide sufficient sensitivity or specificity to be used alone as a diagnostic test.39 The presence of an ε4 allele should not alter either the diagnostic evaluation or certainty. People with ε4 have been found to have other dementing disorders. Lastly, when APOE genotyping is performed, clinicians need to decide whether they will discuss the risk of other diseases having an association with APOE (e.g, cardiovascular disease, macular degeneration etc).
CLINICAL DECISION MAKING
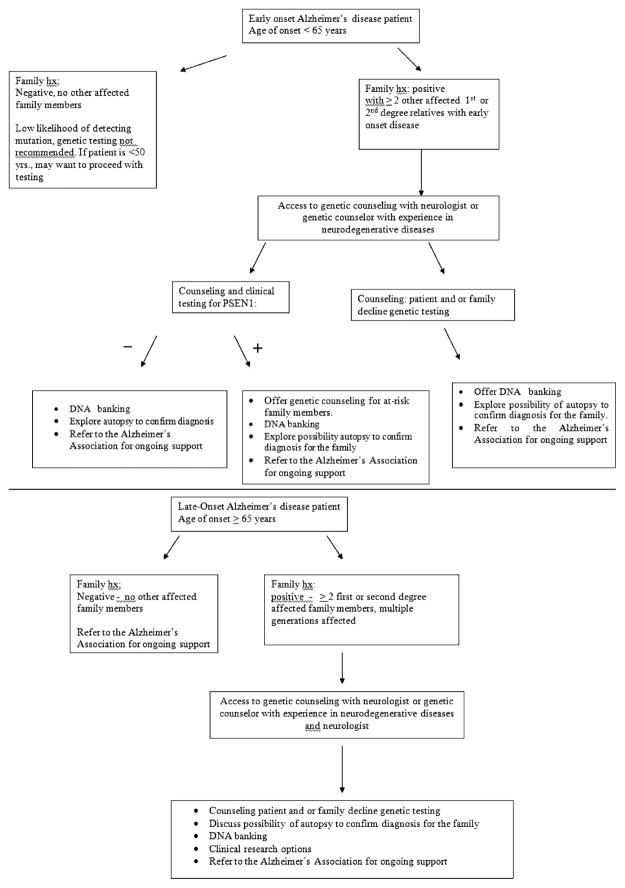
If the most recent discoveries have not yet made their way into diagnostic or predictive testing, the 4 known genes associated with early and late-onset AD do have clinical application. When considering genetic testing for AD, age-of-onset and family history are the 2 most important factors to consider. AD can be divided into early-onset sporadic and early-onset familial disease occurring prior to 65 years of age, and late-onset sporadic and late-onset familial disease occurring after 65 years of age. A majority of AD cases are associated with a late-onset clinical presentation. The ability to offer genetic testing can be directly related to the age-of-onset of disease in the patient and/or family, and the number of affected individuals in the family.40 In the United States, for a patient with early-onset AD (onset 65 or younger) and/or a family history of early onset dementia, clinical genetic testing may be considered. If at-risk family members wish testing, confirmatory testing of a symptomatic patient is suggested to interpret the results properly. Thus, if a family mutation is unknown and tissue from an affected person is unavailable, presymptomatic testing should not be offered. Such patients may be told that they are likely to be at 50% risk of inheriting the disease but that without a definitive family mutation, genetic testing would not be able to confirm a true negative test (Fig. 1).
FIGURE 1.
Clinical genetic gecision making for Alzheimer disease.
CLINICAL TESTING FOR LOAD
The more prevalent, late-onset form of AD is associated with a susceptibility gene, the APOE ε4 polymorphism. APOE genotyping is available for symptomatic individuals as part of the diagnostic evaluation; however, APOE genotyping has not been found to provide sufficient sensitivity or specificity when used alone as a diagnostic test. Although the presence of the ε4 allele in symptomatic individuals does lead to a small improvement in the predictive value of an AD diagnosis, several consensus statements recommend against diagnostic genotyping (Table 2). APOE genotyping in symptomatic individuals may not be beneficial primarily because the presence of an ε4 allele by itself will not alter the diagnostic evaluation or medical management of the patient and APOE genotyping has implications for other family members. Currently, APOE genotyping for presymptomatic individuals with a family history of late-onset AD is not clinically recommended and is possible only through research studies. However, the REVEAL study, a randomized controlled multicenter study assessing risk assessment and APOE genotyping in asymptomatic individuals to examine the impact of APOE genotyping in the predictive testing situation, has not reported any significant psychologic burden to the information.41 The American College of Medical Geneticists and the American Society of Human Genetics do not recommend APOE genotyping for presymptomatic identification of AD,42 but the longitudinal follow-up of individuals participating in the REVEAL study may influence this practice.
TABLE 2.
Genetic Testing Recommendations for Alzheimer Disease
| Organization | Recommendation |
|---|---|
| National Institute on Aging/Alzheimer’s Association34 | Discretionary use of APOE genotyping as an adjunct to other AD diagnostic procedures |
| Routine clinical use not recommended | |
| Pre-test and post-test counseling and psychosocial support necessary | |
| American College of Medical Genetics/American Society of Human Genetics38 | Genotyping is also not recommended for use as a predictive genetic test |
| Not recommended for use in routine clinical diagnosis |
Genetic testing, whether diagnostic or predictive, should not be done without extensive genetic counseling. Genetic counselors specializing in adult-onset neurodegenerative diseases or working with neurologists who treat patients with dementia may be best suited to offer this counseling, including the benefits and limitations of testing, the impact on both the patient and family including the possibility of genetic discrimination, psychosocial impact, impact on family, and partner relationships and family planning. When considering diagnostic testing, the physician needs to assess the patient’s ability to give informed consent and the surrogate’s potential conflict of interest (since that person may be at risk for inheriting the disease). We suggest that predictive testing should be done through a protocol similar to that used in Huntington chorea.43 The Huntington’s protocol recommends several pre- and post-test genetic counseling sessions, baseline neurologic and neuropsychological/psychiatric assessments, and the presence of a support person. The physician can refer patients to a genetic counselor familiar with this protocol. (www.nsgc.org).
CONCLUSION
AD is one of the most challenging disorders of the century and is considered a looming public health crisis. Understanding the genetics of late-onset AD may lead to early detection, prevention and treatment. The genetics of the rare, early-onset familial form of AD has resulted in a better understanding of the pathophysiology of the disease. The more common, late-onset form of AD has a substantial genetic component that remains unexplained, despite the identification of the susceptibility allele, APOEε4. Genome-wide association and linkage studies and candidate gene studies have produced multiple genes of interest that may also play a role in the pathophysiology of and susceptibility to AD. Until these finding have been replicated and their contribution to risk elucidated, current clinical practice is limited to genetic testing in the 4 confirmed genes, PSEN1, PSEN2, APP, and APOE.
References
- 1.Association, A.s. [Accessed 2007];Alzheimer’s Disease Facts and Figures. 2007 Available at: www.alz.org/national/documents/
- 2.Tabaton M, Tamagno E. The molecular link between beta- and gamma-secretase activity on the amyloid beta precursor protein. Cell Mol Life Sci. 2007;64:2211–2218. doi: 10.1007/s00018-007-7219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherrington R, Froelich S, Sorbi S, et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum Mol Genet. 1996;5:985–988. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 4.Tanzi RE, Vaula G, Romano DM, et al. Assessment of amyloid beta-protein precursor gene mutations in a large set of familial and sporadic Alzheimer disease cases. Am J Hum Genet. 1992;51:273–282. [PMC free article] [PubMed] [Google Scholar]
- 5.Schellenberg GD, Bird TD, Wijsman EM, et al. Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science. 1992;258:668–671. doi: 10.1126/science.1411576. [DOI] [PubMed] [Google Scholar]
- 6.Van Broeckhoven C, Backhovens H, Cruts M, et al. Mapping of a gene predisposing to early-onset Alzheimer’s disease to chromosome 14q24.3. Nat Genet. 1992;2:335–339. doi: 10.1038/ng1292-335. [DOI] [PubMed] [Google Scholar]
- 7.St George-Hyslop P, Haines J, Rogaev E, et al. Genetic evidence for a novel familial Alzheimer’s disease locus on chromosome 14. Nat Genet. 1992;2:330–334. doi: 10.1038/ng1292-330. [DOI] [PubMed] [Google Scholar]
- 8.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 9.Pericak-Vance MA, Bebout JL, Gaskell PC, Jr, et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48:1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 10.Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 11.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 12.Green RC, Cupples LA, Go R, et al. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA. 2002;287:329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- 13.Romas SN, Santana V, Williamson J, et al. Familial Alzheimer disease among Caribbean Hispanics: a reexamination of its association with APOE. Arch Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 14.Bertram L, McQueen MB, Mullin K, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 15.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 16.Gatz M, Fratiglioni L, Johansson B, et al. Complete ascertainment of dementia in the Swedish Twin Registry: the HARMONY study. Neurobiol Aging. 2005;26:439–447. doi: 10.1016/j.neurobiolaging.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Grupe A, Rowland C, et al. DAPK1 variants are associated with Alzheimer’s disease and allele-specific expression. Hum Mol Genet. 2006;15:2560–2568. doi: 10.1093/hmg/ddl178. [DOI] [PubMed] [Google Scholar]
- 19.Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto M, Takahashi H, Nakamura T, et al. Developmental changes in distribution of death-associated protein kinase mRNAs. J Neurosci Res. 1999;58:674–683. doi: 10.1002/(sici)1097-4547(19991201)58:5<674::aid-jnr8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Mah AL, Perry G, Smith MA, et al. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slifer MA, Martin ER, Haines JL, et al. The ubiquilin 1 gene and Alzheimer’s disease. N Engl J Med. 2005;352:2752–2753. doi: 10.1056/NEJM200506303522618. author reply 2752–2753. [DOI] [PubMed] [Google Scholar]
- 23.Luchsinger JA, Tang MX, Shea S, et al. Hyperinsulinemia and risk of Alzheimer disease. Neurology. 2004;63:1187–1192. doi: 10.1212/01.wnl.0000140292.04932.87. [DOI] [PubMed] [Google Scholar]
- 24.Bertram L, Blacker D, Mullin K, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 25.Vepsalainen S, Parkinson M, Helisalmi S, et al. Insulin-degrading enzyme is genetically associated with Alzheimer’s disease in the Finnish population. J Med Genet. 2007;44:606–608. doi: 10.1136/jmg.2006.048470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunther C, von Hadeln K, Müller-Thomsen T, et al. Possible association of mitochondrial transcription factor A (TFAM) genotype with sporadic Alzheimer disease. Neurosci Lett. 2004;369:219–223. doi: 10.1016/j.neulet.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 27.Reiman EM, Webster JA, Myers AJ, et al. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Rowland C, Catanese J, et al. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng Y, Lee JH, Cheng R, et al. Association between SORL1 and Alzheimer’s disease in a genome-wide study. Neuroreport. 2007;18:1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster JA, Myers AJ, Pearson JV, et al. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 32.Mayeux R. Mapping the new frontier: complex genetic disorders. J Clin Invest. 2005;115:1404–1407. doi: 10.1172/JCI25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cupples LA, Farrer LA, Sadovnick AD, et al. Estimating risk curves for first-degree relatives of patients with Alzheimer’s disease: the REVEAL study. Genet Med. 2004;6:192–196. doi: 10.1097/01.gim.0000132679.92238.58. [DOI] [PubMed] [Google Scholar]
- 34.Williamson J, LaRusse S. Genetics and genetic counseling: recommendations for Alzheimer’s disease, frontotemporal dementia, and Creutzfeldt-Jakob disease. Curr Neurol Neurosci Rep. 2004;4:351–357. doi: 10.1007/s11910-004-0081-x. [DOI] [PubMed] [Google Scholar]
- 35.Athan ES, Williamson J, Ciappa A, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. JAMA. 2001;286:2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 36.Campion D, Dumanchin C, Hannequin D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Post SG, Whitehouse PJ, Binstock RH, et al. The clinical introduction of genetic testing for Alzheimer disease. An ethical perspective. JAMA. 1997;277:832–836. doi: 10.1001/jama.277.10.832. [DOI] [PubMed] [Google Scholar]
- 38.Relkin NR, Kwon YJ, Tsai J, et al. The National Institute on Aging/Alzheimer’s Association recommendations on the application of apolipoprotein E genotyping to Alzheimer’s disease. Ann N Y Acad Sci. 1996;802:149–176. doi: 10.1111/j.1749-6632.1996.tb32608.x. [DOI] [PubMed] [Google Scholar]
- 39.Mayeux R, Saunders AM, Shea S, et al. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer’s disease. Alzheimer’s Disease Centers Consortium on Apolipoprotein E and Alzheimer’s Disease. N Engl J Med. 1998;338:506–511. doi: 10.1056/NEJM199802193380804. [DOI] [PubMed] [Google Scholar]
- 40.Tanzi RE. A genetic dichotomy model for the inheritance of Alzheimer’s disease and common age-related disorders. J Clin Invest. 1999;104:1175–1179. doi: 10.1172/JCI8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts JS, Cupples LA, Relkin NR, et al. Genetic risk assessment for adult children of people with Alzheimer’s disease: the Risk Evaluation and Education for Alzheimer’s Disease (REVEAL) study. J Geriatr Psychiatry Neurol. 2005;18:250–255. doi: 10.1177/0891988705281883. [DOI] [PubMed] [Google Scholar]
- 42.Statement on use of apolipoprotein E testing for Alzheimer disease. American College of Medical Genetics/American Society of Human Genetics Working Group on ApoE and Alzheimer disease. JAMA. 1995;274:1627–1629. [PubMed] [Google Scholar]
- 43.Guidelines for the molecular genetics predictive test in Huntington’s disease. International Huntington Association (IHA) and the World Federation of Neurology (WFN) Research Group on Huntington’s Chorea. Neurology. 1994;44:1533–1536. [PubMed] [Google Scholar]