Abstract
Insulin-degrading enzyme (IDE) degrades insulin and other peptides, including the Aβ peptide of Alzheimer's disease. However, the mechanism by which IDE acts on its substrates in vivo is unclear, and its role in pathogenesis of type 2 diabetes and Alzheimer's disease is controversial. Here, we show that in Drosophila knocking down IDE in insulin-producing cells (IPCs) of the brain results in increased body weight and fecundity, decreased circulating sugar levels and reduced lifespan. Moreover, knocking down and overexpressing IDE in IPCs have opposite physiological effects. As misregulated insulin signaling in peripheral tissues is known to cause similar phenotypes, our data suggest a role for Drosophila IDE in determining the level of insulin-like peptides made by IPCs that systemically activate insulin signaling.
Key words: insulin proteolysis, signaling pathway, Drosophila, lifespan, beta cell
Introduction
Insulin plays a critical role in a wide range of physiological processes, including growth control, metabolic regulation, fertility and aging.1,2 Its physiological action is regulated by multiple mechanisms, including cellular removal and degradation, which can be disrupted in type 2 diabetes and other disorders.3 The major activity in mammalian cells that degrades insulin is insulin-degrading enzyme (IDE), a metalloprotease of 110 kDa found in most tissues, including insulin responsive tissues such as liver and muscle.4 IDE also degrades a variety of small peptides besides insulin in vitro, including the Aβ peptide that forms the hallmark amyloid plaques of Alzheimer's disease. Since type 2 diabetes could be a risk factor for Alzheimer's disease, IDE is a possible link between the two disorders.5
While IDE's biochemical properties have been extensively studied, its physiological role remains poorly understood. An IDE knock out mouse was described to have elevated plasma insulin and brain Aβ peptide, consistent with a physiological role for IDE in degrading insulin and Aβ.6 However, an independent study reported increased Aβ peptide, but not insulin, in the same IDE knock out mouse.7 Whether IDE is important in the pathogenesis of type 2 diabetes and Alzheimer's disease is also controversial, as multiple human genetic studies have reached opposite conclusions on this issue.8,9 Moreover, the cellular location where IDE degrades insulin is unclear.10 IDE is most often found in the cytosol, but is also found in mitochondria and peroxisomes, on the cell surface and outside the cell. Since IDE can degrade a variety of peptides in vitro, perhaps it is localized in different subcellular compartments where it degrades different physiological substrates depending on the tissue.
To better understand IDE's physiological role, we decided to study IDE function in Drosophila, a powerful model system for studying the insulin signaling pathway. Drosophila and human IDE have 48% amino acid sequence identity and many enzymatic properties in common, including the ability to degrade insulin in vitro.11,12 Moreover, like mouse IDE, Drosophila IDE appears to be non-essential for organism viability, according to a recent study using a putative Ide gene knockout.13 This study also provided evidence that Drosophila IDE antagonizes the insulin signaling pathway. Potential endogenous substrates for Drosophila IDE are the insulin-like peptides (DILPs), which are structurally homologous to human insulin and agonists of the insulin receptor.14–16 Of the seven known DILPs in Drosophila, DILP1, 2, 3 and 5 are expressed in a cluster of brain neuroendocrine cells called insulin-producing cells (IPCs).14,17 Deletion of DILPs or ablation of IPCs results in defects in glucose metabolism, organism growth and lifespan.17–20 These and other studies suggest that IPCs, as a major source of DILPs that act systemically on peripheral target tissues, are functionally analogous to mammalian pancreatic islet β-cells.
Here, we report that altering IDE expression level in IPCs perturbed organism size, circulating sugar, fecundity and lifespan in Drosophila. As earlier studies have shown that misregulation of insulin signaling in peripheral tissues results in similar phenotypes, our results suggest that IDE expression in IPCs is important for controlling the level of DILPs that act systemically. We further speculate about the broader implication of our findings in the context of mammalian IDE and the control of insulin production by pancreatic islet β cells.
Results and Discussion
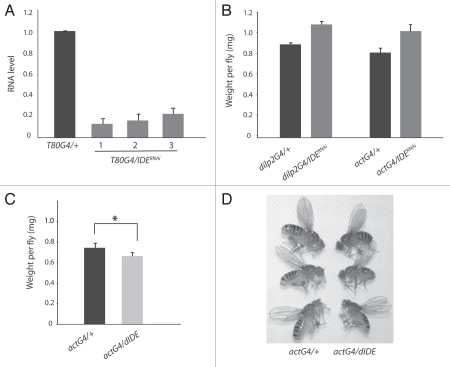
We decided to use RNAi to perturb IDE activity in Drosophila and therefore generated transgenic lines that can express double-stranded RNA targeting endogenous Ide mRNA under Gal4-UAS system control.21,22 Using a bioinformatics off-target search tool (available at http://flyRNAi.org), no potential off-target of this Ide RNAi construct was identified.23 To test knock down efficiency, expression of the Ide RNAi construct was activated with the T80-Gal4 driver, which is ubiquitously expressed in the embryo and larva.24 Quantitative RT-PCR analysis of three different Ide RNAi lines revealed that the Ide mRNA level was significantly reduced by 77–87% in RNAi larva when compared to control larva carrying the T80-Gal4 driver but not the Ide RNAi construct (Fig. 1A). Ide RNAi larvae developed into viable adults with no obvious morphological abnormality. In all subsequent experiments, flies carrying two copies of the Ide RNAi construct were used in order to maximize knock down efficiency.
Figure 1.
IDE is important for body weight. (A) The broadly expressed T80-Gal4 driver was used to activate Ide RNAi in virtually all tissues during larval development. Quantitative RT-PCR analysis, using the same amount of total RNA isolated from 3rd instar larvae of the same age, indicated that Ide mRNA was reduced by 77–87% in three independent Ide RNAi lines (T80G4/IDERNAi) compared to the control where the driver was crossed into the w1118 background lacking the Ide RNAi construct (T80G4/+). (B) The broadly expressed act-Gal4 driver or the IPC-specific dilp2-Gal4 driver was used to activate Ide RNAi. The body weight increased by 25% in actG4/IDERNAi flies (n = 195) and by 21% in dilp2G4/IDERNAi flies (n = 134) when compared to the respective controls (actG4/+, n = 188; dilp2G4/+, n = 134). 7 day-old adult females were weighed. (C) Flies in which Ide expression is broadly activated (actG4/dIDE, n = 618) weighed 11% less than control flies (actG4/+, n = 878). Seven-day-old adult females were weighed. *p < 0.0002, two-tailed t-test. (D) IDE overexpressing flies (actG4/dIDE) were generally smaller than control flies (actG4/+). Flies of the two different genotypes were cultured separately but in parallel under identical conditions as possible to avoid over-crowding. The flies shown in this figure were randomly selected from culture vials and photographed about 1 day after eclosion.
Earlier studies showed that overexpression of DILPs results in a 10–50% increase in the body weight of flies depending on the DILP isoform that is overexpressed.14,15 If IDE degrades DILPs in vivo, then flies with reduced IDE activity would be expected to have a greater than normal DILP level and body weight. Indeed, adult flies in which Ide RNAi was broadly activated with the act-Gal4 driver weighed 25% more than control flies (Fig. 1B). IDE overexpression with the same driver, which reduced adult fly viability by up to 92%, had the opposite effect. Surviving IDE overexpressing flies weighed 11% less and were visibly smaller than control flies (Fig. 1C and D). As a comparison, flies lacking DILPs 1–5 were reported to have around 50% of the normal body weight.20 The opposing effects on body weight from RNAi and overexpression approaches argue that altering IDE activity is responsible for the observed changes in body weight.
Insulin producing cells (IPCs) of the Drosophila brain are a major source of systemically acting DILPs.17–20 By RNA in situ hybridization analysis, IDE appears to be broadly expressed in Drosophila tissues, including the brain (data not shown). To examine whether IDE expression is required in IPCs, we activated Ide RNAi using the dilp2-Gal4 driver, which is specific for IPCs during late larval to adult stages.15 Surprisingly, such IPC-specific Ide RNAi resulted in flies that weighed more than control flies by 21%, nearly comparable to the increase seen when Ide RNAi was more broadly activated (Fig. 1B). This result suggests that IDE expression is required in IPCs for normal body weight in Drosophila.
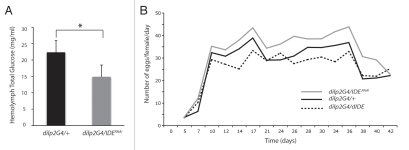
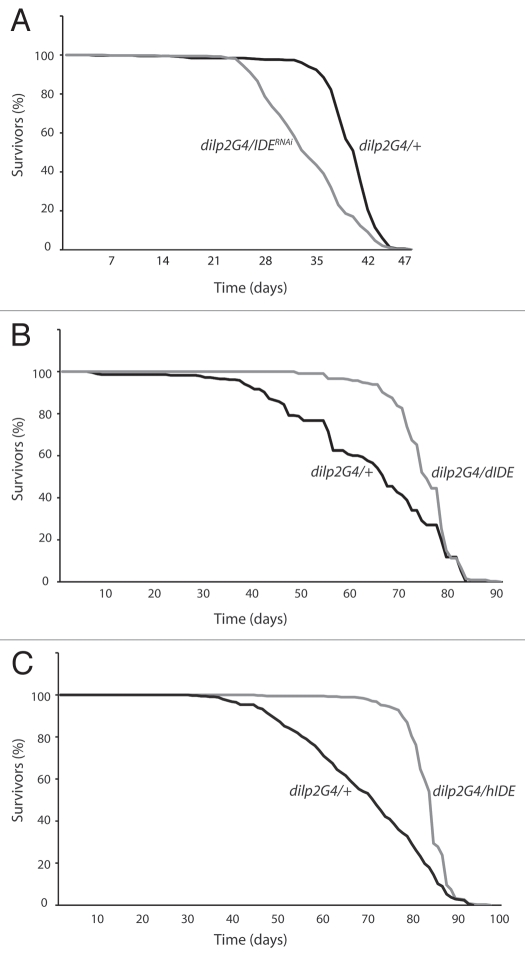
In Drosophila, reducing DILPs or ablating IPCs results in elevated circulating sugar, reduced fecundity and increased lifespan, all phenotypes associated with lowered insulin signaling.15,17–20 We therefore investigated whether changing the IDE level in IPCs, by RNAi or overexpression with the dilp2-Gal4 driver, affects these physiological parameters. To assay circulating sugar, we measured the hemolymph level of trehalose, a glucose disaccharide representing the predominant circulating sugar in Drosophila and other insects.17,20 We found that IPC-specific Ide RNAi flies had about 30% lower hemolymph trehalose when compared to control flies (Fig. 2A), as would be predicted if these flies had higher than normal DILP levels. In addition, when compared to the control, IPC-specific Ide RNAi females laid more eggs while IPC-specific IDE overexpressing females produced fewer eggs (Fig. 2B). For example, in terms of the average number of eggs laid per female per day, IPC-specific Ide RNAi flies laid 14% more and IPC-specific IDE overexpressing flies laid 11% fewer than control flies; by comparison, flies lacking DILP1–5 were reported to lay 90% fewer eggs than normal.20 While modest, the differences that we observed were reproducible and consistent with the changes that would be predicted from increasing or decreasing DILP levels. Finally, we found that IPC-specific Ide RNAi female flies had a median lifespan 18% shorter than control female flies (Fig. 3A), and a similar result was obtained with male flies (data not shown). Conversely, IPC-specific IDE overexpressing female flies had a median lifespan 14% greater than control females (Fig. 3B), while no consistently reproducible lifespan extension was observed for IPC-specific IDE overexpressing males. In comparison, ablation of IPCs was reported to increase the median lifespan of females and males by 33.5% and 10.5%, respectively, also revealing a substantial difference in effect between females and males.18 Ectopic expression of human IDE in IPCs resulted in an 18% extension of median lifespan, again only in female flies, demonstrating that Drosophila and human IDE have similar in vivo activities (Fig. 3C).
Figure 2.
IDE is important for circulating sugar level and fecundity. (A) Hemolymph total glucose, derived mostly from trehalose and representing ≤1% free glucose, was about 30% lower for flies in which Ide RNAi was activated with the dilp2-Gal4 driver (dilp2G4/IDERNAi) when compared to the control (dilp2G4/+). *p = 0.0127, one-tailed t-test. The circulating sugar level was not determined for IDE overexpressing flies. (B) Using the dilp2-Gal4 driver to activate Ide RNAi (dilp2G4/IDERNAi) or IDE overexpression (dilp2G4/dIDE) in IPCs resulted in flies with increased and decreased fecundity, respectively, compared to control flies (dilp2G4/+). Eggs laid by individual virgin females were counted, with a plot showing the average of data from 50–60 females per genotype minus any flies that died during the experiment. The dilp2G4/+, dilp2G4/IDERNAi and dilp2G4/dIDE females, respectively, laid per female an average of about 1,090, 1,250 and 970 eggs over 39 days, or about 28, 32 and 25 eggs per day. Preliminary experiments with mated females showed similar results.
Figure 3.
IDE is important for lifespan. (A) Flies in which IDE was knocked down in IPCs with the dilp2-Gal4 driver (dilp2G4/IDERNAi, n = 381) had 18% lower median lifespan compared to control flies carrying the driver alone (dilp2G4/+, n = 383). Results are shown for female flies cultured at 29°C to enhance knockdown efficiency. Similar results were obtained with male flies. (B) Flies in which Drosophila IDE was overexpressed in IPCs with the dilp2-Gal4 driver (dilp2G4/dIDE, n = 328) had 14% greater median lifespan compared to control flies carrying the driver alone (dilp2G4/+, n = 288). Results are shown for female flies cultured at 25°C. (C) Flies in which human IDE was overexpressed in IPCs with the dilp2-Gal4 driver (dilp2G4/hIDE, n = 374) had 18% greater median lifespan compared to control flies carrying the driver alone (dilp2G4/+, n = 344). Results are shown for female flies cultured at 25°C.
In conclusion, we found that knocking down IDE in IPCs resulted in phenotypes previously associated with the upregulation of DILPs and insulin signaling, while overexpressing IDE in IPCs resulted in opposite phenotypes. Thus, our findings are consistent with the idea that IDE degrades DILPs made by IPCs that activate insulin signaling. A role for IDE in degrading DILPs is also supported by a recent study showing that, upon coexpression, IDE can antagonize the activity of DILP2 in promoting tissue growth.13 However, the same study reported that, while IDE overexpression lowered the body weight of flies as we observed, a putative Ide knockout mutation did not alter body weight, or other physiological parameters, in the manner that we observed with Ide RNAi. One possible explanation for the apparent discrepancy between the Ide knockout and RNAi results is that Ide knockdown causes an upregulation of DILPs, whose phenotypic effect is masked by upregulation of a distinct substrate or multiple substrates to a level only achieved upon complete elimination of Ide activity. Drosophila IDE likely degrades other substrates besides DILPs, just as mammalian IDE degrades multiple substrates.3
Further analysis is required to understand how IDE might degrade DILPs in vivo. Given that mammalian IDE rapidly degrades insulin but not proinsulin, Drosophila IDE would be expected to degrade mature and active DILPs rather than the pro form.3 Thus, an antibody capable of detecting processed DILPs would be an important reagent in order to directly test IDE-dependent degradation of DILPs in vivo. However, the only western blot analysis of DILPs in Drosophila tissues reported so far detected the pro but not the processed form of DILP2, suggesting that processed DILPs are non-abundant.19,25 Within IPCs, IDE could function in the secretory pathway during the secretion of DILPs. Alternatively, IDE could function in an endocytic pathway to degrade DILPs after they are secreted and internalized by the insulin receptor in IPCs, perhaps as part of an autocrine feedback loop as hypothesized to occur via insulin signaling in IPCs and in mammalian β cells.19,26 By either mechanism, IDE could play an important role in IPCs to determine the level of DILPs that ultimately activate insulin signaling in peripheral tissues. Given the functional homologies between Drosophila and mammalian IDE, as well as between Drosophila IPCs and mammalian β-cells, we speculate that mammalian IDE could have a similarly important role in β cells in controlling insulin levels.
Materials and Methods
Fly strains.
Transgenic lines carrying constructs encoding full-length Drosophila IDE (UAS-dIDE) on the second and third chromosomes, full-length human IDE (UAS-hIDE) on the third chromosome and double-stranded RNA specific for Drosophila IDE (UAS-IDERNAi) on the second and third chromosomes were made by standard P-element transformation of w1118 (Fbal0018186) flies.27 The dilp2-Gal4/CyO line was from D. Bohmann (Univ. of Rochester). T80-Gal4/CyO (FBst0001878) and act-Gal4/CyO (FBst0004414) were from the Bloomington Stock Center (Bloomington, IN). In all RNAi and overexpression experiments using a given driver (e.g., dilp2-Gal4), parallel crosses between driver bearing flies and w1118 flies were used to generate control flies (e.g., dilp2-Gal4/+).
Molecular biology.
The UAS-dIDE and UAS-hIDE constructs were made using Drosophila IDE cDNA (RE17458) and the pPWF vector from the Drosophila Genomics Resource Center (Bloomington, IN) and human IDE cDNA (BC096336) from Open Biosystems (Huntsville, AL). The UAS-IDERNAi construct contains sequences 1,343–1,856 from the Drosophila IDE cDNA in the pWIZ vector.21 RT-PCR was performed using the Roche LightCycler® on total RNA isolated from third instar larvae with TRIzol (GIBCO-BRL), using ribosomal protein 49 as the internal normalization standard for both RNAi and control flies.
Hemolymph sugar assay.
Hemolymph was isolated from adult females aged 2–4 days after eclosion essentially as described in reference 28. About 1.5 µl of hemolymph obtained from 40 flies was mixed with 20 µl PBS and incubated with 1 µl trehalase (3.7 units/ml, Sigma) overnight at 37°C in order to digest trehalose to glucose. After incubation, 5 µl of trehalase reaction in duplicate was incubated with 100 µl of Glucose Assay Reagent (Sigma) in a 96-well plate for 15 minutes at room temperature. The glucose concentration in these samples was determined by spectroscopic measurement against a glucose standard curve using a Bio-Rad microplate reader, and then used to determine the concentration of total glucose in hemolymph. Free glucose, the glucose in hemolymph prior to trehalase treatment, represented ≤1% of total glucose.
Fecundity measurements.
Fecundity was determined by counting the number of eggs laid by females during a period of 3–42 days after eclosion. Females of a given genotype were placed in ten 100 ml beakers, each containing 5–6 females. Each beaker was inverted over an apple juice agar plate with yeast paste, which was exchanged every 2–3 days. The eggs laid on the plate were counted, with data presented as described in reference 19.
Lifespan measurements.
Survival rate was determined as described in reference 19. Freshly eclosed adult flies were collected into vials, with each vial containing 25 flies. Flies were transferred to a fresh vial every other day and the dead flies remaining in the old vial were counted. This process was repeated until all flies of a given genotype had died. Flies were maintained at 25°C on standard medium.
Acknowledgements
We thank Jonathan Bogan and Soumya Vemuganti for helpful comments on the manuscript. The authors gratefully acknowledge support of NIH grants F32GM84651 to Joogyung Hyun and R01GM49370 to Carl Hashimoto.
Abbreviations
- IDE
insulin-degrading enzyme
- IPC
insulin-producing cell
- DILP
Drosophila insulin-like peptide
References
- 1.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis and life span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 3.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 4.Hulse RE, Ralat LA, Wei-Jen T. Structure, function and regulation of insulin-degrading enzyme. Vitam Horm. 2009;80:635–648. doi: 10.1016/S0083-6729(08)00622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu WQ, Folstein MF. Insulin, insulin-degrading enzyme and amyloid-β peptide in Alzheimer's disease: review and hypothesis. Neurobiol Aging. 2006;27:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid β-protein and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, et al. Amyloid-β peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc Natl Acad Sci USA. 2003;100:6221–6226. doi: 10.1073/pnas.1031520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowotny P, Hinrichs AL, Smemo S, Kauwe JS, Maxwell T, Holmans P, et al. Association studies between risk for late-onset Alzheimer's disease and variants in insulin degrading enzyme. Am J Med Genet B Neuropsychiatr Genet. 2005;136:62–68. doi: 10.1002/ajmg.b.30186. [DOI] [PubMed] [Google Scholar]
- 9.Vepsalainen S, Parkinson M, Helisalmi S, Mannermaa A, Soininen H, Tanzi RE, et al. Insulin-degrading enzyme is genetically associated with Alzheimer's disease in the Finnish population. J Med Genet. 2007;44:606–608. doi: 10.1136/jmg.2006.048470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh LB. The insulysin (insulin degrading enzyme) enigma. Cell Mol Life Sci. 2006;63:2432–2434. doi: 10.1007/s00018-006-6238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duckworth WC, Garcia JV, Liepnieks JJ, Hamel FG, Hermodson MA, Frank BH, et al. Drosophila insulin degrading enzyme and rat skeletal muscle insulin protease cleave insulin at similar sites. Biochemistry. 1989;28:2471–2477. doi: 10.1021/bi00432a018. [DOI] [PubMed] [Google Scholar]
- 12.Kuo WL, Gehm BD, Rosner MR. Cloning and expression enzyme. Mol Endocrinol. 1990;4:1580–1591. doi: 10.1210/mend-4-10-1580. [DOI] [PubMed] [Google Scholar]
- 13.Tsuda M, Kobayashi T, Matsuo T, Aigaki T. Insulin-degrading enzyme antagonizes insulin-dependent tissue growth and Aβ-induced neurotoxicity in Drosophila. FEBS Lett. 2010;584:2916–2920. doi: 10.1016/j.febslet.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 15.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 17.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 18.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broughton S, Alic N, Slack C, Bass T, Ikeya T, Vinti G, et al. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One. 2008;3:3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci USA. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 22.Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 23.Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, et al. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:489–494. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, et al. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis. 2002;34:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- 25.Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leibiger IB, Leibiger B, Berggren PO. Insulin signaling in the pancreatic β cell. Annu Rev Nutr. 2008;28:233–251. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 27.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 28.Lung O, Wolfner MF. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem Mol Biol. 1999;29:1043–1052. doi: 10.1016/s0965-1748(99)00078-8. [DOI] [PubMed] [Google Scholar]