Usher syndrome is an autosomal, recessively inherited disorder involving progressive retinitis pigmentosa and hearing loss with or without vestibular dysfunction. Usher type I (USH1) is the severest form (1). It involves profound deafness, vestibular areflexia and onset of retinitis pigmentosa in early childhood (1). To date seven genetic loci for USH1 have been mapped (USH1B-USH1H) and five of the genes have been identified. USH1G (MIM #606943) is associated with mutations in SANS (2). The encoded 461 amino acid protein, SANS, is predicted to have three ankyrin-like domains, a PDZ binding motif and a Sterile alpha motif (SAM) domain (2). SANS is expressed in many tissues including the inner ear and retina (2, 3). Jackson shaker (js) mice have a mutation in Sans. They exhibit disorganized stereocilia and are profoundly deaf, but do not exhibit a retinal phenotype (3).
USH1G appears to be a rare cause of USH1 as only five mutations in SANS have been reported to cause Usher syndrome in four families (2, 4, 5) (Table 1). All mutations, except one, cause classic symptoms of USH1. A mutation in exon 2 substituting p.V458D in SANS is associated with atypical Usher syndrome. It is therefore hypothesized that missense mutations in SANS may result in hypomorphic alleles and cause a less severe phenotype as compared to frameshift mutations (4).
Table 1.
List of all mutations in SANS
List of all known mutations identified in SANS.
| Country | Exon | Mutationa | Consequence | Hearing Loss | Reference |
|---|---|---|---|---|---|
| Germany | 1 | c.143 T>C* | p.L48P | Profound | 2 |
| 2 | c.186_187delCA | Frameshift | |||
| Tunisia | 2 | c.393insG | Frameshift | Profound | 2 |
| Jordan | 2 | c.832_851del20** | Frameshift | Profound | 2 |
| U.S.A | 1 | c.113 G>A | p.W38X | Profound | 5 |
| Turkey | 2 | c.1373 A>T | p.D458V | Variable (moderate to profound) | 4 |
| Pakistan | 1 | c.163_164+13del15 | Frameshift | Moderate to severe | This report |
Numbers with respect to the open reading frame of SANS, corresponding to cDNA sequence from GenBank (AK091243) with the first nucleotide in the initiation codon designated as “+1”.
Originally reported as c.142 C>T.
Originally reported as 829_848del20.
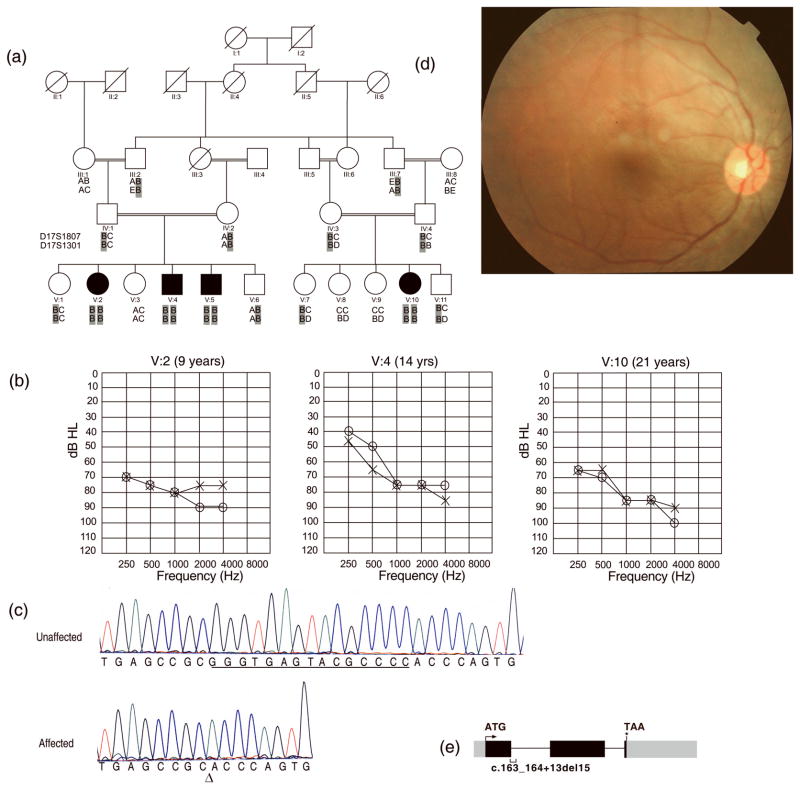
We report a consanguineous family with four affected individuals with moderate to severe hearing loss, mild retinitis pigmentosa and normal vestibular function, a phenotype which resembles an USH2 diagnosis (1). Family HLRB12 (Fig. 1a) was recruited from Sheikhupura, Pakistan with Institutional Review Board approval at University of the Punjab, and written informed consent was obtained from all participants. Three affected children were subjected to audiometric examinations at the age of 12, 14 and 21 years, in ambient noise conditions. Clinical examination revealed moderate to severe hearing loss (Fig. 1b). Vestibular function was evaluated by Romberg and tandem gait tests. Additionally, inquiries were made about whether affected children felt insecure while walking in darkness or had motion sickness. There was no delay in independent ambulation and none of the affected individuals had difficulty with balance suggesting vestibular function was normal. Additionally, none of the patients reported problems with eyesight including night vision. However, funduscopy revealed mild symptoms of retinitis pigmentosa in three of the older affected individuals examined at 5, 13, 15, and 22 years respectively. No bone spicules were observed in the retinal epithelium (Fig. 1d). The optic discs were pale as compared to those in normal individuals. Electroretinography could not be performed. Optical testing revealed a mild loss of near sight vision, which was not noted by the patients.
Figure 1.
(a) Pedigree of family HLRB12. Haplotypes for two closest markers to SANS are shown for the genotyped members. Alleles of the two markers were homozygous for all affected individuals. The ancestral chromosome with the SANS mutation is shaded in gray (b) Audiograms of three affected individuals of family HLRB12 indicate moderate to severe hearing loss. Age at the time of audiometry is indicated on tops of the audiograms. “o” indicates air conduction for right ear, while “x” indicates air conduction for left ear (c) A fifteen nucleotide deletion was observed in SANS at the first exon-intron junction in DNA of affected individuals. “Δ” indicates the start of deletion. Deleted bases are underlined in the trace from a control. (d) A representative funduscopic image from a patient with atypical Usher syndrome in family HLRB12. No bone spicules were observed (e) Diagrammatic depiction of SANS. Black boxes represent translated exons while gray boxes denote untranslated parts of the exons. The two introns are represented by horizontal lines. The position of the deletion of 15 nucleotides identified in family HLRB12 is shown by a bracket under the depicted exon 1 and the following intron. The arrow and the asterisk denote the start and stop codons in SANS, respectively.
Genotyping with fluorescently labeled markers for linkage analyses excluded all Usher syndrome loci except USH1G. Markers D17S1807 and D17S1301 lie close to SANS and showed homozygosity by descent in all affected individuals of family HLRB12 (Fig. 1a). Using a disease allele frequency of 0.001 and coding the phenotype as a fully penetrant autosomal recessive disorder, maximum two-point LOD scores of 4.2 and 3.9 were obtained at recombination fraction θ = 0 with the markers D17S1807 and D17S1301 respectively. We PCR amplified and sequenced the three exons and flanking intronic regions of SANS. In the DNA of affected individuals of family HLRB12 we identified a homozygous 15bp deletion (c.163_164+13del15) involving nucleotides in the first exon and intron of SANS (Fig. 1c, e). We did not detect this mutation in 200 chromosomes from ethnically matched controls assayed by Tetra primers ARMS PCR (6).
In order to identify the effect of c.163_164+13del15 on the SANS transcript, we obtained RNA from whole blood and generated cDNA. However we found that SANS is not expressed sufficiently in blood samples (data not shown). Usually mutations that delete donor splice sites in first exons result in retention of the following introns or use of a cryptic splice site within the affected exons or introns (7, 8) . Indeed, in silico analysis for cryptic splice sites in wild type and mutant genomic sequences of SANS with GeneSplicer (http://cbcb.umd.edu/software/GeneSplicer/gene_spl.shtml) predicted retention of the first intron of in the RNA resulting from mutant, but not from the normal, SANS genomic sequence (data not shown). The retention of the first intron will introduce a frameshift and a premature stop codon in the SANS open reading frame. The presence of premature stop codons are known to mark some mRNAs for nonsense mediated decay (9) and it is possible that no mutant mRNA will be produced in patients with the deletion mutation. However, if the mRNA escapes this surveillance mechanism, the frameshift will result in a truncated nonfunctional protein of 58 amino acids.
Our work indicates that both missense and deletion mutations in SANS can result in atypical Usher syndrome. Thus the location or type of mutation does not predict the severity of the disorder and the phenotypic course can be modified by unknown genetic or epigenetic factors. Furthermore, some patients with no mutations in genes which cause USH2 may have mutations in SANS.
Acknowledgments
We thank the family for participation in this research and the Layton Rehmatullah Benevolent Trust (LRBT) Lahore for funduscopic examination. We express our gratitude to Dr. Thomas B Friedman for his valuable comments on the manuscript and for generously providing primers for PCR amplification of SANS cDNA. We are grateful to Dr. Saadia S Alam for providing technical assistance. This work was supported by grant number R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest
References
- 1.Kimberling WJ, Moller C. Clinical and molecular genetics of Usher syndrome. J Am Acad Audiol. 1995;6:63–72. [PubMed] [Google Scholar]
- 2.Weil D, El-Amraoui A, Masmoudi S, et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 3.Kikkawa Y, Shitara H, Wakana S, et al. Mutations in a new scaffold protein Sans cause deafness in Jackson shaker mice. Hum Mol Genet. 2003;12:453–461. doi: 10.1093/hmg/ddg042. [DOI] [PubMed] [Google Scholar]
- 4.Kalay E, de Brouwer AP, Caylan R, et al. A novel D458V mutation in the SANS PDZ binding motif causes atypical Usher syndrome. J Mol Med. 2005;83:1025–1032. doi: 10.1007/s00109-005-0719-4. [DOI] [PubMed] [Google Scholar]
- 5.Ouyang XM, Yan D, Du LL, et al. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum Genet. 2005;116:292–299. doi: 10.1007/s00439-004-1227-2. [DOI] [PubMed] [Google Scholar]
- 6.Ye S, Dhillon S, Ke X, et al. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:E88–88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balemans W, Cleiren E, Siebers U, et al. A generalized skeletal hyperostosis in two siblings caused by a novel mutation in the SOST gene. Bone. 2005;36:943–947. doi: 10.1016/j.bone.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez AA, Reyes ML, Carvajal CA, et al. Congenital lipoid adrenal hyperplasia caused by a novel splicing mutation in the gene for the steroidogenic acute regulatory protein. J Clin Endocrinol Metab. 2004;89:946–951. doi: 10.1210/jc.2003-030345. [DOI] [PubMed] [Google Scholar]
- 9.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]