Abstract
Background
Amyotrophic lateral sclerosis (ALS) is characterized by the progressive loss of motor neurons, of unknown etiology. Previous studies showed reverse transcriptase in serum of ALS patients at levels comparable to HIV-infected patients; however, the source and significance of the retroviral elements is uncertain.
Methods
Expression of a human endogenous retrovirus (HERV-K), was determined in autopsy brain tissue of patients with ALS and compared to control populations, by real time polymerase chain reaction followed by sequencing of the amplified genes and confirmed by immunostaining.
Results
HERV-K pol transcripts were increased in patients with ALS compared to those with chronic systemic illness, but could not be detected in Parkinson’s disease or in the accidental death controls. Sequencing revealed several actively transcribed loci in the HML-2 and 3 subfamilies of HERV-K, with a specific pattern of expression including intact open reading frames and the transcription of a unique locus in ALS. The frequency of intact pol transcripts was highest in the motor cortex and the reverse transcriptase protein was localized to cortical neurons of ALS patients. HERV-K expression strongly correlated with TDP-43, a multi-functional protein known to be dysregulated in ALS.
Interpretation
We have identified a specific pattern of HERV-K expression in ALS, which may potentially define the pathophysiology of ALS. Targeting of activated genome-encoded retroviral elements may open new prospects for the treatment of ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the progressive loss of spinal and cortical motor neurons, the cause of which is unknown. Approximately twenty percent of familial ALS is associated with mutations in the superoxide dismutase (SOD1) gene 1; whereas triggers of sporadic ALS have yet to be clearly identified. As the majority of ALS cases are of sporadic nature, viral contributors to some cases of sporadic ALS may be possible. The possibility that viral infections may contribute to the etio-pathogenesis of ALS has been considered for several reasons. Previous studies have identified reverse transcriptase (RT) in serum of patients with ALS at levels comparable to those of human immunodeficiency virus (HIV)-infected patients; however, the source and significance of the retroviral elements are uncertain 2–4.
Retroviruses such as HIV and human T cell leukemia virus type-1 (HTLV-1) can occasionally lead to an ALS-like syndrome 5, 6. Anti-retroviral therapy in these HIV-infected patients can reverse the symptoms of this syndrome 7, 8. Murine retrovirus infection models, specifically murine leukemia virus (MuLV), can also display motor neuron pathology similar to ALS 9. Additional evidence for putative retrovirus involvement in ALS stems from measurement of RT activity, the enzyme that transcribes viral RNA into DNA. Sera and cerebrospinal fluid (CSF) from HIV-seronegative patients with ALS demonstrate RT activity, at comparable levels to that of HIV-infected individuals 3, 4. Increased RT activity is also found in serum of their first degree relatives, which leads to the speculation that RT activity may derive from inherited active human endogenous retroviruses (HERVs) 2.
HERVs are retroviruses that have become integrated into the human genome and are now transmitted in a Mendelian fashion. Some HERV loci retain sufficient capacity to express mRNA, viral proteins and viral particles, under either normal or inducible conditions such as chemical exposure, infection, inflammation or cancer. HERV expression has been identified in several disease states: neurologic disorders, including multiple sclerosis and schizophrenia; viral infections, including HIV and herpesvirus; and multiple types of cancers 10, 11. However, a causal relationship between HERV expression and disease is just beginning to be elucidated and their contribution to pathobiology remains poorly understood.
Despite clinical associations, past approaches to identify a definite causative viral agent in ALS has been unsuccessful. Here, for the first time, we characterized HERV-K polymerase (pol) mRNA and RT protein in brain tissue from patients with ALS and compared it to non-ALS individuals. We show that there is over-expression of select HERVs in patients with ALS and that HERV from these specific cytogenetic loci may be markers of ALS, as well as other motor neuron diseases.
RESULTS
Increased expression of HERV-K pol transcripts in ALS brain tissue
Quantitative real-time PCR was performed using degenerate HERV-K pol primers, which were able to amplify HERV-K pol products from HML-1 to HML-10 families 12, 13. PCR products were cloned into the pCRII vector and recombinant plasmids were used as a positive control. The use of plasmids for absolute quantification yielded similar results to that of relative gene expression, using the endogenous control 18S rRNA.
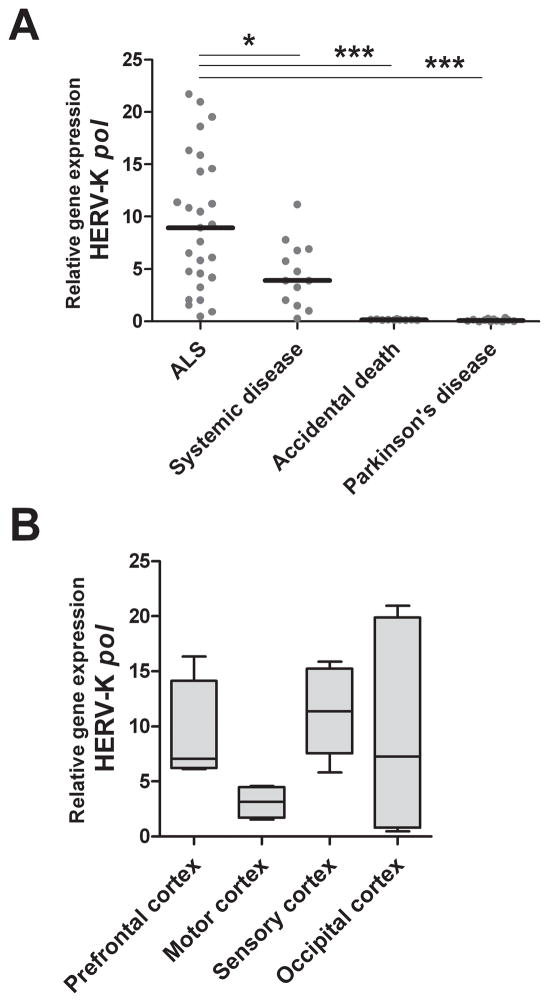
To explore a putative association between HERV expression and ALS, real-time PCR was performed using RNA samples from 62 frozen brain tissues: 28 from patients with pathologically confirmed ALS, 12 from age-matched control patients with chronic systemic disease, 10 from patients who died acutely without any pre-existing systemic illness and 12 from Parkinson’s patients. HERV-K pol mRNA transcripts were detectable in brain samples from patients with ALS and those with systemic disease, but not in patients who had died of Parkinson’s disease or in the accidental death group. Stratification of the population revealed a significant increase in HERV-K pol gene expression in ALS compared to all non-ALS control brain tissues (Figure 1A). Categorizing the data based on brain regions showed that HERV-K pol gene expression was substantially higher in prefrontal cortex and sensory cortex compared to motor cortex (p=0.05 and <0.05 respectively) from patients with ALS (Figure 1B). Regional differences in HERV expression were not seen in the control patient groups, suggesting that the patients with ALS had a unique pattern of HERV expression in the brain.
Figure 1. Increased expression of HERV-K pol in ALS brain tissue compared to non-ALS controls.
Expression of HERV-K pol was detected by real-time PCR using degenerate primers and cDNA from brain tissue samples from patients with ALS, systemic disease, accidental death controls and patients with Parkinson’s disease. Brain tissues from patients with ALS exhibit increased HERV-K pol RNA expression compared to each of the non-ALS control groups (A). Expression of HERV-K pol in patients with ALS was stratified by cortical brain regions: prefrontal, sensory, motor and occipital. There is weaker HERV-K pol expression in motor cortex of patients with ALS (B). Each point represents mean of two replicates for each individual brain sample, with lines indicating median value for each clinical group. Asterisk represents *p<0.05, ***p<0.0001.
Expression of HERV-K pol mRNA sequences from select genomic loci
To determine if sequence variation in the pol region may account for the increased RT activity in patients with ALS, HERV-K pol PCR products from prefrontal, motor, sensory and occipital cortex samples of 20 ALS and 9 patients with systemic disease were cloned into the pCRII vector for amplicon sequencing. Since the transcript levels from the patients with Parkinson’s Disease and the accidental death group were undetectable, they were not analyzed any further.
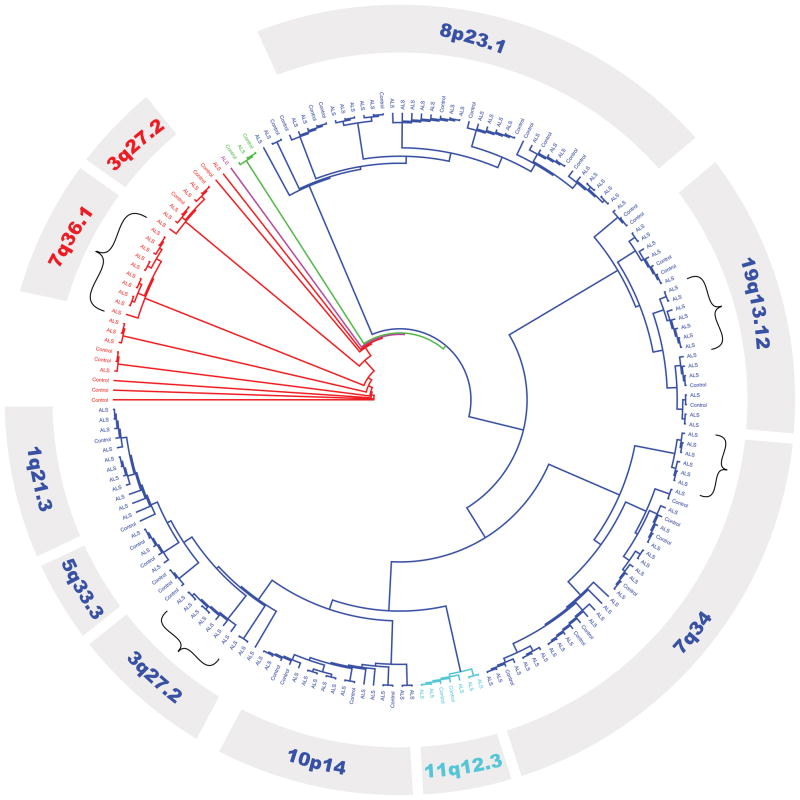
Alignment of >180 clones of HERV-K pol sequences from ALS and systemic disease control patients were examined (Figure S1). Analysis of the alignment of the partial pol sequences showed that within the same individual, multiple variants of HERV-K may be expressed. Phylogenetic tree analysis revealed that the majority of sequences originated from the HERV-K HML-2 family, with concomitant representation by HML-3, HML-6, HML-7 and HML-10 members (Figure 2). The fact that multiple loci from HERV-K are expressed in the same patient suggests that most HERV genes may respond to a common transactivator – provided that the promoter is responsive. More specifically, major branches could be assigned to HERV-K sequences from unique cytogenetic coordinates. Overall, expression from 26 individual cytogenetic regions were identified (Table S1), and the major loci represented among the clones are listed in Table 1. Select minor branches, specifically from 3q27.2 (HML-2), 7q34 (HML-2), 7q36.1 (HML-3) and 19q13.12 (HML-2) demonstrated grouping of only ALS-derived HERV-K clones.
Figure 2. Phylogenetic tree of HERV-K pol transcripts from ALS and systemic disease individuals.
HERV-K pol transcripts from patients with ALS and controls with systemic disease were sequenced and clustered using PhyML. HML-2 (blue), HML-3 (red), HML-6 (purple), HML-7 (cyan) and HML-10 (green) HERV-K families were identified. The most common HERV-K transcripts are identified by their cytogenetic coordinates. Parentheses indicate subgroups populated by transcripts only in patients with ALS.
Table 1.
Predominant HERV-K sequences from human cortical brain tissue of ALS and control patients.
| Cytogenetic coordinates | ALS patients n=20 | Percentage of ALS patients | Non-ALS patients n=9 | Percentage of Non-ALS patients | Fisher’s p value |
|---|---|---|---|---|---|
| 1q21.3 | 7 | 35.0% | 1 | 11.1% | NS |
| 3q27.2 (HML-2) | 4 | 20.0% | 5 | 55.6% | NS |
| 3q27.2 (HML-3) | 4 | 20.0% | 1 | 11.1% | NS |
| 5q33.3 | 4 | 20.0% | 3 | 33.3% | NS |
| 7q34 | 17 | 85.0% | 3 | 33.3% | <0.05 |
| 7q36.1 | 6 | 30.0% | 0 | 0.0% | Unique |
| 8p23.1 | 13 | 65.0% | 5 | 55.6% | NS |
| 10p14 | 9 | 45.0% | 3 | 33.3% | NS |
| 11q12.3 | 5 | 25.0% | 2 | 22.2% | NS |
| 19q13.12 | 14 | 70.0% | 4 | 44.4% | NS |
| Other | 11 | 55.0% | 4 | 44.4% | NS |
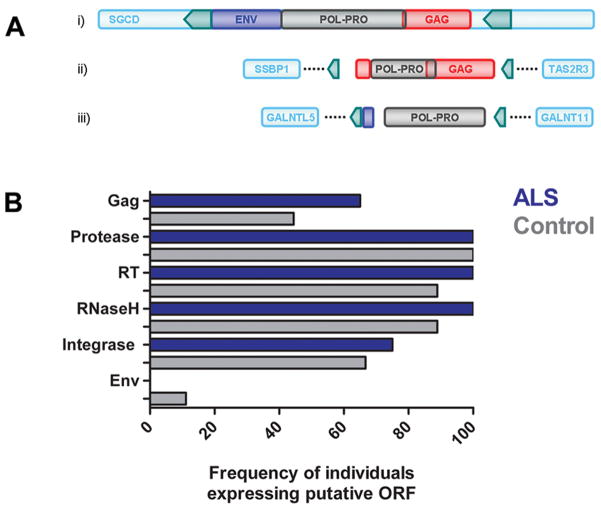
A prototypical HERV-K genome was identified in 5q33.3 (Figure 3A); in contrast, a large number of HERV-K pol sequences originated from loci that harbored partial HERV-K genomes, such as 7q34 and 7q36.1. Nonetheless, many of the truncated proviral genomes had putative ORFs in the pol gene (RT, RNaseH, integrase), and to a lesser extent in the gag gene (matrix and nucleocapsid proteins). All motor cortex samples from patients with ALS contained transcripts originating from genomic loci containing ORFs in the pol gene. Figure 3B shows the frequency of individuals expressing putative ORFs for retroviral proteins, based on the HERV-K mRNA expression pattern within each individual. Patients with ALS more frequently expressed mRNA from loci containing ORF for RT, RNaseH, integrase and gag proteins (65%) than controls (44%), whereas loci expressing ORF for protease were ubiquitously expressed in both groups (Figure 3B). Transcripts originating from loci with intact gag and pol ORF transcripts were present in all patients with ALS from which motor cortex samples were obtained, and predominantly originated from cytogenic regions 3q27.2 and 5q33.3 (Table S1, RetroSearch loci 14677 and 22748, respectively). This suggests that despite lower overall HERV-K pol mRNA levels in motor cortex; there is increased expression of intact gag and pol ORF transcripts in the ALS-affected tissue.
Figure 3. HERV-K gag and pol ORFs are more prevalent in ALS.
Both prototypical HERV-K genome in 5q33.3 (Panel A, i) and partial HERV-K genomes, such 7q36.1 (Panel A, iii) can encode for RT. In contrast, 7q34 has no ORF for RT, but contributes gag ORFs (Panel A, ii). HERV-K loci encode potential ORFs for retroviral proteins Gag, protease, RT, RNaseH, integrase and Env. Patients with ALS more frequently express combinations of loci encoding gag and pol ORFs, as compared to controls with systemic disease (Panel B).
HERV-K sequences from 7q34 were more frequently expressed in patients with ALS compared to controls (p<0.05). Importantly, the adjacent region 7q36.1 was uniquely expressed in patients with ALS and was not detected in any of the other patient groups (Table 1). Interestingly, this genomic region has previously been associated with motor neuron disease (MND) 14; however, the contributing genes have not been identified to date. The sequence translation of 7q34 transcripts reveals multiple stop codons and does not appear to have an ORF for RT, but has putative Gag matrix (p10) protein ORF 15, 16. Conversely, 7q36.1 which belongs to the HML-3 subfamily of HERV-K has a putative ORF for RT, with a unique insertion of two codons that would potentially translate to two amino acids inserted prior to the conserved RT motif LPQG (Figure S1)17, 18. The HML-3-derived 7q36.1 HERV-K ORF for RT resides in the probable 14.3 cM candidate interval (between D7S2511 and D7S798) for MND 14, and may thus represent a putative disease marker in ALS and other types of MND. These observations show a disease specific pattern of expression of HERV-K in ALS from novel loci in the human genome and intact large ORFs in the motor cortex, while patients with systemic illness had a non-specific pattern of expression with truncated transcripts and there was no expression of HERV-K in patients with Parkinson’s disease or those with accidental death. Together, these identified HERV-K sequences, particularly in the genomic region of 7q34 and 7q36.1, are potential candidate genes for ALS and MND.
Cellular localization of HERV-K reverse transcriptase in ALS brain tissue
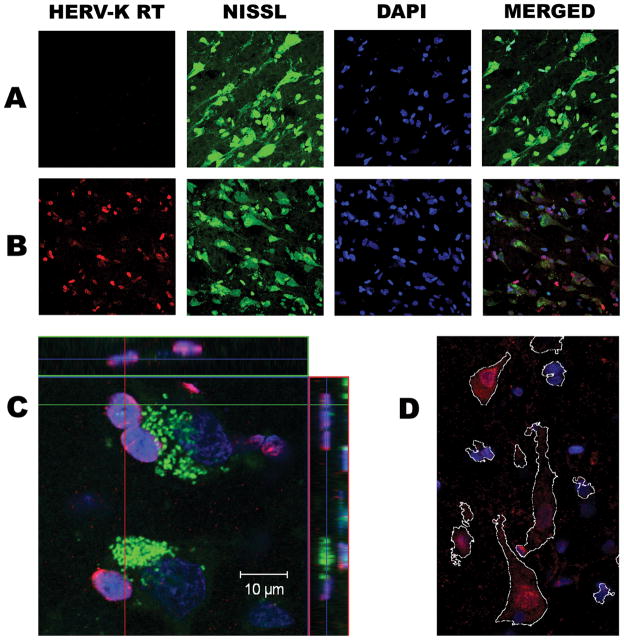
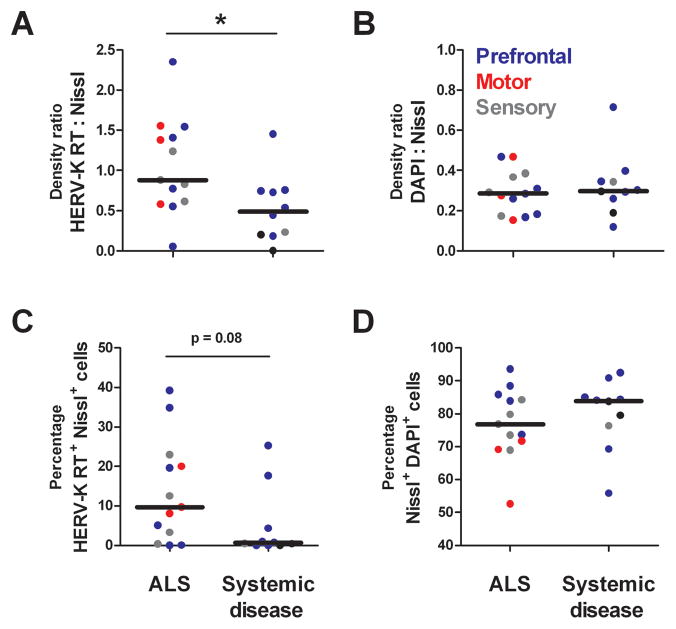
To determine the cell types in which the HERV-K transcripts were expressed, sections from ALS (n=13) and systemic disease (n=10) cortical brain tissue samples were immunostained for HERV-K RT. Figure 4 shows representative images of HERV-K RT protein detection in systemic disease (Panel A) and ALS (Panel B) cortical brain tissue. HERV-K RT expression was limited to neurons, as identified by fluorescent Nissl stain. At higher magnification, HERV-K RT staining appears nuclear, with most prominent staining in the perinuclear region, in smaller neurons (Panel C) and exhibits both nuclear and punctuate cytoplasmic staining in larger, pyramidal-like neurons (Panel D). Quantification of HERV-K RT staining was analyzed using two independent methods, adjusted density measurements and cell counting. The adjusted density measurement accounts for non-specific autofluorescence of human brain tissue by subtracting the background fluorescent density (with no primary antibody for HERV-K) from that of the stained tissue. Patients with ALS exhibited significantly more HERV-K RT protein, as measured by staining density, than cortical tissue from patients with systemic disease (Figure 5A). The same results were evident when cell counting was used to quantify HERV-K RT protein (Figure 5C). The HERV-K RT protein over-expression was especially evident in the ALS-affected prefrontal and motor cortex tissue samples. Overall, 10 of 13 patients with ALS and only 3 of 10 patients with systemic disease showed imunostaining for RT (Figure 5C), demonstrating a higher frequency of patients with ALS exhibiting HERV-K RT protein detection(Fisher’s test p<0.05). As an experimental control, the Nissl to DAPI ratios demonstrated that there was similar neuronal staining in both ALS patients and tissue from patients with systemic disease (Figure 5, panels B and D). These findings show a unique pattern of RT protein expression in patients with ALS that is consistent with the above observation of expressed transcripts with intact open reading frames for HERV-K RT in ALS brain tissue.
Figure 4. Representative images of HERV-K RT protein detection in cortical brain tissue.
HERV-K RT protein was detected by immunostaining in patients with systemic disease (Panel A) and ALS (Panel B) cortical brain tissue. High magnification of Nissl stained neuronal cells reveals peri-nuclear HERV-K RT staining (Panel C) and both nuclear and cytoplasmic staining in larger neurons (Panel D) in a patient with ALS.
Figure 5. Quanitative analysis of HERV-K RT immunostaining in cortical brain tissue of patients with ALS or systemic disease.
Patients with ALS exhibit significantly more HERV-K RT protein expression, as measured by staining density, than cortical tissue from patients with systemic disease (Panel A). The same trend was evident when cell counting was used to quantify HERV-K RT expression (Panel C). As an experimental control, the Nissl to DAPI ratios demonstrate that there is similar neuronal staining in both ALS and patients with systemic disease (Panels B and D). Asterisk represents p<0.05.
Corelation and co-localization of HERV-K RT and TDP-43 in neurons in patients with ALS
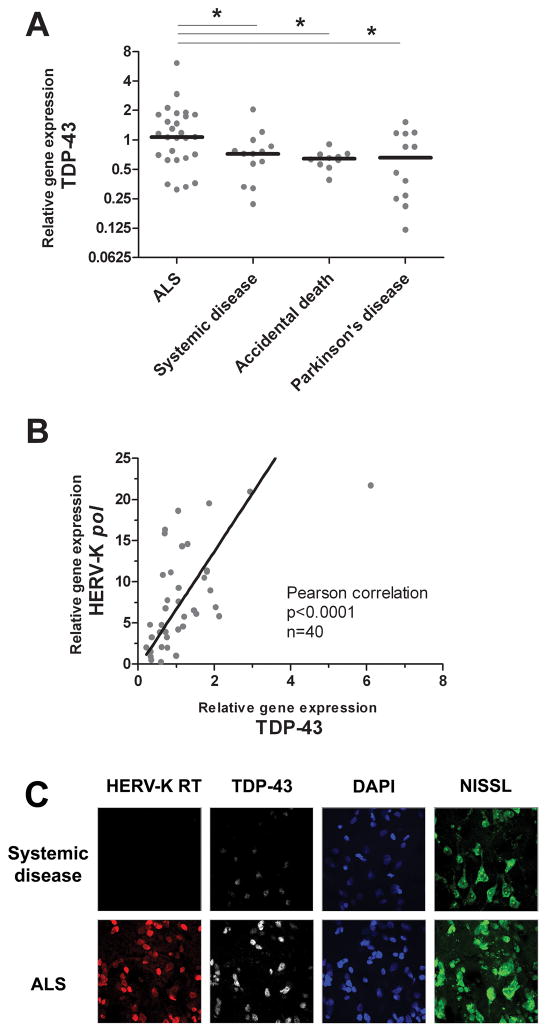
TDP-43 is a multi-functional protein involved in many cellular processes, including RNA binding, splicing and stability, microRNA biogenesis, provirus silencing and ssDNA binding; see reviews 19–21. In ALS, as well as other neurological diseases 20, TDP-43 has been shown to form cytoplasmic inclusions resulting from abberant ubiquitination, hyperphosphorylation, degradation and trafficking of the protein. Overexpression of total TDP-43 is also a hallmark of frontotemporal lobar degeneration 22, and ALS brain tissue as shown in Figure 6A. Interestingly, both HERV-K pol (RT RNA) and TDP-43 transcripts were positively and strongly correlated in ALS brain tissue (Figure 6B, p<0.0001). To determine whether the observed HERV-K RT expression was occurring simultaneously in neurons undergoing aberrant cellular function, ALS and control prefrontal cortex brain tissue was immunostained for both HERV-K RT and TDP-43 proteins. Figure 6C demonstrates that HERV-K RT and TDP-43 colocalize in neurons of patients with ALS. .
Figure 6. HERV-K pol (RT RNA) and RT protein expression correlates with TDP-43 in cortical neurons of patients with ALS.
Patients with ALS exhibit significantly more TDP-43 RNA expression, as measured by real-time PCR, than each of the groups of non-ALS controls (Panel A). A strong positive correlation between HERV-K pol and TDP-43 transcripts is observed in cortical brain tissue (Panel B). HERV-K RT protein expression colocalizes in neurons overexpressing TDP-43 (Panel C). Asterisk represents p<0.05.
DISCUSSION
In this novel study of HERV gene regulation and protein expression in brain tissue from ALS and non-ALS individuals, we show that there an ALS specific pattern of HERV-K subtypes expressed in neurons within the frontal lobe. HERV-K pol mRNA and RT demonstrate preferential regional expression within brain tissue, whereby prefrontal cortex exhibited the highest levels. For the first time, we demonstrate preferential expression of distinctive HERV-K pol sequences in patients with ALS compared to non-ALS individuals.
Multiple reports have previously demonstrated increased retroviral RT activity in the serum and CSF of patients with ALS compared to matched controls 2–4. In most of these patients RT was functionally active however, a clinical trial with a HIV protease inhibitor, Indinavir was without clinical benefit 3. Here we show that there is increased expression of HERV-K pol transcripts and RT protein in patients with ALS, which may explain the prior observations of RT activity in these patients. Moreover, differential regional HERV-K expression was seen in brain tissue from patients with ALS. Increased amounts of HERV-K transcripts were detected in prefrontal and sensory cortex, areas adjacent to the motor cortex, while the motor cortex itself displayed minimal RNA expression of HERV-K. A limitation of using brain samples in patients with terminal disease was that the neuronal cells producing pol mRNA were likely degenerated and replaced by glial cells, therefore the transcripts were detectable at lower levels in the motor cortex. Yet, HERV-K RT expression as detected by immunostaining remained robust in the surviving neurons of the motor cortex. Interestingly, RT was detected only in neurons and not in other cell types. This is in contrast to HERV-W which is expressed in glial cells of patients with Multiple Sclerosis 23 and has been associated with macrophage activation 24. HERV-W gag protein has also been found in neurons of normal brain, but has striking accumulation in axons of patients with Multiple Sclerosis in demylinated regions 25. While the role of HERV-K proteins in causing neuronal damage is unknown, previous studies have shown that HERV-W can cause inflammation in the brain leading to T cell-dependent hemorrhages in rodents 26. The env protein of HERV-W mediates its proinflammatory properties via the toll like receptor 427. The env protein of HIV, which is an exogenous human retrovirus, has been shown to be a potent neurotoxin 28. Therefore, increased RT protein expression in neurons may serve as a marker of ALS-involved brain tissue and may be of pathophysiological significance. It remains to be determined if similar findings can be found in the motor neurons of the brainstem and spinal cord. One possibility is that HERV-K expression precedes tissue atrophy. There is extensive evidence of prefrontal involvement in ALS; upwards of one-third of patients with ALS demonstrate frontal executive deficits 29. The observation that higher HERV-K expression occurs frequently in prefrontal cortex raises the possibility that RT over-expression may contribute to cognitive deficits attributed to the frontal lobe, although premortem cognitive assessments were not available on the patients studied. Moreover, HERV-K RT expression correlates with increased TDP-43 levels in neurons from frontal cortex tissue of ALS patients, suggesting that RT expression occurs in combination with other abberant cellular processes characteristic of ALS 19, 20. Although it is possible that the increased levels of these proteins may be an epiphenomenon, it seems unlikely since there is a very strong correlation in the expression of boith genes; and the two proteins colocalize in the same neurons in the motor cortex. This pattern was absent from all the other control groups. TDP-43 has been shown to act as a retroviral restriction factor by repressing the transactivation of HIV 21. Typical of most retroviral restriction factors, the TDP-43 promoter is likely responsive to interferon and inflammation associated transcription factors, as it contains binding sites for IRF1, IRF3 and NFκB. Hence the possibility that TDP-43 may also play a role in the innate immune response to HERV expression needs to be considered 30.
While some expression of HERV-K pol transcripts was noted in an age-matched group of patients who had prolonged systemic illnesses such as atherosclerotic disease and cancer, the frequency and pattern of protein expression was clearly different compared to patients with ALS. In these patients, the HERV-K transcripts were mostly defective and did not cluster in motor neurons, transcribe into protein or correlate with TDP-43 expression. In contrast, HERV-K pol transcripts were undectable in patients with Parkinson’s disease who were much older or in a much younger group of individuals who had died acutely without preexisting systemic illnesses. It is possible that systemic diseases, by virtue of generalized immune dysfunction, may lead to non-specific activation of HERV transcripts, as has been shown following macrophage activation 24 and in patients with HIV infection 31. Since the brain samples were obtained from different brain banks the possibility that some differences may occur to differences in handling of tissues cannot be entirely ruled out. However, this is unlikely since the postmortem intervals were similar in all groups (<24 hrs), they were all stored at −80°C until tested and other control genes could be amplified from all tissues.
Differing frequency of HERV-K pol mRNA transcripts in ALS versus non-ALS brain tissue may reflect either insertional polymorphism of select HERV-K sequences in the genome 11 or a global epigenetic change leading to cellular expression of a selection of HERV-K sequences not expressed in non-ALS patients 32. Examination of expressed HERV-K sequences suggests that a proportion is non-coding RNA which is unable to contribute to RT levels. Therefore, the degree of RT protein expression in patients with ALS is not due to non-specific increase in mRNA levels in affected tissue. Select HERV-K sequences must contribute to RT expression, and may exhibit differential enzymatic activity depending on amino acid fidelity to RT active sites. This was further confirmed by detection of RT protein by immunostaining in neurons of patients with ALS. Clearly, the ALS-specific HERV-K mRNA derived from 7q36.1 encodes an RT with amino acid insertions prior to the LPQG active site which may result in altered RT activity. The overall relevance of RT activity within cortical neurons is unknown; however, there is evidence that endogenous RT activity (and retrotransposition) can affect cell proliferation, differentiation and gene expression patterns 33, 34. Thus, RT expression in cortical neurons of patients with ALS may contribute to neurodegeneration.
The analysis of HERV expression in ALS brain tissue shows that there are specific loci of gene activation. Analysis of these loci suggest that there were ORFs for several HERV-K proteins besides RT. The expression of transcripts from the HERV-K loci in 7q34 and 7q36.1 in patients with ALS was specific for this population and may thus serve as markers of disease. HERV long terminal repeats (LTRs) can act as alternate promoters and enhancers for nearby gene expression 35, 36, regulating activation of adjacent genes. This may have pathophysiological significance for a gene such as DPP6 in 7q36.2 which has been associated with ALS susceptibility 37. Moreover, HERV-K 7q34 and 7q36.1 reside within a candidate interval for MND in which the susceptibility genes were not identified 14. This suggests that cytogenetic identification of HERV transcripts can prove to be a useful tool to identify disease-associated genes and sites of altered transcriptional activity.
The prototypic HERV-K family HML-2 has been previously studied in association with disease, however here we report substantial expression of HML-3 transcripts in cortical brain tissue from patients with ALS. Interestingly, HML-3 is believed to have amplified its presence in the human genome using retrotransposition in cis, a process that requires functional gag and pol but not env 38. It is estimated that there are 150 HML-3 loci within the human genome compared to only 60 HML-2 copies 39, 40. Yet, HML-2 loci are transcriptional more active than their HML-3 counterparts, largely due to HML-3’s longer period of integration. Phylogenetic analysis of HERV-K transcripts from patients with ALS confirms that at least some of HML-3 loci coding for RT, such as 7q36.1, remain actively transcribed during disease. It remains unclear if recombination of various HERV-K proteins originating from multiple loci may activate cycles of retrotransposition (or reinfection) and result in DNA damage precipitating cell death.
Evidence of RT activity has led to the search for novel retroviruses in ALS with no success 4. Here, for the first time, we show that HERV-K expression may account for the RT activity in ALS. In contrast to previous studies, we evaluated the expression of all HERV-K families at the mRNA and sequence level. Expression of the HERV-K RT protein, derived from the specific genomic loci 7q34-7q36.1 and HERV-K HML2 and HML3 subtypes, was detected in cortical and motor neurons of patients with ALS. Viral RNA and protein expression strongly correlated with expression of TDP-43, defining a specific phenotype for ALS. This pattern was not seen in patients with chronic systemic illness, nor in patients with Parkinson’s disease or younger patients with accidental death without evidence of disease. , These findings identify novel HERV-K and genomic markers of ALS, which may have important implications for defining the pathophysiology of sporadic forms of this disease. However, the mechanisms by which these viruses cause or contribute to pathological changes requires further study.
MATERIALS AND METHODS
Patients Brain Samples
Brain tissue from patients with ALS (n=28) and age-matched controls with chronic systemic illness (n=12) were obtained from the Johns Hopkins School of Medicine Brain Bank. The mean age of the study population was 63.6 (ALS) and 59.2 (systemic illness) years. Most patients with ALS were diagnosed as sporadic ALS, except for 3 patients with familial ALS. Pathologic examination was used to confirm the clinical diagnosis of ALS. All except one patient with ALS were Caucasian. The brain regions analyzed included prefrontal, motor, sensory and occipital cortex. The cause of death among the age-matched control patients was cardiac arrest related to coronary artery disease (n=7), systemic cancer (n=4; lung, pancreas, stomach and Hodgkins), hepatic failure (n=1). The brain regions analyzed included prefrontal, sensory and occipital cortex. Brain tissue (prefrontal and motor cortex) from another set of controls without any preexisting illnesses (n=10, mean age 32) who died from traffic accidents without CNS injury (n=7), or severe asthma (n=1), drowning (n=1), gastric perforation following laproscopy (n=1) were obtained from the National Institute for Child Health Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. Brain tissue (inferior parietal cortex) from patients with Parkinson’s Disease (n=12, mean age 79) was obtained from the Johns Hopkins School of Medicine, Department of Pathology, Brain Resource Center Brain Bank. The post-mortem interval of all patients was <24 hours.
Real-Time PCR and Sequencing
Total RNA was purified from frozen brain tissues using RNeasy® Lipid Tissue Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instruction. Optional On-Column DNase digestion was performed during RNA isolation using the RNase-Free DNase Set (Qiagen). Each 1 μg sample of purified RNA was treated with RNase-free DNase I (Invitrogen, Carlsbad, CA) at room temperature for 15 min to eliminate any genomic DNA contamination. DNase I digested RNA samples were reverse transcribed using random hexamers and a gene-specific HERV-K pol primer (5′-GTTGAAGAGCTCGACCTACAAAA- 3′) using SuperScript III First-Strand Synthesis System for real-time-PCR (Invitrogen).
Real-time PCR using SYBR Green was used to determine the level of HERV-K pol transcripts. The degenerate primer pair, HERV-K pol forward primer (5′-TCCCCTTGGAATACTCCTGTTTTYGT-3′) and HERV-K pol reverse primer (5′-CATTCCTTGTGGTAAAACTTTCCAYTG-3′), had been previously used to amplify HML-1 to HML-10 of HERV-K subfamilies 12, 13. 18S rRNA primer pairs and its competitors were used in a 3 to 7 ratio (Ambion Inc., Austin, TX). TDP-43 primers were: forward primer (5′-GTACGGGGATGTGATGGATG-3′) and reverse primer (5′-CTGCGCAATCTGATCATCTG-3′). PCR reactions (in duplicate) consisted of 12.5 μl of RT2 SYBR Green/ROX qPCR Master mix (Superarray Bioscience Corporation, Frederick, MD) and 0.2 μM of primer pairs and 5 μl cDNA (diluted 10 times from reverse transcription reaction) in a final 25 μL volume. The optimal PCR program was as follows: 94°C for 20 sec; 50°C for 1 min; 72°C for 30 sec and ran 45 cycles for both HERV-K pol and the endogenous control 18S rRNA. The ΔΔCt ratio method was used to determine relative gene expression as previously described 41, using a HERV-K pol positive ALS cDNA sample as a calibrator.
Using cDNA samples from cortical tissue, HERV-K pol amplicons (≈ 300 bp each, depending on sequence) of 29 patients (20 ALS and 9 non-ALS patients with systemic disease) were cloned into the TA cloning vector pCR II (Invitrogen, CA). Competent DH5α E. coli was transformed with plasmid and seven clones were randomly picked from each patient sample and sequenced (Johns Hopkins Core DNA facility, Baltimore, MD). Sequence alignment and phylogenetic analysis was performed using Geneious software 42 and verified with NCBI BLAST searches. Retroviral sequences and ORFs were identified using the RetroSearch database 15, and verified using NCBI Conserved Domains 16.
Immunohistochemistry and Imaging
Fresh frozen brain tissue was fixed with 4% paraformaldehyde at 4°C for 6 hours, cryopreserved at 4°C in increasing concentration (10%, 20% and 30%) of sucrose and 0.05 M PBS solution at 24 hour intervals and sectioned into 40 μm slices. Floating sections were washed with TBS-T (TBS with 0.05% Triton X-100), and blocked in 5% normal donkey serum and 0.25% Triton X-100 in TBS. Primary HERV-K RT antibody was (#H00002087-A01, AbNova, Taiwan) and incubated overnight at 4 °C. Secondary antibody, anti-mouse IgG (H + L)-Alexa Fluo594 conjugate was applied for 2 hours at room temperature (1:250, #A11005, Molecular Probes). NeuroTrace 500/525 Fluorescent Nissl Stain (Molecular Probes) was used as per manufacturer’s instructions. Nuclei were counter-stained with DAPI (Sigma). Controls were prepared by immunostaining without the primary antibody. Images of the slides were acquired using a Zeiss LSM 510 confocal microscope. The images were processed and analyzed using Zeiss LSM Image Browser, Zeiss Zen 2008 Light Edition and Image J software.
Statistical analysis
Associations between HERV-K pol and RT expression in ALS and non-ALS groups were determined using 2-tailed Mann-Whitney tests (non-parametric data). Fisher’s exact test was used to compare differences in frequencies of responsive individuals. All statistical analysis was performed using GraphPad Prism 5.0 (http://www.graphpad.com).
Supplementary Material
HERV-K pol transcripts from patients with ALS and those with systemic disease were sequenced and aligned using Geneious software. Consensus sequences and sequence logos are shown above individual alignments for HML-2 and HML-7, HML-3 and HML-6, as well as HML-10 HERV-K families. Overlap of sequences indentified in both ALS and patients with systemic disease are illustrated by grey bars.
Acknowledgments
The authors would like to thank Dr. Myoung Hwa Lee for providing expertise in confocal microscopy technique and Dr. Stuart Ray for advice on bioinformatics analysis. Funding was provided by NINDS, NIH.
References
- 1.Vucic S, Kierna MC. Pathophysiology of neurodegeneration in familial amyotrophic lateral sclerosis. Curr Mol Med. 2009;9:255–272. doi: 10.2174/156652409787847173. [DOI] [PubMed] [Google Scholar]
- 2.Steele AJ, Al-Chalabi A, Ferrante K, et al. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology. 2005;64:454–458. doi: 10.1212/01.WNL.0000150899.76130.71. [DOI] [PubMed] [Google Scholar]
- 3.MacGowan DJ, Scelsa SN, Imperato TE, et al. A controlled study of reverse transcriptase in serum and CSF of HIV-negative patients with ALS. Neurology. 2007;68:1944–1946. doi: 10.1212/01.wnl.0000263188.77797.99. [DOI] [PubMed] [Google Scholar]
- 4.McCormick AL, Brown RH, Jr, Cudkowicz ME, et al. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology. 2008;70:278–283. doi: 10.1212/01.wnl.0000297552.13219.b4. [DOI] [PubMed] [Google Scholar]
- 5.Verma A, Berger JR. ALS syndrome in patients with HIV-1 infection. J Neurol Sci. 2006;240:59–64. doi: 10.1016/j.jns.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki T, Nakagawa M, Nagai M, et al. HTLV-I-associated myelopathy (HAM)/tropical spastic paraparesis (TSP) with amyotrophic lateral sclerosis-like manifestations. J Neurovirol. 2000;6:544–548. doi: 10.3109/13550280009091955. [DOI] [PubMed] [Google Scholar]
- 7.von Giesen HJ, Kaiser R, Koller H, et al. Reversible ALS-like disorder in HIV infection. An ALS-like syndrome with new HIV infection and complete response to antiretroviral therapy. Neurology. 2002;59:474. doi: 10.1212/wnl.59.3.474. author reply 474–475. [DOI] [PubMed] [Google Scholar]
- 8.Moulignier A, Moulonguet A, Pialoux G, Rozenbaum W. Reversible ALS-like disorder in HIV infection. Neurology. 2001;57:995–1001. doi: 10.1212/wnl.57.6.995. [DOI] [PubMed] [Google Scholar]
- 9.Jolicoeur P. Neuronal loss in a lower motor neuron disease induced by a murine retrovirus. Can J Neurol Sci. 1991;18:411–413. doi: 10.1017/s031716710003256x. [DOI] [PubMed] [Google Scholar]
- 10.Dolei A, Perron H. The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: a biological interface between virology, genetics, and immunology in human physiology and disease. J Neurovirol. 2009;15:4–13. doi: 10.1080/13550280802448451. [DOI] [PubMed] [Google Scholar]
- 11.Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326–333. doi: 10.1016/j.tig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Contreras-Galindo R, Gonzalez M, Almodovar-Camacho S, et al. A new Real-Time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J Virol Methods. 2006;136:51–57. doi: 10.1016/j.jviromet.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 13.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopinath S, Blair IP, Kennerson ML, et al. A novel locus for distal motor neuron degeneration maps to chromosome 7q34-q36. Hum Genet. 2007;121:559–564. doi: 10.1007/s00439-007-0348-9. [DOI] [PubMed] [Google Scholar]
- 15.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A, Anderson JB, Chitsaz F, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma B, Kaushik N, Singh K, et al. Substitution of conserved hydrophobic residues in motifs B and C of HIV-1 RT alters the geometry of its catalytic pocket. Biochemistry. 2002;41:15685–15697. doi: 10.1021/bi026311z. [DOI] [PubMed] [Google Scholar]
- 18.Berkhout B, Jebbink M, Zsiros J. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 1999;73:2365–2375. doi: 10.1128/jvi.73.3.2365-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, Baralle FE. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv Genet. 2009;66:1–34. doi: 10.1016/S0065-2660(09)66001-6. [DOI] [PubMed] [Google Scholar]
- 20.Geser F, Martinez-Lage M, Kwong LK, et al. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: the TDP-43 diseases. J Neurol. 2009;256:1205–1214. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou SH, Wu F, Harrich D, et al. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra M, Paunesku T, Woloschak GE, et al. Gene expression analysis of frontotemporal lobar degeneration of the motor neuron disease type with ubiquitinated inclusions. Acta Neuropathol. 2007;114:81–94. doi: 10.1007/s00401-007-0240-7. [DOI] [PubMed] [Google Scholar]
- 23.Mameli G, Astone V, Arru G, et al. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J Gen Virol. 2007;88:264–274. doi: 10.1099/vir.0.81890-0. [DOI] [PubMed] [Google Scholar]
- 24.Johnston JB, Silva C, Holden J, et al. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann Neurol. 2001;50:434–442. doi: 10.1002/ana.1131. [DOI] [PubMed] [Google Scholar]
- 25.Perron H, Lazarini F, Ruprecht K, et al. Human endogenous retrovirus (HERV)-W ENV and GAG proteins: physiological expression in human brain and pathophysiological modulation in multiple sclerosis lesions. J Neurovirol. 2005;11:23–33. doi: 10.1080/13550280590901741. [DOI] [PubMed] [Google Scholar]
- 26.Firouzi R, Rolland A, Michel M, et al. Multiple sclerosis-associated retrovirus particles cause T lymphocyte-dependent death with brain hemorrhage in humanized SCID mice model. J Neurovirol. 2003;9:79–93. doi: 10.1080/13550280390173328. [DOI] [PubMed] [Google Scholar]
- 27.Perron H, Lang A. The Human Endogenous Retrovirus Link between Genes and Environment in Multiple Sclerosis and in Multifactorial Diseases Associating Neuroinflammation. Clin Rev Allergy Immunol. 39:51–61. doi: 10.1007/s12016-009-8170-x. [DOI] [PubMed] [Google Scholar]
- 28.Power C, McArthur JC, Nath A, et al. Neuronal death induced by brain-derived human immunodeficiency virus type 1 envelope genes differs between demented and nondemented AIDS patients. J Virol. 1998;72:9045–9053. doi: 10.1128/jvi.72.11.9045-9053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomen-Hoerth C, Murphy J, Langmore S, et al. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 30.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nature Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 31.Garrison KE, Jones RB, Meiklejohn DA, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS pathogens. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura A, Okazaki Y, Sugimoto J, et al. Human endogenous retroviruses with transcriptional potential in the brain. J Hum Genet. 2003;48:575–581. doi: 10.1007/s10038-003-0081-8. [DOI] [PubMed] [Google Scholar]
- 33.Spadafora C. A reverse transcriptase-dependent mechanism plays central roles in fundamental biological processes. Syst Biol Reprod Med. 2008;54:11–21. doi: 10.1080/19396360701876815. [DOI] [PubMed] [Google Scholar]
- 34.Muotri AR, Chu VT, Marchetto MC, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 35.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol. 2006;80:10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. Human specific modulation of transcriptional activity provided by endogenous retroviral inserts. J Virol. 2009 doi: 10.1128/JVI.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Es MA, van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40:29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 38.Belshaw R, Katzourakis A, Paces J, et al. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol. 2005;22:814–817. doi: 10.1093/molbev/msi088. [DOI] [PubMed] [Google Scholar]
- 39.Mayer J, Meese EU. The human endogenous retrovirus family HERV-K(HML-3) Genomics. 2002;80:331–343. doi: 10.1006/geno.2002.6839. [DOI] [PubMed] [Google Scholar]
- 40.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 41.Laderoute MP, Giulivi A, Larocque L, et al. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. Aids. 2007;21:2417–2424. doi: 10.1097/QAD.0b013e3282f14d64. [DOI] [PubMed] [Google Scholar]
- 42.Drummond AJAB, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious. (v4.6) 2009 www.geneious.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HERV-K pol transcripts from patients with ALS and those with systemic disease were sequenced and aligned using Geneious software. Consensus sequences and sequence logos are shown above individual alignments for HML-2 and HML-7, HML-3 and HML-6, as well as HML-10 HERV-K families. Overlap of sequences indentified in both ALS and patients with systemic disease are illustrated by grey bars.