Abstract
Lower levels of global DNA methylation in white blood cell (WBC) DNA have been associated with adult cancers. It is unknown whether individuals with a family history of cancer also have lower levels of global DNA methylation early in life. We examined global DNA methylation in WBC (measured in three repetitive elements, LINE1, Sat2 and Alu, by MethyLight and in LINE1 by pyrosequencing) in 51 girls aged 6–17 years. Compared to girls without a family history of breast cancer, methylation levels were lower for all assays in girls with a family history of breast cancer and statistically significantly lower for Alu and LINE1 pyrosequencing. After adjusting for age, body mass index (BMI) and Tanner stage, only methylation in Alu was associated with family history of breast cancer. If these findings are replicated in larger studies, they suggest that lower levels of global WBC DNA methylation observed later in life in adults with cancer may also be present early in life in children with a family history of cancer.
Key words: Alu, DNA global methylation, early life exposure, epigenetics, LINE1, methylight, pyrosequencing, Sat2
Introduction
Lower levels of global DNA methylation (demethylation) have been observed in breast cancer tissues1 and white blood cell (WBC) DNA of cases compared to controls.2 Global DNA methylation in WBC has also been associated with a number of other cancers including head and neck and bladder cancers.3,4 Some epidemiological studies suggest that nutritional, chemical and physical factors might alter methylation levels in WBC DNA.5–9 Greater differences in global DNA methylation levels between older monozygotic twins compared with differences in global DNA methylation in younger monozygotic twins suggest that DNA methylation levels change over the lifecourse and that these changes cannot be explained by genetics.6 Decreases in global DNA methylation with increases in age suggest that endogenous and/or exogenous exposures may be associated with DNA methylation. For example, an occupational study found that benzene exposure was associated with lower levels of global methylation of peripheral blood DNA.5 Global DNA methylation has also been associated with other environmental exposures including cigarette smoking.10
Less is known about whether exposures early in life are associated with global DNA methylation. Early life exposures may be particularly important to hormonal cancers like breast cancer.11–14 For example, prenatal exposure to diethylstilbestrol, a synthetic nonsteroidal estrogen, has been associated with an increase in breast cancer.13 In animal models, neonatal exposure to diethylstilbestrol alters the expression of DNA methyltransferases and methylation of global DNA in the epididymis of mice.15 Persistent dysregulation of global DNA methylation may be a plausible molecular mechanism underlying the association between early life exposures and disease development later in life, including breast cancer.10,16–18 However, data on the association between early life exposures and global methylation levels in WBC DNA are limited. Using information from girls ages 6–17 years enrolled in a prospective study of early-life exposures and pubertal development, we conducted a pilot study in 51 girls to examine whether global DNA methylation measured in childhood and adolescence differed in girls with and without a family history of breast cancer.
Results
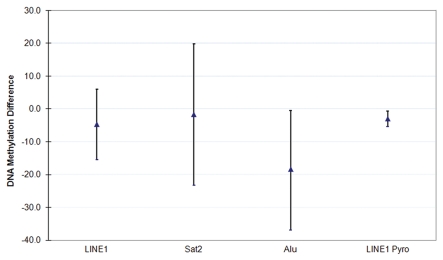
The average ages of girls with and without a family history of breast cancer were 13.3 [standard deviation (SD0 = 2.9] and 12.3 (SD = 3.2) years, respectively. Age was not statistically significantly associated with global DNA methylation. The Spearman correlations with age for LINE1, Sat2 and Alu by MethyLight are −0.08 (p = 0.57), 0.15 (p = 0.31) and 0.09 (p = 0.53), respectively. The correlation between age and LINE1 by pyrosequencing was −0.19 (p = 0.18). Table 1 presents the mean and SD of global DNA methylation using the MethyLight and pyrosequencing assays by different participant characteristics. Differences by family history are reported in Table 1 and illustrated in Figure 1 (Sup. Table 1). Figure 1 presents the mean difference in global DNA methylation between girls with and without a family history of breast cancer. Compared to girls without a family history of breast cancer, global DNA methylation levels were lower for all assays in girls with a family history of breast cancer, and statistically significantly lower as measured by Alu MethyLight and LINE1 pyrosequencing.
Table 1.
Mean levels of WBC DNA methylation in young girls using the MethyLight and LINE1 pyrosequencing assays by participant characteristics
| MethyLight | Pyrosequencing | ||||
| LINE1 | Sat2 | Alu | LINE1 | ||
| Breast cancer family history | |||||
| Yes (n = 31) | Mean, (SD) | 93.7, (16.9) | 125.7, (40.6) | 151.4, (35.9)* | 75.0, (4.4)* |
| No (n = 20) | Mean, (SD) | 98.4, (21.2) | 127.4, (31.6) | 169.8, (25.1) | 78.0, (3.5) |
| Breast Tanner stage | |||||
| 1–3 (n = 22) | Mean, (SD) | 99.4, (21.1) | 128.2, (37.5) | 155.5, (28.9) | 76.6, (4.2) |
| 4–5 (n = 28) | Mean, (SD) | 93.3, (16.3) | 125.7, (37.6) | 160.9, (36.9) | 75.7, (4.5) |
| Menarche | |||||
| No (n = 20) | Mean, (SD) | 97.0, (21.7) | 120.0, (36.5) | 155.1, (29.8) | 77.1, (3.4) |
| Yes (n = 31) | Mean, (SD) | 94.7, (16.7) | 130.5, (37.2) | 160.8, (35.4) | 75.6, (4.7) |
| Age at menarche (years) | |||||
| <12 (n = 16) | Mean, (SD) | 99.7, (16.1) | 128.9, (23.5) | 153.4, (23.4) | 77.2, (3.8) |
| ≥12 (n = 15) | Mean, (SD) | 89.3, (16.1) | 132.2, (48.7) | 168.7, (44.3) | 73.9, (5.1) |
| BMI (kg/m2) | |||||
| <23.8 (n = 31) | Mean, (SD) | 95.5, (21.4) | 130.4, (43.4) | 159.3, (36.6) | 75.6, (4.5) |
| ≥23.8 (n = 20) | Mean, (SD) | 95.6, (13.9) | 120.2, (23.6) | 157.5, (27.6) | 77.0, (3.9) |
p < 0.05.
Figure 1.
Mean differences (and 95% CI) in global DNA methylation between girls with a family history of breast cancer compared to girls without a family history of breast cancer.
Girls with Tanner scores of 4–5 had slightly lower LINE1 and Sat2 methylation by MethyLight compared to girls with Tanner scores of 1–3. LINE1, but not Sat2 or Alu, levels were also lower among girls who had already experienced menarche versus girls who had not. And among girls who had already experienced menarche, LINE1 methylation levels measured by MethyLight and pyrosequencing were lower among girls with menarche ≥12 years compared to girls who started menstruation at <12 years. Among girls who had already experienced menarche, age at menarche was inversely correlated with LINE1 methylation by both MethyLight and pyrosequencing (r = −0.41, p = 0.02; and r = −0.48, p = 0.007, respectively).
In the multivariable linear models (Table 2), LINE1 methylation measured by MethyLight was inversely associated with Tanner score. In addition, family history of breast cancer was inversely associated with Alu methylation but not with the other DNA methylation markers after adjusting for age, BMI and Breast Tanner Staging.
Table 2.
Multivariable* age-adjusted linear regression of genomic DNA methylation
| Parameter estimate (standard error) | MethyLight | Pyrosequencing | ||
| LINE1 | Sat2 | Alu | LINE1 | |
| Family history (Yes vs. No) | 0.04 | −0.02 | −0.14 | −0.03 (0.13) |
| (0.55) | (0.80) | (0.03) | ||
| BMI (kg/m2) | 0.01 | 0.005 | 0.001 | 0.002 (0.15) |
| (0.07) | (0.94) | (0.87) | ||
| Breast Tanner Staging | −0.07 | 0.003 | 0.02 | −0.01 (0.21) |
| (0.0008) | (0.93) | (0.52) | ||
adjusted for age, family history, BMI and Breast Tanner Staging.
Discussion
We observed lower levels of global methylation in WBC DNA in girls with a family history of breast cancer than in girls without a family history, although this association was only statistically significant with global DNA methylation as measured by Alu MethyLight and LINE1 pyrosequencing. In utero exposures and early-life factors have been associated with increased risk of breast cancer.11,12,14 Changes in DNA methylation level may be one mechanism linking early-life exposures to adult health.10,16,21 We have previously reported that in utero exposure to maternal cigarette smoking and other risk factors across the life course, such as age at menarche, birth length and pregnancy history, were associated with adult levels of methylation measured in the peripheral blood of women.16 Here we observed an inverse association between LINE1 methylation and age at menarche and breast development (Tanner score). The association with Tanner staging remained after adjusting for age, family history and body mass index. The molecular mechanisms by which pubertal status may be associated with methylation patterns needs to be further investigated in larger samples.
Studies of global DNA methylation patterns in families have found positive correlations in DNA methylation levels within families pointing to shared environment and/or genetics in explaining DNA methylation patterns.8 However, twin studies also support greater differences in DNA methylation patterns as twins age suggesting a role for the environment in explaining differences even within families.6 Lower 5-methyldeoxycytosine (m5C) in leukocyte DNA was observed in breast cancer patients compared with cancer-free controls, and the association between global demethylation and breast cancer risk was more pronounced among women with a family history of breast cancer.2 We found that girls with a family history of breast cancer had lower methylation levels in Alu and LINE1 elements compared with girls without a family history; the association with Alu and family history remained after further adjusting for Tanner stage and BMI.
DNA methylation of centromeric repeats and repetitive sequences accounts for the bulk of global DNA methylation levels in the genome. LINE sequences account for at least 34% of the human genome and Alu repeats comprise at least 10%.22,23 Loss of DNA methylation in these sequences is thought to cause chromosomal instability, reactivation of transposable elements and loss of imprinting resulting in the initiation of carcinogenesis.24
If replicated in much larger studies, our study suggests that DNA methylation levels may differ between families with and without breast cancer, and that these differences may be observed early in life. Prospective studies measuring global DNA methylation over time within the same individuals are needed in order to understand the potential role of environmental factors in altering global DNA methylation patterns, even early in life.
Materials and Methods
Study participants.
This pilot study includes girls participating in LEGACY (Lessons in Epidemiology and Genetics of Adult Cancer from Youth), a prospective study of early-life exposures, pubertal development and other endpoints relevant to breast cancer etiology. The study included girls aged 6–17 years with a family history of breast cancer whose mothers are enrolled in the California (N = 22) and New York (N = 9) sites of the Breast Cancer Family Registry (BCFR).19 Girls from families without breast cancer in first or second degree relatives were identified from participating BCFR mothers who were asked to provide names and contact information for friends or acquaintances who have daughters of similar ages (N = 20). The study was approved by the Institutional Review Boards of Columbia University and the Cancer Prevention Institute of California. The girls' mothers were interviewed by trained interviewers about their daughters' health, pubertal development and exercise. The girls completed a self-administered questionnaire about their pubertal status. Weight and height measurements and blood for the isolation of plasma and WBC DNA were collected from the girls at the time of interview in the clinic (NY) or the girls' home (CA).
DNA extraction and bisulfite treatment.
Genomic DNA was extracted from total WBC by a salting out procedure. Cells were lysed with SDS in a nuclei lysis buffer and treated with RNase A (final 133 µg/mL) and RNase T1 (final 20 units/mL) to remove RNA. Proteins were coprecipitated with NaCl (330 µL of saturated NaCl added per 1 mL solution) by centrifugation. Genomic DNA was recovered from the supernatant by precipitation with 100% ethanol, washed in 70% ethanol and dissolved in the Tris-EDTA buffer. The laboratory investigator who performed the assays was blinded to the epidemiologic data.
For the MethyLight and LINE1 pyrosequencing assays, aliquots of DNA (500 ng) were bisulfite-treated with the EZ DNA methylation kit (Zymo Research, Orange, CA) to convert unmethylated cytosines to uracils while leaving methylated cytosines unmodified. The DNA was resuspended in 20 µL of distilled water and stored at −20°C until used.
MethyLight assay.
We used the sequences of probes and forward and reverse primers designated as LINE1-M1, Alu-M2 and Sat2-M1 in Weisenberger et al.20 Polymerase chain reaction (PCR) was performed in a 10 µl reaction volume with 0.3 µM forward and reverse PCR primers, 0.1 µL probe, 3.5 µM MgCl2, using the following PCR program: 95°C for 10 min, then 55 cycles of 95°C for 15 sec, followed by 60°C for 1 min. Standard curves for the AluC4 repeat control reaction were generated from 1:25 serial dilutions of bisulfite-converted, CpGenome universal methylated and unmethylated DNAs. A pooled sample of DNA from five controls was used as a quality control and analyzed with each batch of test samples. Assays were run on an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). The inter assay coefficient of variation (CV) was 1.2%.
The MethyLight data were expressed as percent of methylated reference (PMR) values.
PMR = 100% * 2 exp − [δ Ct (target gene in sample − control gene in sample) − δ Ct (100% methylated target in reference sample − control gene in reference sample)].
LINE1 amplification and pyrosequencing.
The methylation status of LINE1 was also measured by pyrosequencing. The sequences of primers and PCR condition have been described in detail previously.5 The biotinylated PCR products were purified and made single-stranded to act as a template in the pyrosequencing reaction as recommended by the manufacture using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Westborough, MA). Then, 0.3 nmol/L of pyrosequencing primer was annealed to the purified single-stranded PCR product and pyrosequencing was run on a PyroMark Q24. We used non-CpG cytosine residues as internal controls to verify efficient sodium bisulfite DNA conversion and universal unmethylated and methylated DNAs were run as controls. Methylation quantification was performed using the PyroMark Q24 1.010 software. The degree of methylation was expressed for each DNA locus as percentage methylated cytosine over the sum of methylated and unmethylated cytosine. The inter assay CV was 1.1%.
Statistical methods.
We used ANOVA to test for differences in global methylation by participant characteristics. We calculated Spearman correlation coefficients to determine the correlation of each marker with age at menarche and used multivariable linear regression models to examine associations between each methylation marker and participant characteristics, including family history of breast cancer (yes vs. no), age (years), body mass index (BMI, kg/m2), reached menarche (yes vs. no), age at menarche (<12 vs. ≥12) and Tanner breast score (1–3 vs. 4–5). We also examined supplemental models which adjusted for age at menarche and whether a girl started menstruation (yes vs. no). All analyses were performed with SAS software 9.0 (SAS Institute, Cary, NC).
Acknowledgements
This work was supported by an award from the Breast Cancer Research Foundation and NIH grants U01 CA69398, P30 CA13696 and P30 ES009089. The recruitment of girls in California was supported by a Stanford Cancer Center Development grant and NIH grant R03 CA141528. This work was also supported by the National Cancer Institute, National Institutes of Health under RFA # CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR.
Abbreviations
- BMI
body mass index
- LINE1
long interspersed nucleotide element-1
- m5C
5-methylcytosine
- Sat2
satellite 2
- SINEs
short interspersed nucleotide elements
- WBC
white blood cell
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/13393
Supplementary Material
References
- 1.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:5–9. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 2.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis E, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation is a biomarker for bladder cancer susceptibility in the Spanish bladder cancer case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 5.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose Benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 6.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar MK, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–1697. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 8.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change in DNA methylation over time with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 10.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Titus-Ernstoff L, Egan KM, Newcomb PA, Ding J, Trentham-Dietz A, Greenberg ER, et al. Early life factors in relation to breast cancer risk in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2002;11:207–211. [PubMed] [Google Scholar]
- 12.Trichopoulos D. Hypothesis: Does breast cancer originate in utero? Lancet. 1990;335:939–940. doi: 10.1016/0140-6736(90)91000-z. [DOI] [PubMed] [Google Scholar]
- 13.Palmer JR, Wise LA, Hatch EE, Troisi R, Titus-Ernstoff L, Strohsnitter W, et al. Prenatal diethylstilbestrol exposure and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1509–1514. doi: 10.1158/1055-9965.EPI-06-0109. [DOI] [PubMed] [Google Scholar]
- 14.Michels KB, Xue F. Role of birthweight in the etiology of breast cancer. Int J Cancer. 2006;119:2007–2025. doi: 10.1002/ijc.22004. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, Fukata H, Kogo Y, Ohgane J, Shiota K, Mori C. Neonatal exposure to diethylstilbestrol alters the expression of DNA methyltransferases and methylation of genomic DNA in the epididymis of mice. Endocr J. 2006;53:331–337. doi: 10.1507/endocrj.k06-009. [DOI] [PubMed] [Google Scholar]
- 16.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–2310. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on Adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annual Review of Nutrition. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 19.John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, et al. The breast cancer family registry: An infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:375–389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucl Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA Methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 24.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.