Abstract
Objective
To determine susceptibility genes for high myopia in Singaporean Chinese.
Design
A meta-analysis of two genome wide association (GWA) datasets in Chinese and a follow-up replication cohort in Japanese.
Participants and Controls
Two independent datasets of Singaporean Chinese individuals aged 10–12 years (SCORM -- Singapore Cohort Study of the Risk factors for Myopia: cases=65, controls=238) and aged > 21 years (SP2 -- Singapore Prospective Study Program: cases=222, controls=435) for GWA studies, and a Japanese dataset aged >20 years (cases=959, controls=2128) for replication.
Methods
Genomic DNA samples from SCORM and SP2 were genotyped using various Illumina Beadarray platforms (> HumanHap 500). Single-locus association tests were conducted for each dataset with meta-analysis using pooled z-scores. The top-ranked genetic markers were examined for replication in Japanese dataset. Fisher’s P was calculated for the combined analysis of all three cohorts.
Main outcome measures
High myopia, defined by spherical equivalent (SE) ≤ −6.00 diopters (D); controls defined by SE between −0.50D and +1.00D.
Results
Two SNPs (rs12716080 and rs6885224) in the gene CTNND2 on chromosome 5p15 ranked top in the meta-analysis of our Chinese datasets (meta- P = 1.14×10−5 and meta- P = 1.51×10−5, respectively) with strong supporting evidence in each individual dataset analysis (Max P = 1.85.x10−4 in SCORM: Max P = 8.8×10−3 in SP2). Evidence of replication was observed in Japanese dataset for rs6885224 (P = 0.035, meta-P of three datasets: 7.84×10−6).
Conclusion
This study identified strong association of CTNND2 for high myopia in Asian datasets. The CTNND2 gene maps to a known high myopia linkage region on chromosome 5p15.
Keywords: myopia, genome wide association, CTNND2, single nucleotide polymorphism, genetics
Myopia is a common eye disorder and a major public health concern in urban East Asian populations, affecting nearly 40% of Chinese persons aged 40 to 79 years1–3. High myopia, defined by spherical equivalent (SE) ≤ −5.00 diopter (D) or SE ≤ −6.00 D for at least one eye, is associated with significant ocular morbidity, including retinal detachment and myopic macular degeneration4;5.
The genetic etiologic basis of myopia and high myopia is supported by data from familial aggregation, segregation, and twin studies5–12. The relative risk of myopia in siblings of a person with myopia (λs) has been estimated to be strongest in high myopia (SE ≤ −6.00 D; λs = 5 – 20), and moderate for lower degrees of myopia (SE: −1.00 to −3.00D; λs=1.5–3)5;12. To date, more than 15 chromosomal regions (or genetic loci, designated as MYP loci) have been mapped for myopia-related phenotypes by genome wide linkage scans, and many candidate genes have been reported by association and sequencing studies13. However, no gene implicated in myopia has been consistently replicated.
Genome wide association (GWA) studies have become an important and unbiased approach to aid in the search for causal sequence variants by screening upwards of a million single nucleotide polymorphisms (SNPs) spaced across the genome. This is exemplified by the recent GWA studies of seven complex trait disorders by the Wellcome Trust Case-Control Consortium14. Recently, Nakanishi et al15 reported the first GWA study for pathological myopia (axial length > 26 mm of both eyes; equivalent to refractive error<−6.00 D16), and detected a novel susceptibility locus at chromosome 11q24.1. At the time of writing this paper, no GWA studies have been reported for myopia in a Chinese population.
The aim of the present study is to identify genetic variants that may account for individual susceptibility to high myopia using a GWA approach in two well-characterized population studies of Chinese in Singapore: the Singapore Cohort Study of the Risk factors for Myopia (SCORM) and the Singapore Prospective Study Program (SP2) study. The Chinese in Singapore are primarily immigrants from southern provinces of China, and a recent study on the genomic variations of Singapore Chinese demonstrates a homogenous group with minimal substructure17. Utilizing GWA approach in this homogenous population provides a viable means to discover susceptibility genes for high myopia in Chinese persons. We therefore performed a meta analysis using the SCORM and SP2 genotyped datasets to identify top-ranked ‘susceptibility’ markers for high myopia. The Nakanishi et al15 Japanese dataset (Japan) was used as a replication cohort to confirm these top-ranked markers.
Patients and Methods
Study populations
SCORM and SP2 are the primary datasets in this study with genome-wide high-density SNP data. SCORM is one of few cohorts with precise longitudinal ocular phenotypic data from predominantly Chinese Singaporean children18. SP2 is a population-based study of primarily Chinese adults aged 21 years and above with refractive error data19–22. Our study adhered to the Declaration of Helsinki. SCORM was approved by the Institutional Review Boards of the National University of Singapore and the Singapore Eye Research Institute, while SP2 was approved by the Singapore Eye Research Institute and Singapore General Hospital. Written informed consent was obtained from all parents (SCORM) and participants (SP2). The Japan adult dataset is enriched for high myopia and control samples, and was used as the replication dataset15.
SCORM
A total of 1979 children in grades 1, 2, and 3 from three schools were recruited from 1999 to 200118;23. The children were examined on the school premises every year by a team of eye care professionals. Three drops of 1% cyclopentolate were administered 5 minutes apart. At least 30 minutes after the third drop, the refractive error was measured using a stand-alone autorefractor (Canon RK-F1, Japan). Contact ultrasound biometry measures were performed using one of two biometry machines (Echoscan model US-800; Nidek Co, Ltd, Tokyo, Japan). To reduce genetic heterogeneity derived from different racial groups in SCORM, the GWA study was conducted in a subset of children, specifically 1116 Chinese, comprising 56% of the whole cohort,. The high myopia phenotype used in this study was based on the refractive error obtained on the 4th annual examination of the study (children at age 10 to 12 years).
SP2
Samples of SP2 were from a revisit protocol of two prior population-based surveys, the 1992 National Health Survey and the 1998 National Health Survey19–22. Both studies recruited a random sample of individuals from the Singapore population. Disproportionate sampling was stratified by ethnicity to increase the number of minority ethnic groups (Malaysians and Asian Indians). A total of 8266 subjects were invited to participate in the follow-up survey. 6301 (76.1% response rate) subjects completed the questionnaire and of these, 4056 also attended the health examination and donated blood specimens (64.4% of those who completed the questionnaire). The protocol for obtaining refractive error measurements was identical to that of SCORM. Refractive error was measured using the stand-alone autorefractor (Canon RK-F1, Japan). The GWA genotyping for SP2 involved only individuals of Chinese descent (n=2867).
Japan
The Japan dataset consists of 959 high myopia cases and 2128 population controls. Details of the Japan data have been reported15. Briefly, the high myopia cases were primarily selected based on axial length > 26 mm of both eyes. It is known that excessive increase in axial length of the eye ball is the key contributor to myopic refractive. For instance, an axial length > 26 mm is equivalent to SE < −6.00D in general16, which corresponds to the criteria for high myopia used in SCORM and SP2. Cases were recruited at the Center for Macular Disease of Kyoto University Hospital, the High Myopia Clinic of Tokyo Medical and Dental University, and Fukushima Medical University Hospital. All subjects underwent comprehensive ophthalmologic examinations, including dilated pupillary indirect ophthalmoscopy of the fundus, slit-lamp biomicroscopy of the anterior chamber, automatedrefraction evaluation, and measurement of the axial length by applanation A-scan ultrasonography (UD-6000, Tomery, Nagoya, Japan) or partial coherence interferometry (IOLMaster, Carl Zeiss Meditec, Dublin, CA). Controls were obtained from the JSNP database24;25 and recruited at the Aichi Cancer Center Research Institute. The Institutional Review Board and the Ethics Committee of each institution approved the study protocols.
Phenotype studied
The threshold state of high myopia was determined by SE [refractive sphere + cylinder/2 (in plus cylinder)]. For both SCORM and SP2, high myopia cases were defined as SE ≤ −6.00 D in at least one eye, and controls were defined as SE between −0.50D and +1.00D in both eyes (emmetropia).
Genotyping
SCORM & SP2
Genome-wide SNP genotyping was conducted for both SCORM and SP2 using Illumina Beadarrays (http://www.illumina.com/, December 2007). For SCORM, a total of 1116 DNA samples (1037 from buccal swab and 79 from saliva) were genotyped using Illumina HumanHap 550 or 550 Duo Beadarrays®. For SP2, a total of 2867 blood-derived samples were genotyped using Illumina HumanHap 550v3, 610Quad, and 1Mduov3 platforms. That is, 392 samples were genotyped by 550v3, 1459 samples by 610Quad, 817 samples by 1Mduov3, 191 samples by both 550v3 and 1Mduov3, and 8 samples by 610 Quad and 1Mduov3. Genotyping consistency across platforms was addressed using 199 samples placed on the two different Beadarrays.
Japan
The SNPs to be validated were genotyped in the Japanese samples. Genotyping was performed with the Taqman SNP assay using the ABI PRISM 7700 system (Applied Biosystems, Foster City, CA).
Quality control (QC) criteria
The Illumina BeadStudio program (Illumina Inc., San Diego CA) was used for genotyping calls of each marker. To ensure high quality genotype data, we instituted a series of marker filtering criteria. Markers were excluded if they significantly deviated from Hardy-Weinberg equilibrium (HWE) in the control dataset (P<10−5), had a minor allele frequency (MAF) < 1%, or had missing genotype calls >10% across samples. Samples were excluded from further analysis if the overall genotype call rate was <98%, and more than 6 standard deviation in the population structure analysis using EIGENSTRAT26 programs.
Association analysis
Since the genotype data were not generated from a single Illumina Beadarray platform (two for SCORM and three for SP2), we analyzed all common markers in the dataset(s) tested. The software PLINK27 served as the primary analytical tool. Single-locus association tests were performed under a logistic regression framework for all SNPs in SCORM and SP2. Genotypes of each marker were coded as 0, 1, and 2 for the number of minor alleles carried, and a trend test for association was conducted within a logistic regression framework. Age and gender covariates were included in the logistic regression model.
The z-test28 was used for meta-analysis of both the SCORM and SP2 cohorts. Markers were considered as genome wide significant if P < 5×10−8 (individual p-value or meta p-value), which is the mostly commonly accepted significance threshold for GWA studies29;30. In addition to this stringent criteria, a relaxed significance threshold was applied for choosing markers for the replication analysis in the Japan dataset, That is, markers need to satisfy both (1) a meta p-value of P < 5×10−5, and a combination of study-specific p-values of P < 10−3 and P < 0.01 in either dataset. For the Japan dataset analysis, the trend test for association was performed for high myopia31. P < 0.05 was considered significant for replication in the Japan dataset. To assess the effect size of the target marker, the per-allele odds ratio (OR) and its accompanying 95% confidence interval (CI) was calculated from the trend test for the replicated SNP(s) for all datasets.
Results
GWAS analysis datasets
The basic demographic data for both SCORM and SP2 datasets is summarized in Table 1, and the description of SNPs selection process for meta-analysis of both cohorts is shown in Figure 1.
Table 1.
Summary statistics for spherical equivalent and sample size for each phenotypic category from the Singapore Cohort Study of Risk Factors for Myopia and Singapore Prospective Study Program datasets.
| Cohorts | Singapore Cohort Study of Risk Factors for Myopia (SCORM) |
Singapore Prospective Study Program (SP2) |
||
|---|---|---|---|---|
| Male/Female | 481/448 | 921/1087 | ||
|
Mean Age* (SD§) |
10.83 (0.83) | 47.9 (11.18) | ||
| Phenotypes |
No. Samples (Percentage) |
Both Eye Avg SE Mean (SD§) |
No. Samples (Percentage) |
Both Eye Avg SE Mean (SD§) |
| SE*,§ | 929 | −2.02 (2.26) |
1931 | −1.67 (2.89) |
| High myopia** | 65 (6.9%) |
−6.88 (1.06) |
222 (11.5%) |
−7.51 (2.02) |
|
Emmetropia** (Controls) |
238 (25.6%) |
0.42 (0.53) |
455 (23.6%) |
0.19 (0.35) |
For the SCORM dataset, age and spherical equivalent were based on the 4th annual examination of the study.
SE = Spherical equivalent; SD = Standard deviation
High myopia: SE ≤ −6.00D for at least one eye; Control: −0.50D< SE< +1.00D for both eyes; D=Diopter
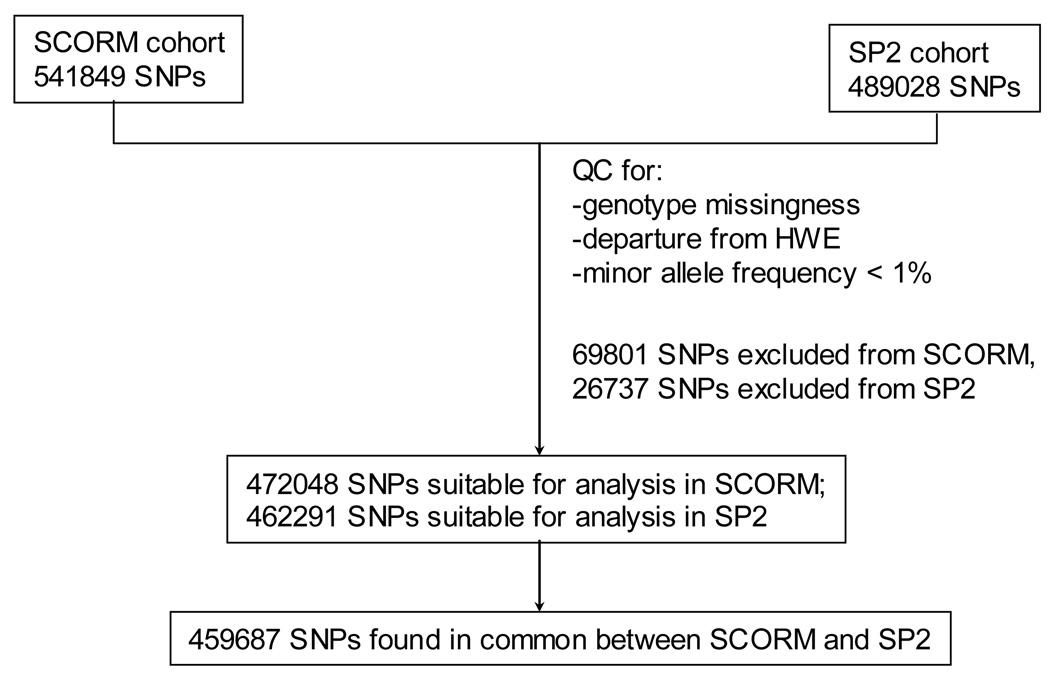
Figure 1.
The flowchart of the number of markers genotyped, excluded, and analyzed for Singapore Cohort Study of the Risk factors for Myopia (SCORM) and the Singapore Prospective Study Program (SP2) datasets. SNPs = single nucleotide polymorphisms; QC = quality control; HWE = Hardy-Weinberg equilibrium.
The post-QC SCORM GWA dataset was comprised of 929 subjects (481 males and 448 females) with refractive error data, of which 65 subjects have high myopia and 238 subjects are emmetropic controls. Due to two types of Illumina chips used in SCORM, only 541,849 autosomal SNPs were investigated. We excluded 69,801 markers based on the marker exclusion criteria described in the Methods section. After implementation of the exclusions and filtering, 472,048 SNPs remained in the SCORM dataset for the analysis.
The post-QC SP2 GWA dataset comprised of 2008 subjects (921 male, 1087 female), for which 222 have high myopia and 455 are emmetropic controls (Table 1). Three Illumina chips were used in the SP2 GWA study, and 489,028 common autosomal SNPs were evaluated. A total of 26,737 SNPs were excluded due to the violation of genotype missingness > 10%, gross departure from HWE, and monomorphism, which left 462,291 SNPs for GWA analysis.
For the meta-analysis of the SCORM and SP2 GWA results, we analyzed 459,687 markers genotyped in common for both datasets.
Meta-analyses and replication studies
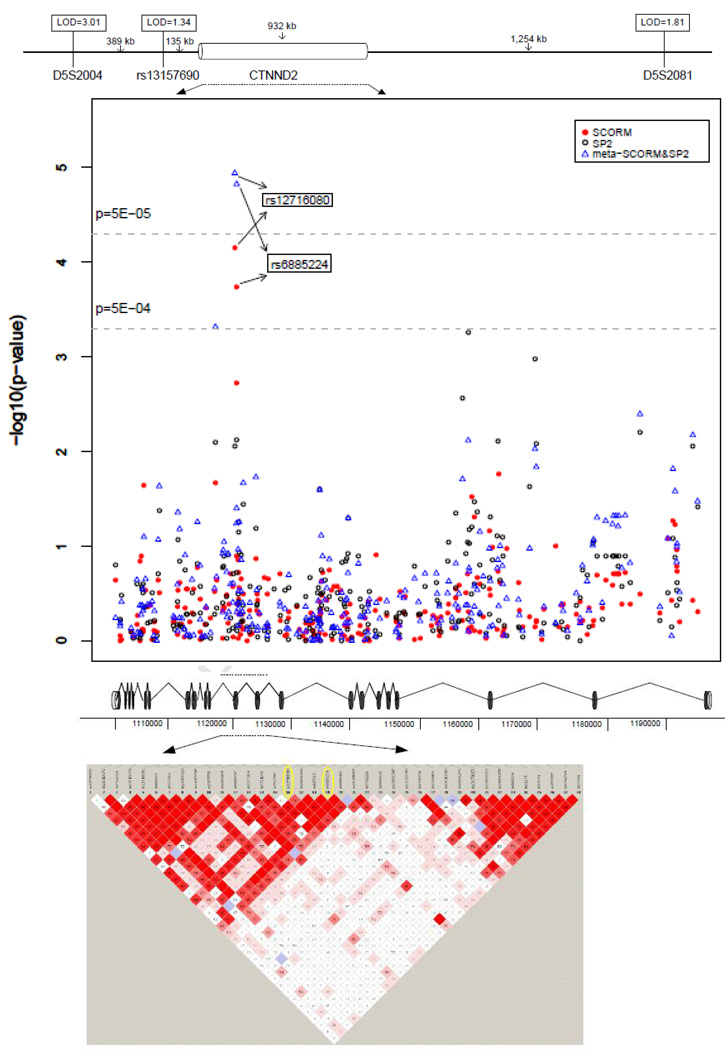
The full list of markers with p-value less than 0.001 from the GWA analyses of SCORM, SP2, and meta-analysis of SCORM and SP2, respectively, is listed in Tables 2, 3 and 4 (available at http://aaojournal.org). Figure 2 provides an overview of the meta-analysis of SCORM and SP2 for high myopia. Although no markers met the genome wide significance level (P<5×10−8), three markers, rs10508626 (chromosome 10 (chr10), meta-P=9.24×10−6), rs6885224 (chr5, meta-P=2.25×10−5), and rs12716080 (chr5, meta-P=2.61×10−5), met our marker selection criteria (see methods) to be brought forward for replication in the Japan dataset. Among the three markers, rs12716080 and rs6885224 (both from chromosome 5) showed evidence of association in the Japan dataset (Table 5). That is, we observed nominal significant for rs6885224 (P=0.035) in Japan, thus supporting association evidence of this marker in SCORM and SP2 meta-analysis. The second SNP (rs12716080) showed weaker evidence of association at P=0.11, in the same direction as that of rs6885224 (Table 5). When these two markers were examined in detail, we found that both displayed statistically significant association even when analysed within the specific SCORM and SP2 cohorts: rs12716080 (SCORM: P=7.06×10−5; OR = 2.41, 95% CI: 1.56 – 3.72, SP2: P = 8.8×10−3; OR = 1.48, 95% CI:1.1 – 1.97), and rs6885224 (SCORM: P=1.85×10−4, OR=2.25, 95% CI: 1.47 – 3.43, SP2: P=7.6×10−3, OR=1.5, 95% CI:1.11 – 2.01). Figure 3 depicts the p-values from the meta-analysis of SCORM and SP2, and p-values of individual dataset for all markers within CTNND2 region. The linkage disequilibrium (LD) plot clearly shows that both rs6885224 and rs12716080 are in the same LD block.
Figure 2.
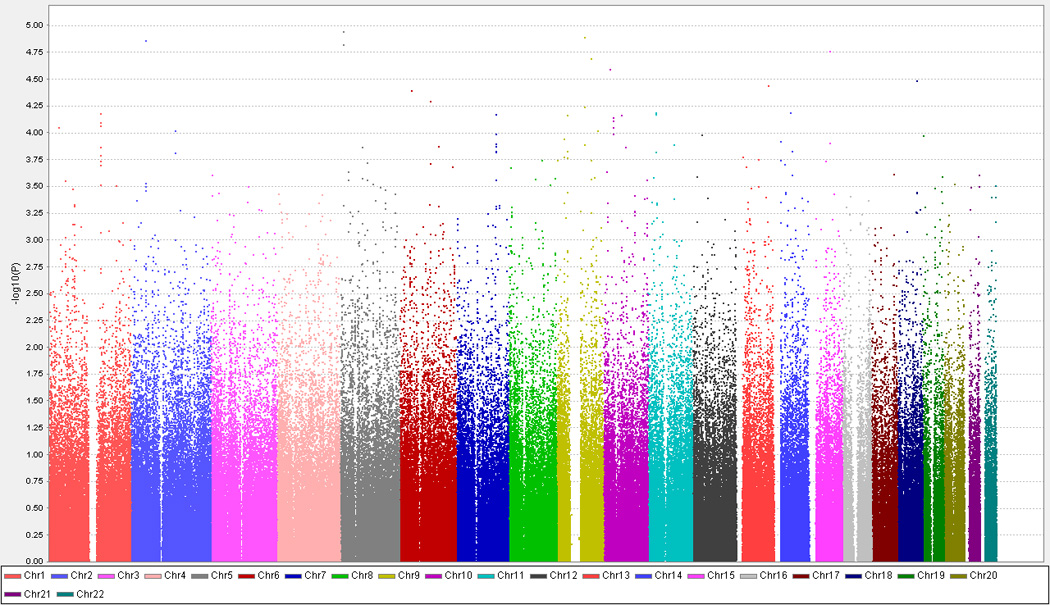
Manhattan plots of meta-analyses of Singapore Cohort Study of the Risk factors for Myopia (SCORM) and Singapore Prospective Study Program (SP2) datasets for high myopia. Chr = chromosome.
Table 5.
Summary of top single nucleotide polymorphisms from the Singapore Cohort Study of the Risk factors for Myopia* and Singapore Prospective Study Program* datasets, and the replication results in Japan dataset for high myopia.
| Chr* | SNP* | BP* | Gene | Minor Allele |
Meta- SCORM* and SP2* |
SCORM* | SP2* | Japan | Meta- SP2* SCORM* and Japan |
|
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | rs6885224 | 11222945 | CTNND2 | C | Sample | |||||
| sizes | 1070 | 303 | 767 | 3083 | 4153 | |||||
| MAF*,§ | 0.28 (0.21) | 0.38 (0.20) | 0.25 (0.21) | 0.26 (0.23) | ||||||
| p-values | ||||||||||
| 1.51E-05 | 1.85E-04 | 7.60E-03 | 0.035 | 7.84E −06** | ||||||
| OR* | 1.71 | 2.25 | 1.50 | 1.14 | 1.24 | |||||
| [95% CI*] | [1.34,2.18] | [1.47,3.43] | [1.11,2.01] | [1.02,1.27] | [1.11,1.39] | |||||
| 5 | rs12716080 | 11219948 | CTNND2 | G | Sample | |||||
| sizes | 1070 | 303 | 767 | 3085 | 4155 | |||||
| MAF*,§ | 0.31 (0.23) | 0.42 (0.22) | 0.28 (0.23) | 0.29 (0.27) | ||||||
| p-values | ||||||||||
| 1.14E-05 | 7.06E-05 | 8.80E-03 | 0.11 | 1.05E-05* | ||||||
| OR* | 1.72 | 2.41 | 1.48 | 1.10 | 1.20 | |||||
| 95% CI*] | ||||||||||
| [1.35,2.19] | [1.56,3.72] | [1.10,1.97] | [0.98,1.22] | [1.08,1.34] | ||||||
SCORM = Singapore Cohort Study of the Risk factors for Myopia; SP2 = Singapore Prospective Study Program; Chr = chromosome; SNP = single nucleotide polymorphisms; BP = basepair position; MAF = minor allele frequencies; OR = per-allele odds ratio for high myopia; 95% CI = 95% confidence interval of OR.
MAF is listed for high myopia cases and controls, respectively, where the one for controls is in parenthesis.
Fisher p values39 by adding Japan dataset.
Figure 3.
Location, Association Results, and linkage disequilibrium (LD) Pattern of CTNND2. The relative location of CTNND2 to the target markers for the chromosome 5 linkage region reported in Lam et al. (2005) (D5S2004 and D5S2081) and Li et al. (2009) (rs13157690). Two single nucleotide polymorphisms (SNPs) (rs12716080 and rs6885224) shown promising replication evidence are located in the same LD block. LOD=logarithm (base 10) of odds.
Discussion
This study utilized two GWA datasets of Singaporean Chinese with 287 high myopia cases and 693 controls out of 2937 GWA samples, and a follow-up replication study in 3087 (959 high myopia cases and 2128 controls) Japanese. As we combined refractive error data from children and adults, high myopia is likely the more robust phenotype, as children with high myopia are very likely to remain highly myopic for life and phenotype reversal is extremely rare.
We found significant association of the CTNND2 gene on chromosome 5p15 to high myopia. The minor allele of rs6885224 was consistently associated with increased susceptibility to high myopia in SCORM (OR = 2.25, 95% CI:1.47–3.43) and SP2 (OR = 1.5, 95% CI:1.11–2.01), with evidence of replication in Japan dataset (OR = 1.14, 95%CI:1.02–1.27), thus suggesting that this gene is a potential candidate for high myopia across pediatric and adult age groups in two East Asian populations. A second CTNND2 SNP rs12716080 also had evidence of association to high myopia for the SCORM and SP2 cohorts (Figure 2), with weaker evidence in the Japan cohort (Table 5). Both SNPs are in LD with r2 of 0.89 in Chinese and 0.937 in Japanese. In the HapMap and Human Genome Diversity projects, Chinese and Japanese have high similarity in population structure32;33. Intuitively, CTNND2 may also be a genetic determinant for childhood high myopia, leading to high-grade myopia in adulthood. This finding may be useful for developing interventions in high risk children who carry the CTNND2 risk allele.
The CTNND2 gene spans 933 Kb with 22 coding exons and resides within a 17.45 cM region on chromosome 5p15 previously found to be linked to high myopia in a family segregation study of three Hong Kong Chinese pedigrees (LOD = 4.68)34. More recently, evidence of linkage replication for that region was determined in a small Asian subset of families with high-grade myopia (N = 10 families) (LOD=1.34)4 (Figure 2). Of note, in non-human models, CTNND2 has been documented to play a crucial role in retinal morphogenesis, adhesion, and retinal cell architectural integrity via regulation of adhesion molecules35;36. Interestingly, CTNND2 was one of five biologically plausible genes for high myopia examined by Lam et al.34, who conducted direct sequencing of their coding regions to determine possible segregation with high myopia. In their study, five SNPs in the vicinity of CTNND2 were genotyped in a case-control association analysis using 94 cases with high myopia (SE at least −6.00 D) and 94 non-myopic controls. Evidence of segregation or association was not determined for CTNND2 in the Lam et al. study. It should be noted that both rs6885224 and rs12716080 were not assessed by Lam et al., and that the underpowered small sample size may have influenced the ability to detect evidence of association.
The strengths of our GWA studies are the reporting of association of unique genetic variants in two datasets of Singapore Chinese with very high rates of myopia, followed by the availability of the Japanese dataset for direct replication. The epidemic of myopia, especially in Chinese populations in Asia, may be due to either environmental factor (e.g., competitive educational systems with intensive near-work activity at an early age, limited outdoor activity)37;38, genetic susceptibility, or both. We are, therefore, mindful that myopia is a complex disease with (up till now) no major, striking susceptibility locus. Rather, the disease susceptibility may be accounted for by multiple loci each exerting very small effect sizes. Therefore, it was not surprising that genome wide significance for single-marker analysis was not observed in our SCORM and SP2 datasets due to the small sample sizes, which have insufficient power to detect markers with small genetic effects (e.g., OR < 1.2). In order to reach the formal genome-wide threshold (P < 5×10−8), we estimate that we would need a sample size of > 9000, which we are currently working actively to achieve.
Despite missing this formal threshold, our observations with the CTNND2 SNPs are significantly consistent across all 3 cohorts (2 Chinese and 1 Japanese), and this argues against it being a false positive finding. Overall, our study suggests that although genetic factors could be influential (e.g., CTNND2), they are in all likely to be very modest 1;2;23.
In summary, we report novel association to high myopia of a common polymorphism of the gene CTNND2 in Chinese and Japanese cohorts. This new locus may inform functional variants for myopia, and provide insights into the eventual development of early intervention strategies to retard the progression of myopia in high risk populations.
Supplementary Material
Acknowledgments
We would like to specifically thank the following Duke University affiliates: Mrs. Carol Haynes, A.B. for her participation in the databasing of genotype data from the SCORM GWA study, Dr. Andrew Dellinger, Ph.D. for his participation in providing initial assistance with the bioinformatics, and Dr. Dana Hornbeak, M.D. for her participation in generating the initial summary tables for the SCORM GWA study.
Financial Support: The SCORM GWA study is supported by the Singapore BioMedical Research Council (BMRC), grant 06/1/21/19/466 to SSM and the US National Institute of Health (NIH) grant (1R21-EY-019086-01) to YJL. The SP2 GWA study is supported by BMRC, grant 03/1/27/18/216 to EST. TLY is supported by research funding from the Duke-NUS Graduate Medical School, and Research to Prevent Blindness, Inc. Additional support was provided by the Singapore Tissue Network. The sponsor or funding organization had no role in the design or conduct of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure(s):
The authors have made the following disclosures:
E-Shyong Tai – Consultant, Glaxo-Smith Kline; Consultant, Merck Sharp and Dohme (IA) Corp
References
- 1.Seet B, Wong TY, Tan DT, et al. Myopia in Singapore: taking a public health approach. Br J Ophthalmol. 2001;85:521–526. doi: 10.1136/bjo.85.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong TY, Foster PJ, Hee J, et al. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000;41:2486–2494. [PubMed] [Google Scholar]
- 3.Saw SM, Shankar A, Tan SB, et al. A cohort study of incident myopia in Singaporean children. Invest Ophthalmol Vis Sci. 2006;47:1839–1844. doi: 10.1167/iovs.05-1081. [DOI] [PubMed] [Google Scholar]
- 4.Li YJ, Guggenheim JA, Bulusu A, et al. An international collaborative family-based whole genome linkage scan for high-grade myopia. Invest Ophthalmol Vis Sci. 2009;50:3116–3127. doi: 10.1167/iovs.08-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guggenheim JA, Kirov G, Hodson SA. The heritability of high myopia: a reanalysis of Goldschmidt's data [letter] J Med Genet. 2000;37:227–231. doi: 10.1136/jmg.37.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein AP, Duggal P, Lee KE, et al. Support for polygenic influences on ocular refractive error. Invest Ophthalmol Vis Sci. 2005;46:442–446. doi: 10.1167/iovs.04-0794. [DOI] [PubMed] [Google Scholar]
- 7.Teikari JM, O'Donnell J, Kaprio J, Koskenvuo M. Impact of heredity in myopia. Hum Hered. 1991;41:151–156. doi: 10.1159/000153994. [DOI] [PubMed] [Google Scholar]
- 8.Sorsby A, Sheriden M, Leary GA. Refraction and its components in twins. Spec Rep Ser Med Res Counc (G B) 1962;303:1–43. [PubMed] [Google Scholar]
- 9.Sorsby A, Leary GA, Fraser GR. Family studies on ocular refraction and its components. J Med Genet. 1966;3:269–273. doi: 10.1136/jmg.3.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–1236. [PubMed] [Google Scholar]
- 12.Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45:2873–2878. doi: 10.1167/iovs.03-1155. [DOI] [PubMed] [Google Scholar]
- 13.Young TL. Molecular genetics of human myopia: an update [report online] [Accessed May 8, 2010];Optom Vis Sci. 2009 86:E8–E22. doi: 10.1097/OPX.0b013e3181940655. Available at: http://journals.lww.com/optvissci/Fulltext/2009/01000/Molecular_Genetics_of_Human_Myopia__An_Update.5.aspx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1 [report online] [Accessed May 8, 2010];PLoS Genet. 2009 5:e1000660. doi: 10.1371/journal.pgen.1000660. Available at: http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobi FK, Zrenner E, Broghammer M, Pusch CM. A genetic perspective on myopia. Cell Mol Life Sci. 2005;62:800–808. doi: 10.1007/s00018-004-4353-z. [DOI] [PubMed] [Google Scholar]
- 17.Teo YY, Sim X, Ong RT, et al. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 2009;19:2154–2162. doi: 10.1101/gr.095000.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saw SM, Tong L, Chua WH, et al. Incidence and progression of myopia in Singaporean school children. Invest Ophthalmol Vis Sci. 2005;46:51–57. doi: 10.1167/iovs.04-0565. [DOI] [PubMed] [Google Scholar]
- 19.Hughes K, Yeo PP, Lun KC, et al. Cardiovascular diseases in Chinese, Malays, and Indians in Singapore. II. Differences in risk factor levels. J Epidemiol Community Health. 1990;44:29–35. doi: 10.1136/jech.44.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan CE, Emmanuel SC, Tan BY, Jacob E. Prevalence of diabetes and ethnic differences in cardiovascular risk factors. The 1992 Singapore National Health Survey. Diabetes Care. 1999;22:241–247. doi: 10.2337/diacare.22.2.241. [DOI] [PubMed] [Google Scholar]
- 21.Hughes K, Aw TC, Kuperan P, Choo M. Central obesity, insulin resistance, syndrome X, lipoprotein(a), and cardiovascular risk in Indians, Malays, and Chinese in Singapore. J Epidemiol Community Health. 1997;51:394–399. doi: 10.1136/jech.51.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cutter J, Tan BY, Chew SK. Levels of cardiovascular disease risk factors in Singapore following a national intervention programme. Bull World Health Organ. 2001;79:908–915. [PMC free article] [PubMed] [Google Scholar]
- 23.Saw SM, Chua WH, Hong CY, et al. Nearwork in early-onset myopia. Invest Ophthalmol Vis Sci. 2002;43:332–339. [PubMed] [Google Scholar]
- 24.Hirakawa M, Tanaka T, Hashimoto Y, et al. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30:158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haga H, Yamada R, Ohnishi Y, et al. Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190,562 genetic variations in the human genome. J Hum Genet. 2002;47:605–610. doi: 10.1007/s100380200092. [DOI] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bakker PI, Ferreira MA, Jia X, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 31.Agresti A. An Introduction to Categorical Data Analysis. New York: Wiley; 1996. pp. 34–39. Wiley Series in Probability and Statistics. [Google Scholar]
- 32.Weir BS, Cardon LR, Anderson AD, et al. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 34.Lam CY, Tam PO, Fan DS, et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–3778. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 35.Duparc RH, Boutemmine D, Champagne MP, et al. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300:647–655. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 36.Paffenholz R, Kuhn C, Grund C, et al. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- 37.Rose KA, Morgan IG, Smith W, et al. Myopia, lifestyle, and schooling in students of Chinese ethnicity in Singapore and Sydney. Arch Ophthalmol. 2008;126:527–530. doi: 10.1001/archopht.126.4.527. [DOI] [PubMed] [Google Scholar]
- 38.Saw SM, Chua WH, Wu HM, et al. Myopia: gene-environment interaction. Ann Acad Med Singapore. 2000;29:290–297. [PubMed] [Google Scholar]
- 39.Fisher RA. Statistical Methods for Research Workers. Edinburgh, Scotland: Oliver and Boyd; 1925. pp. 99–101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.