Synopsis
The past few years have brought new advances in our understanding of the molecular mechanisms underlying the development of Barrett’s esophagus and esophageal adenocarcinoma. Although knowledge of the genetic basis for these conditions has not yet translated into clinically useful biomarkers, the current pace of biomedical discovery holds endless possibilities for molecular medicine to improve the diagnosis and management of patients with these conditions. This article provides a useful conceptual basis for understanding the molecular events involved in the making of Barrett’s metaplasia and in its neoplastic progression and provides a rationale for evaluating studies on the application of molecular medicine to the diagnosis and management of patients with Barrett’s esophagus and esophageal adenocarcinoma.
Keywords: Barrett’s esophagus, metaplasia, esophageal adenocarcinoma
Introduction
While overall cancer incidence in the United States has decreased in recent years1, the number of new cases of esophageal cancer is increasing2. According to American Cancer Society estimates, there were 16,470 new cases and 14,530 deaths in this country in 2009 from esophageal cancer3. Esophageal cancer has two main histologic subtypes: squamous cell carcinoma and adenocarcinoma. In the West, the incidence of the former has remained stable or decreased since the 1970s, while the incidence of the latter has risen steadily during the same time period2. Esophageal adenocarcinoma has now become the more prevalent histologic subtype in the United States2.
Esophageal adenocarcinoma typically arises in the distal one-third of the esophagus, and its main risk factors are gastroesophageal reflux disease (GERD) and Barrett’s esophagus. For patients with Barrett’s esophagus, endoscopic surveillance to detect dysplasia is the primary strategy recommended to decrease morbidity and mortality from esophageal adenocarcinoma4. This strategy has not proven effective, as evidenced by the rising incidence of esophageal adenocarcinoma and the results of a recent study showing that the majority of patients with this cancer have no prior diagnosis of Barrett’s esophagus and, therefore, are not enrolled in surveillance programs5.
Basic investigations that have defined the genetic events underlying colonic carcinogenesis have led to effective strategies for the management and prevention of colorectal cancer6. Analogously, it is important to understand the molecular carcinogenesis of Barrett’s esophagus in order to identify specific targets to guide the development of effective diagnostic strategies and novel therapeutic agents. To do this, we must first understand the molecular events that lead to the replacement of normal esophageal squamous cells by metaplastic Barrett’s cells. Building on this understanding, we can appreciate how the genetic abnormalities acquired by metaplastic Barrett’s cells disrupt their normal properties so they can take on the morphologic and physiologic features of dysplasia and cancer. This report provides a conceptual basis for how normal esophageal squamous cells undergo columnar metaplasia and how metaplastic Barrett’s cells progress to dysplasia and carcinoma. Some of the main genetic alterations involved in the development and neoplastic progression of Barrett’s esophagus will be reviewed; however, the reader should appreciate that these represent a fraction of the genetic changes required for the making of Barrett’s metaplasia, dysplasia, and esophageal adenocarcinoma.
The Making of Barrett’s Metaplasia
Most, if not all, esophageal adenocarcinomas arise from Barrett’s esophagus, the condition in which the normal squamous cells lining the distal esophagus are replaced by intestinal-type columnar cells7. Barrett’s esophagus develops through the process of metaplasia, the replacement of one adult cell type by another. Metaplasia is thought to arise as a protective response to chronic tissue inflammation8, which in the esophagus is thought to be due to GERD. Barrett’s metaplasia can result from either changing fully differentiated esophageal squamous cells directly into intestinal-type columnar cells or from changing the differentiation pattern of esophageal stem cells8.
Metaplasia Through Transdifferentiation
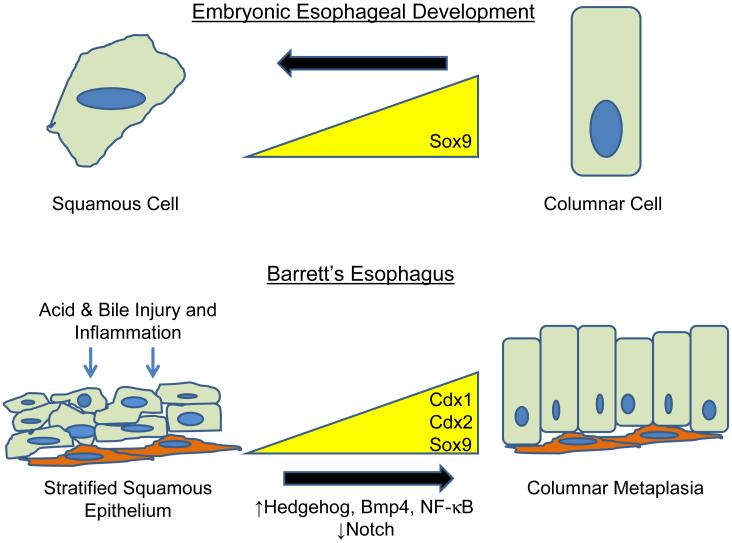
Transdifferentiation is the switch of one fully differentiated cell type directly into another. In general, this switch occurs between cell phenotypes that were present in the organ during embryonic development8. During embryogenesis, the esophagus is initially lined by ciliated, columnar cells which are replaced by stratified squamous cells as maturation proceeds (Figure 1)9-10. Data from ex vivo organ cultures of embryonic mouse esophagus demonstrate direct conversion of the columnar cells lining the esophagus into squamous cells, a process found to be independent of cell proliferation or apoptosis11. In theory, a reversal of this normal developmental switch in cell phenotype may occur during the formation of Barrett’s metaplasia. In support of this hypothesis, studies using scanning electron microscopy have demonstrated a “distinctive cell” at the squamo-columnar junction in Barrett’s mucosa that expresses cytokeratin markers and demonstrates morphologic features of both squamous and columnar epithelium; moreover, this “distinctive cell” has not been detected at the squamo-columnar junction in patients without Barrett’s mucosa12. Once Barrett’s metaplasia is established, the epithelium must undergo maintenance and self-renewal, processes which are not explained by the transdifferentiation hypothesis, however.
Figure 1. Phenotypic changes in esophageal epithelium occur during normal development and Barrett’s esophagus.
During esophageal development (top), the embryonic esophagus is initially lined by columnar epithelial cells expressing the transcription factor Sox9. As the embryo matures, the esophageal epithelium transitions into a stratified squamous epithelium that does not express Sox9. In Barrett’s esophagus (bottom), the stratified squamous epithelium is exposed to acid and bile acids. The ensuing inflammation and injury repair response activate signaling pathways such as Hedgehog, Bmp4, and NF-κB and downregulate Notch signaling. These signals lead to increased expression of Cdx1, Cdx2, and Sox9 which induces columnar metaplasia.
Metaplasia Through Stem Cells
Stem cells can proliferate, self renew, give rise to a variety of cell types, and regenerate tissue following injury13. A stem cell origin would account for the persistence and maintenance of Barrett’s epithelium and could explain the predisposition of this tissue to neoplastic transformation. The stem cell for Barrett’s esophagus may reside in the esophagus itself or originate in the bone marrow. During development, tracheoesophageal progenitor cells express p63, a homologue of p5314. As the esophageal lining forms, p63+ progenitor cells differentiate into ciliated, columnar cells that lack p63 expression14. After stratified squamous epithelium replaces the ciliated, columnar epithelium, cells in the proliferative basal layer of the squamous epithelium continue to stain strongly for p63, whereas cells in the fully differentiated more superficial layers demonstrate no p63 staining14. In mice null for p63, the esophagi completely lack stratified epithelium and are lined by simple columnar epithelium, suggesting that p63+ cells are necessary to establish a stratified squamous epithelium14. Barrett’s epithelium has been found to lack immunostaining for p63, suggesting that the Barrett’s stem cell differs from the p63+ embryonic esophageal progenitor cell and the adult, squamous esophageal stem cell14-15. These findings do not eliminate the possibility that the stem cells for Barrett’s metaplasia reside in esophageal submucosal glands or in glands of the gastric cardia, adjacent to the gastroesophageal junction, as has been suggested by some investigators16-17.
A second potential source of stem cells for the esophagus is the bone marrow. In mice treated with high dose irradiation to induce esophagitis, injection of either esophageal progenitor cells or bone marrow cells was able to repair the injured esophagus through regeneration of new squamous cells18. Using a rat model of severe reflux esophagitis, our group investigated the possibility that bone marrow cells can give rise to metaplastic Barrett’s epithelium. Female rats were lethally irradiated and then rescued with bone marrow from male donor rats. An esophagojejunostomy was then performed on the female rats to induce the reflux of both acid and bile salts19. Eight weeks post-op, the esophagi of the female rats contained squamous and metaplastic cells with a Y chromosome, suggesting that bone marrow cells can hone to the esophagus and give rise to both squamous and columnar epithelium. In humans, cells from male donors have been found within the gastrointestinal tract of females who have undergone bone marrow transplant20.
Regardless of where the stem cell originates, it is likely that the environment in the inflamed, reflux-damaged esophagus mediates the phenotypic switch from a squamous cell to an intestinal-like columnar cell (Figure 1). This phenotypic switch presumably occurs by altering the expression of a few key master genes that regulate cell phenotype. Candidate master genes that are upregulated in Barrett’s esophagus compared to neighboring esophageal squamous epithelium include the transcription factors CDX1, CDX2, and SOX9 (Figure 1)21-23. Not only are these genes normally expressed in the intestine, but target genes of these transcription factors define an intestinal phenotype24-25.
Role for CDX1, CDX2, and SOX9 in Barrett’s Metaplasia
Homeotic genes define the developmental pattern of an organism. Cdx1 and Cdx2 are homeobox genes which specify intestinal epithelial differentiation26. Studies in mice suggest that Cdx2 is required for intestinal differentiation and that Cdx1 may specify a columnar cell27-28. CDX1 and CDX2 mRNA and protein expression have been detected in esophageal biopsy specimens from non-dysplastic Barrett’s metaplasia, Barrett’s metaplasia with dysplasia, and Barrett’s-associated adenocarcinomas, but not in normal esophageal squamous epithelium29-31. Sox9 is another transcription factor, expressed by potential stem cells in intestinal crypts, that plays a role in the formation of goblet cells32-33. Recently, SOX9 protein has been shown to be expressed in Barrett’s metaplasia, Barrett’s with dysplasia, and adenocarcinoma, but not in esophageal squamous epithelium23.
How GERD May Induce Barrett’s Metaplasia
Barrett’s metaplasia is a sequelae of chronic GERD. Components of the refluxed gastric juice (e.g. acid, bile salts) and/or the resulting esophageal inflammation (reflux esophagitis) could cause esophageal metaplasia by inducing transcription factors or activating developmental signaling pathways that determine an intestinal phenotype.
Stimulation of the CDX Transcription Factors by GERD
In mouse esophageal squamous epithelial cells, exposure to bile acids or acid activates Cdx2 expression34-35. In human esophageal squamous cells (HET-1A), exposure to a combination of acid and bile salts increases CDX2 expression and leads to squamous cells forming crypt-like structures and expressing intestinal genes such as Villin, Sucrase-isomaltase, and MUC236-37 In addition, data suggest that GERD-induced inflammation activates CDX expression in esophageal squamous epithelial cells. In a rodent model of esophageal intestinal metaplasia, squamous cells begin to express Cdx2 prior to the development of intestinal metaplasia38. In human esophageal biopsies, CDX2 expression has been found in inflamed esophageal squamous epithelium, but not in non-inflamed squamous epithelium31. In telomerase-immortalized normal esophageal squamous cells established from GERD patients with and without Barrett’s esophagus, exposure to acid and/or bile salts increased CDX2 expression in the squamous cells from Barrett’s patients, but not in those from GERD patients without Barrett’s esophagus39. Inhibition of nuclear factor-κB (NF-κB), a well established mediator of GERD-induced inflammation, prevented the increase in CDX2 expression in esophageal squamous cells from Barrett’s patients in response to acid and/or bile salt exposure, suggesting that inflammatory signaling cascades can also activate CDX2 expression in esophageal squamous cells39.
Stimulation of Developmental Signaling Pathways by GERD
An attractive hypothesis is that esophageal activation of developmental signaling pathways involved in maintaining or developing the normal intestine may lead to Barrett’s metaplasia. These include pathways that are required for normal intestinal development, such as Wnt and Notch, or pathways that are expressed in the embryonic esophagus to maintain a columnar phenotype, such as Hedgehog and Bone Morphogenic Protein (Bmp) 4. Wnt is required to maintain the intestinal crypt progenitor cell population and regulates the expression of Cdx140-41. Wnt pathway activation (as determined by nuclear β-catenin) has not been found in non-dysplastic Barrett’s metaplasia, but has been observed in Barrett’s metaplasia with dysplasia and in esophageal adenocarcinomas42.
The Notch pathway also participates in maintaining the intestinal crypt progenitor pool and perhaps even that of the esophagus43. As intestinal cells begin to differentiate, persistent Notch signaling leads to an absorptive enterocyte fate, while the lack of Notch signaling leads to a secretory fate as an enteroendocrine, goblet, or Paneth cell44-45. Unlike the other developmental signaling pathways, components of the Notch signaling pathway are present in the normal adult esophagus46. As noted above, CDX2 overexpression in squamous HET1A cells causes the cells to form crypt-like structures36. In these same cells, expression of Hes1, a downstream target of Notch, is down-regulated by CDX2 overexpression, suggesting that inhibition of Notch signaling by CDX2 may play a role in metaplasia formation36. Bile salt exposure has also been shown to decrease expression of Notch pathway components in esophageal adenocarcinoma cells47. Recently, in an animal model of reflux and Barrett’s esophagus, inhibitors of Notch signaling caused the proliferative Barrett’s cells to differentiate into goblet cells48.
Bmp4 is normally expressed within the stroma of the embryonic columnar-lined esophagus, but it is absent in the adult squamous-lined esophagus49-50. In a rodent model of reflux esophagitis and Barrett’s esophagus, investigators demonstrated Bmp4 expression in the stroma underlying inflamed esophageal squamous epithelium and specialized intestinal metaplasia, but not in the stroma underlying normal esophageal squamous epithelium50. When human esophageal squamous cells were treated with BMP4 in vitro, the squamous cells began to express cytokeratins characteristic of columnar cells, suggesting that stromal BMP4 expression promotes the change in the esophageal epithelium from squamous to columnar50.
Finally, the Hedgehog signaling pathway likely plays a role in esophageal metaplasia. Sonic hedgehog, the most ubiquitous Hedgehog ligand, is expressed by the embryonic esophagus while it has a columnar epithelium and before it takes on a stratified squamous phenotype51. Recently, Sonic hedgehog expression was observed in Barrett’s metaplasia, but not in normal adult esophageal epithelium23. In a mouse model of reflux esophagitis and Barrett’s esophagus, Sonic hedgehog expression was found in the Barrett’s metaplasia as well as in esophageal squamous cells prior to the development of intestinal metaplasia23. Since Bmp4 is a target of Hedgehog signaling, it was not surprising that stromal BMP4 expression was seen adjacent to Barrett’s epithelium from esophagectomy specimens23. Activation of BMP4 signaling in HET-1A cells induced SOX9 expression and subsequent expression of cytokeratins characteristic of columnar cells23.
The Making Of Barrett’s-Associated Dysplasia and Adenocarcinoma
The histologic diagnoses of dysplasia and cancer are based on a compilation of morphologic features of the tissue which indicate that the cells have acquired “abnormal” physiologic properties. In 2000, Hanahan and Weinberg characterized six physiologic properties of cancer, also called “hallmarks”, that normal cells acquire as cancer ensues52. These hallmarks include the ability of cells to provide their own growth signals, avoid growth inhibitory signals, resist apoptosis, replicate without limit, synthesize new blood vessels, and invade and metastasize52 (Table 1). Surprisingly, studies have shown that these cancer hallmarks can be acquired by normal cells through disruptions in only a few key growth regulatory pathways including the p16/Retinoblastoma (Rb) and p53 pathways, the Ras signaling pathway, and the telomerase-dependent senescence pathway53 (Table 1). Recently, cancer-related inflammation has been proposed as a seventh physiologic hallmark of cancer54.
Table 1.
Cancer Hallmarks and the Key Growth Regulatory Pathways that Contribute to Carcinogenesis in Barrett’s Esophagus. The pathways that cause invasion and metastasis and establish the inflammatory microenvironment are not yet known. The establishment of an inflammatory microenvironment might contribute to the ability of tumor cells to invade and metastasize
| Cancer Hallmark | Key Growth Regulatory Pathway |
|---|---|
| provide growth signals | Ras Pathway |
| Avoid Growth Inhibitory Signals | p16/Rb and p53 Pathways |
| Resist Apoptosis | p53 Pathway, Ras pathway |
| Replicate Without Limit | Telomerase-Dependent Senescence Pathway, p16/Rb pathway, p53 pathway |
| Synthesize New Blood Vessels | Ras Pathway |
| Invade and Metastasize | ?? ?? Inflammatory Microenvironment |
| Inflammatory Microenvironment | ?? |
Key Growth Regulatory Pathways that Contribute to Carcinogenesis
p16/Rb Pathway
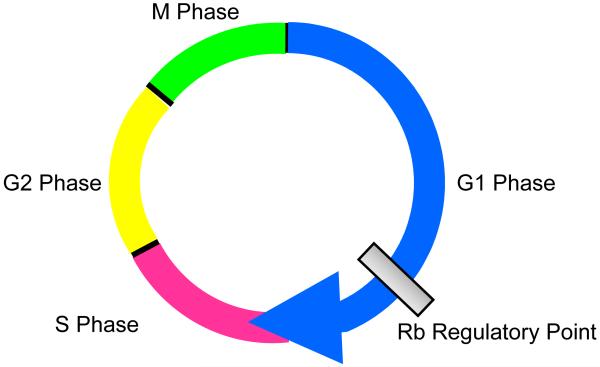
In order to appreciate the contribution of the p16/Rb pathway to carcinogenesis, a brief review of cell proliferation and the cell cycle is in order. The cell cycle encompasses the events that take place in order for a cell to divide. The cycle is partitioned into 4 phases called gap 1 (G1), DNA synthesis (S), gap 2 (G2), and mitosis (M) (Figure 2). The major point of regulation for cell proliferation occurs in the transition from G1 into S phase of the cell cycle, and Rb is the protein that has master control of this critical juncture (Figure 2). The ability of cells to bypass this key regulatory point allows them to avoid growth inhibitory signals and to replicate without limit. Although the data are inconclusive, it appears that Rb itself is targeted for inactivation in the latter stages of Barrett’s carcinogenesis (i.e. dysplasia and carcinoma), but not in non-dysplastic Barrett’s metaplasia55-56. Inactivation of Rb is not the only way to bypass this key regulatory point. p16 is a member of the INK4 family of cell cycle inhibitors. p16 regulates the synthesis of proteins that alter the function of Rb such that cells cannot proceed through the cell cycle. Thus, inactivation of p16 would allow cells to pass unhindered from G1 into S phase and is, in fact, the earliest and most common genetic alteration found in non-dysplastic Barrett’s metplasia57. For example, studies have reported p16 inactivation in 73-87% of biopsy specimens from patients with non-dysplastic Barrett’s esophagus58-59.
Figure 2. Cell Cycle.
There are 4 phases of the cell cycle, gap 1 (G1), DNA synthesis (S), gap2 (G2), and mitosis. The Rb regulatory point controls passage from G1 into S phase.
p53 pathway
p53 is a tumor suppressor gene that inhibits cell proliferation by preventing passage of cells from G1 to S phase of the cell cycle. Like p16, p53 regulates the synthesis of proteins that alter the function of Rb such that cells cannot continue through the cell cycle. p53 also plays a central role in the induction of apoptosis. Therefore, disruption of the p53 pathway gives cells the ability to avoid growth inhibitory signals, to replicate without limit, and to resist apoptosis. Using immunohistochemical staining, mutant p53 expression has been detected in non-dysplastic Barrett’s metaplasia and the frequency of mutant p53 detection increases as dysplasia and adenocarcinoma ensue60-62.
Ras Pathway
The Ras pathway is one of the main intracellular signaling cascades activated following the binding of growth factors to their receptors located on the surface of cells63. Ras-mediated signals regulate the function of proteins that promote passage from G1 into S phase of the cell cycle and proteins that influence apoptosis64. Therefore, disruption of the Ras pathway allows cells to acquire the ability to provide their own growth signals, to resist apoptosis, and to synthesize new blood vessels. The majority of human tumors demonstrate mutations in Ras (i.e. oncogenic Ras) that cause the constant stimulation of downstream signaling cascades independent of growth factor-mediated receptor activation65. Expression of oncogenic K-Ras or H-Ras is rare in non-neoplastic Barrett’s metaplasia; however, expression of both oncogenic Ras proteins has been frequently detected in dysplastic Barrett’s metaplasia and adenocarcinoma66-69. Ras pathway activation does play a role in the early stages of Barrett’s carcinogenesis; however, it does so in a more “physiologic” fashion by transmitting signals downstream of the epidermal growth factor receptor (EGFR) and its ligand, transforming growth factor alpha (TGF-α). Increased levels of both EGFR and TGF-α have been found in biopsy samples of non-dysplastic Barrett’s metaplasia and been proposed to account for increased activation of the mitogenic Ras pathway in the early stages of Barrett’s carcinogenesis70.
As noted above, dysfunction of the Ras pathway also allows cells to acquire the ability to synthesize new blood vessels, a process termed angiogenesis. Binding of vascular endothelial growth factors (VEGFs) to their receptors, the vascular endothelial growth factor receptors (VEGFRs) initiates the proliferation and migration of endothelial cells into the tissue via Ras pathway signaling. Non-dysplastic Barrett’s metaplasia has increased expression of VEGF-A, VEGF-C, and VEGFR-2 as compared to esophageal squamous epithelium71. In fact, an enhanced vascular network has been proposed to account for the salmon color characteristic of Barrett’s esophagus. Esophageal adenocarcinomas have been shown to express even higher levels of VEGF mRNA and protein compared to non-dysplastic or dysplastic Barrett’s metaplasia72.
Telomerase-dependent senescence pathway
Senescence is an intrinsic mechanism of cells that limits their proliferative capacity and is triggered by the progressive loss of telomeres. Telomeres are long stretches of repetitive pieces of DNA located at the ends of chromosomes. With each cell division, some of these telomeric repeats are lost. When telomere loss is such that only a small amount remains, the cell exits the cell cycle into a permanent state of growth arrest which has been termed senescence. In order to overcome senescence, the cell must maintain telomere length. Telomerase is the enzyme that synthesizes and maintains telomeres73. Therefore, disruption of the telomerase-dependent senescence pathway allows cells to replicate without limit and become immortalized. Most normal cells lack telomerase, including normal esophageal squamous cells. Non-dysplastic Barrett’s biopsy specimens express low levels of telomerase which increase as the degree of dysplasia increases74. Esophageal adenocarcinomas also express high levels of telomerase75.
Cancer-Related Inflammation
Cancer-related inflammation can be established through two pathways: 1) an extrinsic pathway in which clinical disorders such as reflux esophagitis cause tissue inflammation that contributes to carcinogenesis, and 2) an intrinsic pathway in which the precancerous cells acquire genetic abnormalities that produce an inflammatory tumor microenvironment54. The intrinsic and extrinsic inflammatory pathways can converge on certain key downstream targets (e.g. cytokines and transcription factors) that promote further inflammation and tumor cell proliferation54. Among the key molecules in cancer-related inflammation are transcription factors such as NF-κB and STAT3. NF-κB is well known to mediate both inflammation and tumor progression. NF-κB expression has been found in 40-60% of biopsy specimens of Barrett’s metaplasia and in 61% to 80% of Barrett’s adenocarcinomas, but in only 13% of biopsy specimens of reflux-injured squamous epithelium76-77. Moreover, NF-κB activation has been found to increase as metaplastic Barrett mucosa develops dysplastic changes of progressive severity, suggesting that an inflammatory response might be contributing to carcinogenesis77.
STAT3 is another transcription factor that is well known to mediate both inflammation and tumorigenesis. In biopsy specimens of Barrett’s epithelium, expression of the active form of STAT3 increases with the severity of dysplasia, also suggesting a link between the inflammatory response and Barrett’s carcinogenesis78. The molecular mechanisms that mediate invasion and metastasis remain unclear; however, data suggest that perhaps the inflammatory response may be playing a role. For example, matrix metalloproteinases (MMPs) are proteolytic enzymes that can degrade the extracellular matrix and contribute to tumor invasion and metastasis79. MMP-1, -2, -7 and -9 expression has been found in non-dysplastic Barrett’s metaplasia and esophageal adenocarcinoma80-82. STAT3 has been found to regulate expression of MMP-2 and -9, potentially linking inflammation with Barrett’s-associated tumor cell invasion and migration83.
CONCLUSION
The rate of increase in the incidence of esophageal adenocarcinoma over the past several decades is quite startling. GERD and Barrett’s esophagus are recognized as major risk factors for esophageal adenocarcinoma. Since most esophageal adenocarcinomas are thought to arise from Barrett’s esophagus, the pathogenesis of esophageal metaplasia at the molecular level has become an area of intense investigation. Molecular markers of metaplasia may soon be used to identify individuals at risk for developing Barrett’s esophagus rather than relying on epidemiologic risk factors alone. Moreover, the use of molecular markers to identify the stem cell of Barrett’s esophagus may allow for the targeting of endoscopic or pharmacologic ablative therapies specifically to the stem cells, thereby eliminating Barrett’s esophagus itself and thus the risk for esophageal adenocarcinoma.
Advances in tumor biology have revealed that the complexity of human tumorigenesis can be boiled down to disruptions in a few key growth regulatory pathways and an inflammatory microenvironment. This approach provides a useful conceptual basis for evaluating studies on molecular markers for detecting cancer progression and for developing chemoprevention and chemotherapeutic strategies. The reader should appreciate however that these are pathways comprised of multiple genes and proteins, and that pathway disruption can be caused by any number of different modifications in genes and/or in proteins within each pathway. Thus panels of molecular markers will likely be used to indicate molecular “signatures” predictive of neoplastic progression, and the molecular characterization of individual tumors will likely be used to tailor therapeutic strategies to an individual patient. Molecular medicine is reshaping our understanding of the biology of Barrett’s metaplasia, dysplasia and adenocarcinoma. Clinicians should stay tuned as molecular medicine unfurls endless possibilities to improve the diagnosis and management of patients with Barrett’s esophagus.
Acknowledgments
This work was supported by the Office of Medical Research, Department of Veterans Affairs (R.F.S., D.H.W.) and the National Institutes of Health (F32-CA123945 to D.H.W. and R01-DK63621 & R01-CA134571 to R.F.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have nothing to disclose.
References
- 1.Jemal A, Thun MJ, Ries LA, et al. Annual report to the nation on the status of cancer, 1975-2005, featuring trends in lung cancer, tobacco use, and tobacco control. J Natl Cancer Inst. 2008 Dec 3;100(23):1672–1694. doi: 10.1093/jnci/djn389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demeester SR. Epidemiology and biology of esophageal cancer. Gastrointest Cancer Res. 2009 Mar;3(2 Suppl):S2–5. [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009 Jul-Aug;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Wang KK, Sampliner RE. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett’s esophagus. Am J Gastroenterol. 2008 Mar;103(3):788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 5.Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett’s esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002 Jan;122(1):26–33. doi: 10.1053/gast.2002.30297. [DOI] [PubMed] [Google Scholar]
- 6.Souza RF. A molecular rationale for the how, when and why of colorectal cancer screening. Aliment Pharmacol Ther. 2001 Apr;15(4):451–462. doi: 10.1046/j.1365-2036.2001.00935.x. [DOI] [PubMed] [Google Scholar]
- 7.Spechler SJ. Clinical practice. Barrett’s Esophagus. N Engl J Med. 2002 Mar 14;346(11):836–842. doi: 10.1056/NEJMcp012118. [DOI] [PubMed] [Google Scholar]
- 8.Tosh D, Slack JM. How cells change their phenotype. Nat Rev Mol Cell Biol. 2002 Mar;3(3):187–194. doi: 10.1038/nrm761. [DOI] [PubMed] [Google Scholar]
- 9.Guillem PG. How to make a Barrett esophagus: pathophysiology of columnar metaplasia of the esophagus. Dig Dis Sci. 2005 Mar;50(3):415–424. doi: 10.1007/s10620-005-2451-x. [DOI] [PubMed] [Google Scholar]
- 10.Johns BA. Developmental changes in the oesophageal epithelium in man. J Anat. 1952 Oct;86(4):431–442. [PMC free article] [PubMed] [Google Scholar]
- 11.Yu WY, Slack JM, Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol. 2005 Aug 1;284(1):157–170. doi: 10.1016/j.ydbio.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 12.Sawhney RA, Shields HM, Allan CH, Boch JA, Trier JS, Antonioli DA. Morphological characterization of the squamocolumnar junction of the esophagus in patients with and without Barrett’s epithelium. Dig Dis Sci. 1996 Jun;41(6):1088–1098. doi: 10.1007/BF02088224. [DOI] [PubMed] [Google Scholar]
- 13.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 14.Daniely Y, Liao G, Dixon D, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004 Jul;287(1):C171–181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 15.Glickman JN, Yang A, Shahsafaei A, McKeon F, Odze RD. Expression of p53-related protein p63 in the gastrointestinal tract and in esophageal metaplastic and neoplastic disorders. Hum Pathol. 2001 Nov;32(11):1157–1165. doi: 10.1053/hupa.2001.28951. [DOI] [PubMed] [Google Scholar]
- 16.Leedham SJ, Preston SL, McDonald SA, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008 Aug;57(8):1041–1048. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colleypriest BJ, Palmer RM, Ward SG, Tosh D. Cdx genes, inflammation and the pathogenesis of Barrett’s metaplasia. Trends Mol Med. 2009 Jul;15(7):313–322. doi: 10.1016/j.molmed.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Epperly MW, Guo H, Shen H, et al. Bone marrow origin of cells with capacity for homing and differentiation to esophageal squamous epithelium. Radiat Res. 2004 Sep;162(3):233–240. doi: 10.1667/rr3224. [DOI] [PubMed] [Google Scholar]
- 19.Sarosi G, Brown G, Jaiswal K, et al. Bone marrow progenitor cells contribute to esophageal regeneration and metaplasia in a rat model of Barrett’s esophagus. Dis Esophagus. 2008;21(1):43–50. doi: 10.1111/j.1442-2050.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 20.Korbling M, Katz RL, Khanna A, et al. Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells. N Engl J Med. 2002 Mar 7;346(10):738–746. doi: 10.1056/NEJMoa3461002. [DOI] [PubMed] [Google Scholar]
- 21.Wong NA, Wilding J, Bartlett S, et al. CDX1 is an important molecular mediator of Barrett’s metaplasia. Proc Natl Acad Sci U S A. 2005 May 24;102(21):7565–7570. doi: 10.1073/pnas.0502031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moons LM, Bax DA, Kuipers EJ, et al. The homeodomain protein CDX2 is an early marker of Barrett’s oesophagus. J Clin Pathol. 2004 Oct;57(10):1063–1068. doi: 10.1136/jcp.2003.015727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang DH, Clemons NJ, Miyashita T, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology. 2010 May;138(5):1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan CW, Wong NA, Liu Y, et al. Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc Natl Acad Sci U S A. 2009 Feb 10;106(6):1936–1941. doi: 10.1073/pnas.0812904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traber PG, Wu GD, Wang W. Novel DNA-binding proteins regulate intestine-specific transcription of the sucrase-isomaltase gene. Mol Cell Biol. 1992 Aug;12(8):3614–3627. doi: 10.1128/mcb.12.8.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000 Oct;119(4):961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 27.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009 Apr;16(4):588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mutoh H, Sakurai S, Satoh K, et al. Cdx1 induced intestinal metaplasia in the transgenic mouse stomach: comparative study with Cdx2 transgenic mice. Gut. 2004 Oct;53(10):1416–1423. doi: 10.1136/gut.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips RW, Frierson HF, Jr., Moskaluk CA. Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. Am J Surg Pathol. 2003 Nov;27(11):1442–1447. doi: 10.1097/00000478-200311000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Groisman GM, Amar M, Meir A. Expression of the intestinal marker Cdx2 in the columnar-lined esophagus with and without intestinal (Barrett’s) metaplasia. Mod Pathol. 2004 Oct;17(10):1282–1288. doi: 10.1038/modpathol.3800182. [DOI] [PubMed] [Google Scholar]
- 31.Eda A, Osawa H, Satoh K, et al. Aberrant expression of CDX2 in Barrett’s epithelium and inflammatory esophageal mucosa. J Gastroenterol. 2003;38(1):14–22. doi: 10.1007/s005350300001. [DOI] [PubMed] [Google Scholar]
- 32.Blache P, van de Wetering M, Duluc I, et al. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004 Jul 5;166(1):37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bastide P, Darido C, Pannequin J, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007 Aug 13;178(4):635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnat G, Rau T, Elshimi E, Hahn EG, Konturek PC. Bile acids induce overexpression of homeobox gene CDX-2 and vascular endothelial growth factor (VEGF) in human Barrett’s esophageal mucosa and adenocarcinoma cell line. Scand J Gastroenterol. 2007 Dec;42(12):1460–1465. doi: 10.1080/00365520701452209. [DOI] [PubMed] [Google Scholar]
- 35.Marchetti M, Caliot E, Pringault E. Chronic acid exposure leads to activation of the cdx2 intestinal homeobox gene in a long-term culture of mouse esophageal keratinocytes. J Cell Sci. 2003 Apr 15;116(Pt 8):1429–1436. doi: 10.1242/jcs.00338. [DOI] [PubMed] [Google Scholar]
- 36.Liu T, Zhang X, So CK, et al. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007 Feb;28(2):488–496. doi: 10.1093/carcin/bgl176. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Jones C, Gellersen O, Williams VA, Watson TJ, Peters JH. Pathogenesis of Barrett esophagus: deoxycholic acid up-regulates goblet-specific gene MUC2 in concert with CDX2 in human esophageal cells. Arch Surg. 2007 Jun;142(6):540–544. doi: 10.1001/archsurg.142.6.540. discussion 544-545. [DOI] [PubMed] [Google Scholar]
- 38.Tatsuta T, Mukaisho K, Sugihara H, Miwa K, Tani T, Hattori T. Expression of Cdx2 in early GRCL of Barrett’s esophagus induced in rats by duodenal reflux. Dig Dis Sci. 2005 Mar;50(3):425–431. doi: 10.1007/s10620-005-2452-9. [DOI] [PubMed] [Google Scholar]
- 39.Huo X, Zhang HY, Zhang XI, et al. Acid and Bile Salt-Induced CDX2 Expression Differs in Esophageal Squamous Cells From Patients With and Without Barrett’s Esophagus. Gastroenterology. 2010 Mar 17; doi: 10.1053/j.gastro.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002 Oct 18;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 41.Lickert H, Domon C, Huls G, et al. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000 Sep;127(17):3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 42.Bian YS, Osterheld MC, Bosman FT, Fontolliet C, Benhattar J. Nuclear accumulation of beta-catenin is a common and early event during neoplastic progression of Barrett esophagus. Am J Clin Pathol. 2000 Oct;114(4):583–590. doi: 10.1309/3QLC-5MF1-JYXU-A5XX. [DOI] [PubMed] [Google Scholar]
- 43.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005 Jun 16;435(7044):964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 44.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005 Aug 30;102(35):12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005 Jun 16;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 46.Sander GR, Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004 Apr;52(4):509–516. doi: 10.1177/002215540405200409. [DOI] [PubMed] [Google Scholar]
- 47.Morrow DJ, Avissar NE, Toia L, et al. Pathogenesis of Barrett’s esophagus: bile acids inhibit the Notch signaling pathway with induction of CDX2 gene expression in human esophageal cells. Surgery. 2009 Oct;146(4):714–721. doi: 10.1016/j.surg.2009.06.050. discussion 721-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menke V, van Es JH, de Lau W, et al. Conversion of metaplastic Barrett’s epithelium into post-mitotic goblet cells by gamma-secretase inhibition. Dis Model Mech. 2010 Jan-Feb;3(1-2):104–110. doi: 10.1242/dmm.003012. [DOI] [PubMed] [Google Scholar]
- 49.Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006 Sep;74(7):422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- 50.Milano F, van Baal JW, Buttar NS, et al. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007 Jun;132(7):2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 51.Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998 Sep;20(1):58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 53.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002 Nov 14;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 54.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009 Jul;30(7):1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Meltzer SJ, Yin J, et al. Altered messenger RNA and unique mutational profiles of p53 and Rb in human esophageal carcinomas. Cancer Res. 1993 Apr 15;53(8):1889–1894. [PubMed] [Google Scholar]
- 56.Coppola D, Schreiber RH, Mora L, Dalton W, Karl RC. Significance of Fas and retinoblastoma protein expression during the progression of Barrett’s metaplasia to adenocarcinoma. Ann Surg Oncol. 1999 Apr-May;6(3):298–304. doi: 10.1007/s10434-999-0298-7. [DOI] [PubMed] [Google Scholar]
- 57.Klump B, Hsieh CJ, Holzmann K, Gregor M, Porschen R. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus. Gastroenterology. 1998 Dec;115(6):1381–1386. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- 58.Wong DJ, Barrett MT, Stoger R, Emond MJ, Reid BJ. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997 Jul 1;57(13):2619–2622. [PubMed] [Google Scholar]
- 59.Maley CC, Galipeau PC, Li X, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004 Oct 15;64(20):7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 60.Younes M, Lebovitz RM, Lechago LV, Lechago J. p53 protein accumulation in Barrett’s metaplasia, dysplasia, and carcinoma: a follow-up study. Gastroenterology. 1993 Dec;105(6):1637–1642. doi: 10.1016/0016-5085(93)91058-p. [DOI] [PubMed] [Google Scholar]
- 61.Hamelin R, Flejou JF, Muzeau F, et al. TP53 gene mutations and p53 protein immunoreactivity in malignant and premalignant Barrett’s esophagus. Gastroenterology. 1994 Oct;107(4):1012–1018. doi: 10.1016/0016-5085(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 62.Ramel S, Reid BJ, Sanchez CA, et al. Evaluation of p53 protein expression in Barrett’s esophagus by two-parameter flow cytometry. Gastroenterology. 1992 Apr;102(4 Pt 1):1220–1228. [PubMed] [Google Scholar]
- 63.Malumbres M, Pellicer A. RAS pathways to cell cycle control and cell transformation. Front Biosci. 1998 Aug 6;3:d887–912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- 64.Inamdar GS, Madhunapantula SV, Robertson GP. Targeting the MAPK pathway in melanoma: Why some approaches succeed and other fail. Biochem Pharmacol. 2010 May 9; doi: 10.1016/j.bcp.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper GM. Cellular transforming genes. Science. 1982 Aug 27;217(4562):801–806. doi: 10.1126/science.6285471. [DOI] [PubMed] [Google Scholar]
- 66.Meltzer SJ, Mane SM, Wood PK, et al. Activation of c-Ki-ras in human gastrointestinal dysplasias determined by direct sequencing of polymerase chain reaction products. Cancer Res. 1990 Jun 15;50(12):3627–3630. [PubMed] [Google Scholar]
- 67.Lord RV, O’Grady R, Sheehan C, Field AF, Ward RL. K-ras codon 12 mutations in Barrett’s oesophagus and adenocarcinomas of the oesophagus and oesophagogastric junction. J Gastroenterol Hepatol. 2000 Jul;15(7):730–736. doi: 10.1046/j.1440-1746.2000.02163.x. [DOI] [PubMed] [Google Scholar]
- 68.Sommerer F, Vieth M, Markwarth A, et al. Mutations of BRAF and KRAS2 in the development of Barrett’s adenocarcinoma. Oncogene. 2004 Jan 15;23(2):554–558. doi: 10.1038/sj.onc.1207189. [DOI] [PubMed] [Google Scholar]
- 69.Abdelatif OM, Chandler FW, Mills LR, McGuire BS, Pantazis CG, Barrett JM. Differential expression of c-myc and H-ras oncogenes in Barrett’s epithelium. A study using colorimetric in situ hybridization. Arch Pathol Lab Med. 1991 Sep;115(9):880–885. [PubMed] [Google Scholar]
- 70.Jankowski J, McMenemin R, Hopwood D, Penston J, Wormsley KG. Abnormal expression of growth regulatory factors in Barrett’s oesophagus. Clin Sci (Lond) 1991 Nov;81(5):663–668. doi: 10.1042/cs0810663. [DOI] [PubMed] [Google Scholar]
- 71.Auvinen MI, Sihvo EI, Ruohtula T, et al. Incipient angiogenesis in Barrett’s epithelium and lymphangiogenesis in Barrett’s adenocarcinoma. J Clin Oncol. 2002 Jul 1;20(13):2971–2979. doi: 10.1200/JCO.2002.09.011. [DOI] [PubMed] [Google Scholar]
- 72.Lord RV, Park JM, Wickramasinghe K, et al. Vascular endothelial growth factor and basic fibroblast growth factor expression in esophageal adenocarcinoma and Barrett esophagus. J Thorac Cardiovasc Surg. 2003 Feb;125(2):246–253. doi: 10.1067/mtc.2003.203. [DOI] [PubMed] [Google Scholar]
- 73.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997 Apr;33(5):787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 74.Morales CP, Lee EL, Shay JW. In situ hybridization for the detection of telomerase RNA in the progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer. 1998 Aug 15;83(4):652–659. [PubMed] [Google Scholar]
- 75.Lord RV, Salonga D, Danenberg KD, et al. Telomerase reverse transcriptase expression is increased early in the Barrett’s metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000 Mar-Apr;4(2):135–142. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 76.Abdel-Latif MM, O’Riordan J, Windle HJ, et al. NF-kappaB activation in esophageal adenocarcinoma: relationship to Barrett’s metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann Surg. 2004 Apr;239(4):491–500. doi: 10.1097/01.sla.0000118751.95179.c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Riordan JM, Abdel-latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005 Jun;100(6):1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 78.Dvorak K, Chavarria M, Payne CM, et al. Activation of the interleukin-6/STAT3 antiapoptotic pathway in esophageal cells by bile acids and low pH: relevance to barrett’s esophagus. Clin Cancer Res. 2007 Sep 15;13(18 Pt 1):5305–5313. doi: 10.1158/1078-0432.CCR-07-0483. [DOI] [PubMed] [Google Scholar]
- 79.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002 Mar 29;295(5564):2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 80.Herszenyi L, Hritz I, Pregun I, et al. Alterations of glutathione S-transferase and matrix metalloproteinase-9 expressions are early events in esophageal carcinogenesis. World J Gastroenterol. 2007 Feb 7;13(5):676–682. doi: 10.3748/wjg.v13.i5.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray GI, Duncan ME, O’Neil P, McKay JA, Melvin WT, Fothergill JE. Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer. J Pathol. 1998 Jul;185(3):256–261. doi: 10.1002/(SICI)1096-9896(199807)185:3<256::AID-PATH115>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 82.Salmela MT, Karjalainen-Lindsberg ML, Puolakkainen P, Saarialho-Kere U. Upregulation and differential expression of matrilysin (MMP-7) and metalloelastase (MMP-12) and their inhibitors TIMP-1 and TIMP-3 in Barrett’s oesophageal adenocarcinoma. Br J Cancer. 2001 Aug 3;85(3):383–392. doi: 10.1054/bjoc.2001.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005 Jun;2(6):315–324. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]