Abstract
The discovery of RNA interference (RNAi) generated considerable interest in developing short interfering RNAs (siRNAs) for understanding basic biology and as the active agents in a new variety of therapeutics. Early studies showed that selecting an active siRNA was not as straightforward as simply picking a sequence on the target mRNA and synthesizing the siRNA complementary to that sequence. As interest in applying RNAi has increased, the methods for identifying active siRNA sequences have evolved from focusing on the simplicity of synthesis and purification, to identifying preferred target sequences and secondary structures, to predicting the thermodynamic stability of the siRNA. As more specific details of the RNAi mechanism have been defined, these have been incorporated into more complex siRNA selection algorithms, increasing the reliability of selecting active siRNAs against a single target. Ultimately, design of the best siRNA therapeutics will require design of the siRNA itself, in addition to design of the vehicle and other components necessary for it to function in vivo. In this minireview, we will summarize the evolution of siRNA selection techniques with a particular focus on one issue of current importance to the field, how best to identify those siRNA sequences likely to have high activity. Approaches to designing active siRNAs through chemical and structural modifications will also be highlighted. As the understanding of how to control the activity and specificity of siRNAs improves, the potential utility of siRNAs as human therapeutics will concomitantly grow.
Introduction
Issues of formulation, stability, delivery, and specificity are crucial for the development of any therapeutic. Nonetheless, it is essential to begin therapeutic development with the most active molecule possible in order to enable use of the minimum dose to achieve a therapeutic effect. For short interfering RNA (siRNA)-based therapeutics, identifying the most active sequences requires a thorough understanding of the molecular-level details of the RNA interference (RNAi) mechanism. Implicit in this understanding is that we would know what chemical and physical features of the siRNA are important for maximal activity. However, to date, many details remain unclear. Here, we will briefly review how siRNA selection approaches have become more sophisticated as mechanistic details have emerged and how further analysis of existing and new data can provide additional insights into further refinement of these approaches. We will conclude with a discussion of how chemical and physical manipulations can be used to enhance the activity of a selected siRNA sequence.
Mechanism of siRNA-initiated RNAi in Humans
Since the discovery and characterization of RNAi in C. elegans [1], the broad mechanistic details for the pathway have been largely characterized. Unlike C. elegans, longer double-stranded RNAs (dsRNAs) cannot be used to initiate RNAi in mammalian cells due to the innate immune response [2]. Therefore, siRNAs are used to initiate RNAi [3, 4], though these still have potential immunogenicity (see the companion minireview by Samuel-Abraham and Leonard [5] and [6, 7]). Nonetheless, siRNAs remain the most viable candidates for application of RNAi as a human therapeutic approach.
The basic mechanism for siRNA-initiated RNAi in humans is as follows. siRNAs are first delivered to the cytoplasm of the cells of interest, a non-trivial task, particularly in vivo (see companion minireview by Shim and Kwon [8] and [9]). The siRNAs are then recognized by the proteins of the human RISC loading complex (RLC), Dicer, Argonaute 2 (Ago2), and TAR RNA binding protein (TRBP) [10]. The RLC then selects one of the two strands to act as the guide strand [11–13], yielding the active RNA induced silencing complex (RISC), which contains at a minimum the single-stranded guide strand RNA and Ago2 [14, 15]. (Recent work suggests that the human RLC may not function in an exactly identical manner to the Drosophila RLC [16], suggesting that careful study of the human system and proteins is essential for the development of therapeutic siRNAs for human disease.) RISC then recognizes its target mRNA by complementarity between the guide strand and a region on the mRNA, cleaving the mRNA at the center of the region of intermolecular hybridization [3]. Silencing results from the normal degradation of previously expressed protein, which cannot be replaced due to the reduced levels of intact mRNA. Thus, RNAi provides a powerful tool for inhibiting the expression of any protein product of relatively short half-life (< 12 h) whose expression level is primarily controlled by transcription rate.
It is important to note that the use of siRNAs for transient control of gene expression leverages an endogenous cellular control mechanism that is naturally used by microRNAs (miRNAs) [17–19]. As such, siRNAs do not have to activate a new pathway to function. While there is some concern that this machinery could be diverted from its normal roles by saturation with exogenous siRNAs [20], this would likely be a concern only for a chronic therapy, for which short, hairpin RNAs (shRNAs), which are related in structure and function to both siRNAs and miRNAs, are more suitable [21]. For further information about miRNAs, their roles in gene expression control, and their unique characteristics relative to siRNAs, see [22].
Evolution in the Rules for Selecting siRNA sequences
As the field of RNAi has grown, the rules for selecting candidate siRNA sequences have become more complex. The initial selection of agents for RNAi was based on complementarity of one strand of the dsRNA to the target mRNA. Subsequent to the discoveries of Dicer and siRNAs, it became clear that the structure of the siRNA, with the internal 19 nt hybridized and 2 nt overhangs at each 3′-end (typically UU or TT), was also important for recognition by the pathway proteins. These structural considerations were combined with uniqueness of the target sequence within the known transcriptome of the organism and the simplicity and purity with which the selected sequence could be synthesized to serve as the initial design considerations for siRNAs (see Figure 1 for other possible design variables for siRNAs that can be considered).
Figure 1. Key design options for siRNAs.
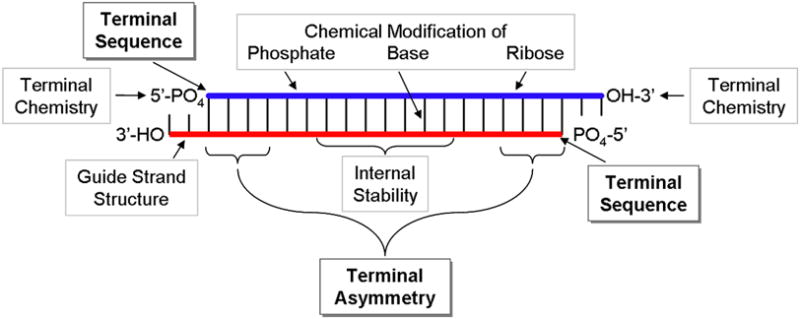
siRNAs can be modified on either their guide (red) or passenger (blue) strands. Multiple chemical modifications can be made along the length of the siRNA or at the termini as discussed elsewhere [50, 51], including modifying the backbone, base, and terminal chemistry, especially of the passenger strand. Formation of intramolecular secondary structure by the guide strand after separation from the passenger strand has also been examined as a design criterion [32–34]. Although positional base preferences have been suggested for multiple locations along the siRNA (e.g. [28]), this review focuses primarily on the characteristics of the siRNA termini, hybridization asymmetry and sequence (i.e., the 5′-nucleotide on each strand), for use in the design of highly active siRNAs.
Another critical feature that subsequently came to light was that siRNAs must possess a 5′-PO3 rather than a 5′-OH [23], which is the typical terminal group for chemically synthesized siRNAs. This 5′-PO3 group is important for recognition of the siRNA by Dicer, as 5′-OCH3 and modified strands are bound far more weakly than phosphorylated strands [24, 25]. Fortunately, unphosphorylated siRNAs are rapidly phosphorylated by Clp1 upon entry to the cytoplasm [26]. As such, modifying the passenger strand with a 5′-methoxy group can prevent its phosphorylation and therefore prevent incorporation into RISC [27].
It soon became clear that not all siRNAs silenced their target with the same efficiency, so the rules for selection of active species were strengthened by the generation and analysis of data on large sets of siRNAs [28–31]. The earliest rules were focused on the siRNA alone with positional base preferences being the dominant factors [28, 31]. It was also proposed that using siRNAs where the guide strand would not form a stable secondary structure would be preferred [32, 33], though this remains in question [34]. The accessibility of the target region on the mRNA, as determined by mRNA secondary structure prediction [35–37], has also been found to be important in determining siRNA function [38, 39]. In general, we found that having accessibility at the 5′-end and 3′-end of the target region, based on the minimum free energy structure prediction, was preferred to accessibility in the center of the target or no accessibility, with the effect being independent of guide strand structure [39]. Regardless of the method used for secondary structure prediction, it is clear that accounting for the target secondary structure is valuable in selecting siRNAs with maximal activity. This is similar to what had been found for the effect of mRNA structure on antisense oligonucleotide activity [40–42].
Terminal Asymmetry Prediction
Because siRNAs are double-stranded, either strand is capable of serving as the guide for active RISC. Thus, to maximize the activity of siRNAs, it is advantageous for one of the two strands of the siRNA to be loaded preferentially into RISC. The preference for loading one strand over the other is referred to as siRNA asymmetry. Based on early studies in Drosophila, it was proposed that siRNAs were asymmetric due to the difference in the hybridization free energy for the terminal four nucleotides on each end of the siRNA [12, 43]. The strand whose 5′-end was located at the less stably hybridized end of the siRNA would preferentially be loaded into active RISC. This was confirmed using sequences with terminal mismatches to induce significant instability at one end of the siRNA. Subsequently, thermodynamic asymmetry was confirmed to be a useful predictor of siRNA function [34].
Though the existence and importance of asymmetry are not in question, the appropriate method for prediction of asymmetry has since received considerable attention, with two primarily parallel viewpoints, terminal sequence or terminal stability, being adopted. With fully-hybridized siRNAs, the thermodynamic stability of the termini is a function of the terminal sequence. Therefore, either the sequence (as suggested by analyses of positional base preferences) or the stability (as suggested by thermodynamic calculations) or both can be the driving force for asymmetry. Moreover, the strategy for calculating thermodynamic asymmetry is not fully settled [44–46], in particular how many base pairs/nearest neighbors to take into account in the calculation. Our previous analyses suggested that ultimate silencing activity could be reliably predicted by simple classification of the 5′ nucleotides on each strand [11]. Supporting the contributions of terminal sequence to eventual function, biochemical and structural studies have demonstrated preferences in terminal nucleotide identity for RNA binding and processing by Dicer and Ago2 [47, 48]. It is therefore beneficial to analyze further the relative utility of terminal thermodynamics and terminal nucleotide sequence for predicting eventual siRNA function.
To do this, we analyzed two available databases of siRNA function [30, 31]. Using information theory, we analyzed the reduction in entropy in the activity data when using terminal nucleotide classification (as in [11]) vs. using the ΔΔG calculations with 1, 2, 3, or 4 terminal nearest neighbors. A reduction in entropy indicates a reduction in the scatter of the data and hence a useful predictor of the data. Examining each variable, all five prediction strategies provide predictive information (Table 1), with the terminal nucleotide classification providing the best predictive accuracy for both datasets. It is worthwhile to note that, among the ΔΔG calculations, using only the terminal nearest neighbor on each end of the siRNA provides the best predictive accuracy (Table 1, bold), echoing the results of others [44]. However, in all cases, the entropy reductions, though statistically significant (via bootstrapping, data not shown), are rather modest.
Table 1.
Entropy Reduction of Activity Data with siRNA Classification Methods
| Dataset | Classification | Entropy | Entropy Reduction |
|---|---|---|---|
| Shabalina [30] | None | 3.32 | N/A |
| Terminal Nucleotide | 2.92 | 0.40 | |
| ΔΔG (1 nn) | 3.07 | 0.25 | |
| ΔΔG (2 nn) | 3.09 | 0.23 | |
| ΔΔG (3 nn) | 3.10 | 0.22 | |
| ΔΔG (4 nn) | 3.16 | 0.16 | |
| Novartis [31] | None | 3.32 | N/A |
| Terminal Nucleotide | 3.07 | 0.25 | |
| ΔΔG (1 nn) | 3.16 | 0.16 | |
| ΔΔG (2 nn) | 3.19 | 0.13 | |
| ΔΔG (3 nn) | 3.23 | 0.09 | |
| ΔΔG (4 nn) | 3.23 | 0.09 |
All ΔΔG calculations include an AU end penalty [63]. nn - nearest neighbor.
We therefore examined the entropy reduction when using both terminal nucleotide classification and ΔΔG calculation to predict siRNA function (Table 2). Use of both approaches greatly reduces the entropy in all cases in a synergistic fashion. The independent information that is possessed by each classification mode is seen in the low (near zero) redundancies between terminal nucleotide classification and any of the ΔΔG calculations (Table 2, column 5). Interestingly, when using the terminal nucleotide classification as well, the best predictive accuracy for both datasets was achieved with the 3 nearest neighbor ΔΔG calculation (Table 2, bold). This analysis, which is consistent between the two datasets, shows that predicting siRNA function using classification by both sequence and asymmetry in terminal stability provides greater accuracy than using either technique independently. This point is emphasized when examining the data sorted using the nucleotide classification and 3 nearest neighbor ΔΔG calculation (Figure 2). There are clear and distinct trends both horizontally and vertically, making those sequences that appear in the upper left-hand corner of the figure most likely to be highly active. This further supports that terminal sequence and terminal stability provide unique, useful information for predicting siRNA activity.
Table 2.
Entropy Reduction and Information Redundancy of Activity Data with Terminal Nucleotide Classification and ΔΔG Calculation
| Dataset | Classification | Entropy | Entropy Reduction | Information Redundancy |
|---|---|---|---|---|
| Shabalina [30] | ΔΔG (1 nn) | 1.68 | 1.64 | 0.22 |
| ΔΔG (2 nn) | 1.51 | 1.81 | 0.14 | |
| ΔΔG (3 nn) | 1.46 | 1.86 | 0.12 | |
| ΔΔG (4 nn) | 1.49 | 1.83 | 0.11 | |
| Novartis [31] | ΔΔG (1 nn) | 2.75 | 0.57 | 0.24 |
| ΔΔG (2 nn) | 2.66 | 0.66 | 0.10 | |
| ΔΔG (3 nn) | 2.64 | 0.68 | 0.07 | |
| ΔΔG (4 nn) | 2.64 | 0.68 | 0.06 |
All ΔΔG calculations include an AU end penalty [63]. Redundancy is a value between 0 and 1 describing the portion of the overlapping information between two features, with 1 being complete overlap. nn - nearest neighbor.
Figure 2. Sorting of siRNA Activity Results (from Shabalina, et. al, [30]).
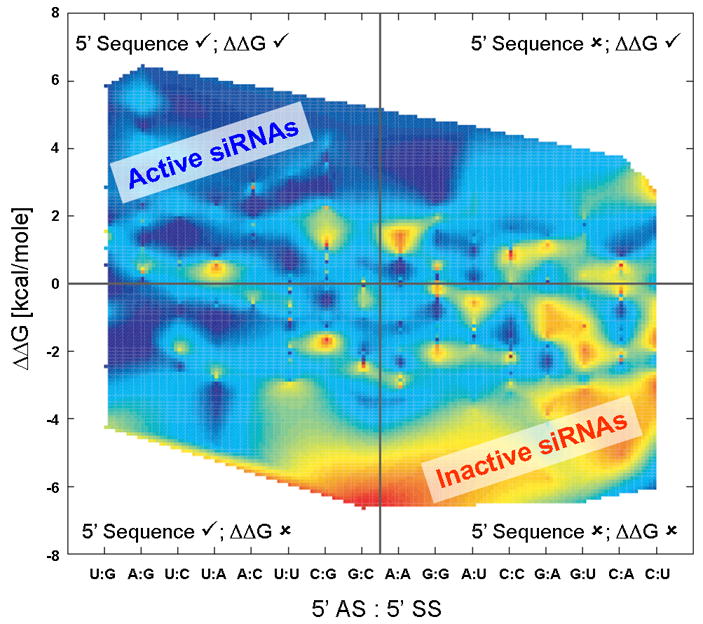
The activities of siRNAs were plotted according to the 5′ nucleotides on their antisense and sense strands (horizontal axis) and the ΔΔG calculated using 3 nearest neighbors (vertical axis). The data points were then interpolated to simplify visualization of data trends. The scale is red (least active siRNAs; highest mRNA concentration) to violet (most active siRNAs; lowest mRNA level). The figure is divided into the four quadrants where check marks (✓) indicate the approach would identify the correct guide strand, and x’s (✘) indicate the approach would identify the incorrect guide strand. Therefore, sequences in the upper left quadrant are those that would be predicted by both methods to prefer the proper guide strand. Plots using the Novartis data [31] with calculations using 1–4 nearest neighbors are available in the supporting information (Figures S1–S8) but are visually similar to this plot.
Internal Thermodynamic Stability
Recent results suggest that highly active siRNAs are likely to have lower internal stabilities than less active siRNAs [46]. Lower internal stabilities were found to be indicative of lower siRNA GC content and limited secondary structure for both the target and guide strand, all of which are known to be important factors in maximizing function. Other results showed that the internal stability of siRNAs can vary along their length [49]. As it is known that the passenger stand of the siRNA is cleaved by Ago2 to free the guide strand [13], the profile of variable internal stability may reflect that the center of the siRNA must be hybridized stably to allow cleavage of the passenger strand by Ago2 but that both 3′-ends of the cleaved passenger strand should be relatively unstable to encourage separation from the guide strand after cleavage [49].
What are the characteristics of the best siRNA therapeutic?
Ultimately, the best siRNA sequence will be determined by the sequence and structure of the target as well as the sequence, structure, and asymmetry of the siRNA. However, once the best siRNA sequence has been selected, the function of the siRNA can still be enhanced through the incorporation of a variety of chemical and structural modifications that improve the performance of the siRNA relative to a particular design variable, e.g., biological half-life. These modifications will be important for generating siRNAs with in vivo efficacies and specificities that are sufficient for therapeutics. The variables that can be manipulated include altering the nucleotide chemistry in the ribose, base, or phosphates; varying the length of the siRNA; and altering the overhang sequence, structure, and chemistry. The first priority for nucleic acid based therapeutics, especially RNAs, is maintaining their integrity in the presence of ubiquitous nucleases. Many chemical modifications that mitigate degradation by RNases have been examined [50, 51], many of these having first been used in the development of antisense oligonucleotides [52–56]. Other important details that can be manipulated using chemical modifications include strand selection, off-target effects, and cellular distribution, as reviewed in [51].
Structural modifications can also be effective at altering siRNA properties. While typical siRNAs have a 19 nt paired region followed by a 2 nt 3′ overhang, longer and shorter siRNAs have been shown to be active at initiating silencing [57]. Longer duplexes that can serve as Dicer substrates can be more efficient at silencing than standard length siRNAs of identical sequence [58], which would be expected from the close contact between Dicer and Ago2 in the structure of the RLC [59]. Mismatches, while useful for inducing asymmetry [12], may not be a practical approach for generating sequences of maximal activity, as mismatched siRNAs are bound by TRBP less strongly than fully-paired sequences [60]. An interesting manipulation of siRNA structure is the use of segmented structures. For instance, small internally segmented interfering RNAs (sisiRNAs) were developed possessing an intact guide strand and two segments of the passenger strand [61]. sisiRNAs, when modified with selected locked nucleic acid (LNA) nucleotides, were found to be more tolerant of chemical modifications than standard siRNAs. Silencing has also been achieved using siRNAs possessing DNA segments on both the guide and passenger strands [62], though it is important to maintain a primarily A-form duplex to ensure recognition by the dsRNA-binding domains of TRBP and Dicer.
Caveat for Development of New Selection Approaches
It is important to note that multiple siRNA manufacturers now have libraries of siRNA sequences available for use in targeting all or nearly all of the known expressed genes in common organisms (e.g., human, mouse, and rat). Many of these contain known or proprietary chemical and structural modifications to enhance their activity or reduce off-target effects. While these libraries are valuable resources for those seeking siRNAs as tools for the laboratory, identification and manipulation of siRNAs to achieve maximal activity is still an important task. The available siRNAs may work adequately, but, without clear mechanistic information about how best to select and modify siRNAs, it is not clear if the available sequences are the best for the specific requirements of a given target and application. When this level of control has been achieved, i.e., when an siRNA can be designed taking into account the uniqueness of a particular target and application, the field will have reached the maturity necessary for general consideration as a therapeutic strategy.
Supplementary Material
Acknowledgments
Financial support for this work was provided in part by Michigan State University, the National Science Foundation (CBET 0941055), the National Institutes of Health (GM079688, RR024439, GM089866), the Michigan Universities Commercialization Initiative (MUCI), and the Center for Systems Biology.
List of Abbreviations
- Ago2
Argonaute 2
- dsRNA
double stranded RNA
- LNA
locked nucleic acid
- miRNA
microRNA
- TRBP
TAR RNA binding protein
- PACT
protein kinase R; activator protein
- PKR
protein kinase R
- RISC
RNA induced silencing complex
- RLC
RISC loading complex
- RNAi
RNA interference
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- sisiRNAs
small internally segmented interfering RNAs
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel-Abraham S, Leonard JN. Staying on Message: Design Principles for Controlling Nonspecific Responses to siRNA. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07905.x. this minireview series. [DOI] [PubMed] [Google Scholar]
- 6.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Sioud M. Deciphering the code of innate immunity recognition of siRNAs. Methods Mol Biol. 2009;487:41–59. doi: 10.1007/978-1-60327-547-7_2. [DOI] [PubMed] [Google Scholar]
- 8.Shim M, Kwon Y. Efficient and Targeted Delivery of siRNA In Vivo. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07904.x. this minireview series. [DOI] [PubMed] [Google Scholar]
- 9.Sioud M. Targeted delivery of antisense oligonucleotides and siRNAs into mammalian cells. Methods Mol Biol. 2009;487:61–82. doi: 10.1007/978-1-60327-547-7_3. [DOI] [PubMed] [Google Scholar]
- 10.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010;49:3148–3155. doi: 10.1021/bi902189s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 13.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 15.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 16.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 18.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 19.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 20.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 21.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol. 2009;487:369–385. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 24.Pellino JL, Jaskiewicz L, Filipowicz W, Sontheimer EJ. ATP modulates siRNA interactions with an endogenous human Dicer complex. RNA. 2005;11:1719–1724. doi: 10.1261/rna.2102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kini HK, Walton SP. In vitro binding of single-stranded RNA by human Dicer. FEBS Lett. 2007;581:5611–5616. doi: 10.1016/j.febslet.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 27.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 29.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shabalina SA, Spiridonov AN, Ogurtsov AY. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 32.Patzel V, Rutz S, Dietrich I, Koberle C, Scheffold A, Kaufmann SH. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 33.Koberle C, Kaufmann SH, Patzel V. Selecting effective siRNAs based on guide RNA structure. Nat Protoc. 2006;1:1832–1839. doi: 10.1038/nprot.2006.206. [DOI] [PubMed] [Google Scholar]
- 34.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 35.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics. 2009;Chapter 12(Unit12):12. doi: 10.1002/0471250953.bi1202s26. [DOI] [PubMed] [Google Scholar]
- 37.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 38.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Gredell JA, Berger AK, Walton SP. Impact of target mRNA structure on siRNA silencing efficiency: A large-scale study. Biotechnol Bioeng. 2008;100:744–755. doi: 10.1002/bit.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walton SP, Stephanopoulos GN, Yarmush ML, Roth CM. Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol Bioeng. 1999;65:1–9. [PubMed] [Google Scholar]
- 42.Ding Y, Lawrence CE. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 44.Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005;579:5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 46.Matveeva OV, Kang Y, Spiridonov AN, Saetrom P, Nemtsov VA, Ogurtsov AY, Nechipurenko YD, Shabalina SA. Optimization of duplex stability and terminal asymmetry for shRNA design. PLoS One. 2010;5:e10180. doi: 10.1371/journal.pone.0010180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010 doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 49.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 50.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chernolovskaya EL, Zenkova MA. Chemical modification of siRNA. Curr Opin Mol Ther. 2010;12:158–167. [PubMed] [Google Scholar]
- 52.Cohen JS. Oligonucleotides as therapeutic agents. Pharmacol Ther. 1991;52:211–225. doi: 10.1016/0163-7258(91)90009-b. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Q, Temsamani J, Iadarola PL, Jiang Z, Agrawal S. Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem Pharmacol. 1996;51:173–182. doi: 10.1016/0006-2952(95)02177-9. [DOI] [PubMed] [Google Scholar]
- 54.Kandimalla ER, Manning A, Zhao Q, Shaw DR, Byrn RA, Sasisekharan V, Agrawal S. Mixed backbone antisense oligonucleotides: design, biochemical and biological properties of oligonucleotides containing 2′-5′-ribo- and 3′-5′-deoxyribonucleotide segments. Nucleic Acids Res. 1997;25:370–378. doi: 10.1093/nar/25.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prakash TP, Kawasaki AM, Johnston JF, Graham MJ, Condon TP, Manoharan M. Antisense properties of 2′-O-dimethylaminooxyethyl (2′-O-DMAOE) oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2001;20:829–832. doi: 10.1081/NCN-100002439. [DOI] [PubMed] [Google Scholar]
- 56.Bell NM, Micklefield J. Chemical modification of oligonucleotides for therapeutic, bioanalytical and other applications. Chembiochem. 2009;10:2691–2703. doi: 10.1002/cbic.200900341. [DOI] [PubMed] [Google Scholar]
- 57.Chu CY, Rana TM. Potent RNAi by short RNA triggers. RNA. 2008;14:1714–1719. doi: 10.1261/rna.1161908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 59.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kini HK, Walton SP. Effect of siRNA terminal mismatches on TRBP and Dicer binding and silencing efficacy. FEBS J. 2009;276:6576–6585. doi: 10.1111/j.1742-4658.2009.07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


