Abstract
Myeloid derived suppressor cells (MDSCs) are one of the main cell populations responsible for regulating immune responses. MDSCs accumulate during tumor progression, autoimmunity, chronic infection and other pathological conditions and can potently suppress T cell function. Recent studies have demonstrated the ability of MDSCs to modulate the activity of NK and myeloid cells and have implicated MDSCs in the induction of regulatory T cells. In this review we discuss recent findings that describe the molecular mechanisms that regulate expansion and function of MDSCs, as well as recent attempts to use MDSCs in cell therapy for different pathologic conditions.
Myeloid-derived suppressor cells (MDSCs) as negative regulators of immune responses
MDSC represent a heterogenic population of immature myeloid cells that consists of myeloid progenitors and precursors of macrophages, granulocytes, and dendritic cells (DC) and are characterized by a strong ability to suppress various T-cell functions [1]. In mice, MDSCs are identified as cells that simultaneous express two markers: CD11b and Gr-11. More recently, MDSCs were subdivided into two different subsets based on their expression of the two molecules Ly-6C and Ly-6G, which can be detected by specific antibodies [2, 3]. CD11b+ Ly-6G− Ly-6Chigh cells have monocytic-like morphology and are termed monocytic-MDSCs (M-MDSCs). CD11b+ Ly-6G+ Ly-6Clow cells have granulocyte-like morphology and are termed granulocytic-MDSCs (G-MDSCs). In cancer patients, MDSCs are defined as cells that express the common myeloid marker CD33 but lack expression of markers of mature myeloid and lymphoid cells [4]. In recent years, more markers were associated with MDSC function in humans. A monocytic MDSC with the phenotype CD14+CD11b+HLA-DRlow/neg has been detected in melanoma patients[5, 6]. In patients with melanoma and colon carcinomas, two main subpopulations functionally suppress the immune response: CD14+ monocytes and CD15+ neutrophils both expressing IL-4 receptor (CD124)[7]. MDSCs are also defined as CD11b+CD14−CD15+CD33+ cells in patients with advanced non-small cell lung cancer [8, 9]. MDSCs from patients with renal cell cancer, express markers of activated granulocytes, including high levels of CD66b and low levels of CD62L, CD16, and vascular endothelial growth factor (VEGF) receptor 1 [10]. More detailed information about MDSC phenotype can be found in several recent reviews [11–13].
MDSCs utilize a number of mechanisms to suppress T-cell function. They include a high level of arginase activity, as well as nitric oxide (NO) and reactive oxygen species (ROS) production [1, 14–16]. These main pathways are linked with different subsets of MDSCs: ROS with G-MDSCs, whereas arginase and NO with M-MDSCs [3, 17]. Several other suppressive mechanisms have recently been suggested: secretion of TGF β [18, 19], induction of regulatory T cells [20–22], depletion of cysteine [23], upregulation of cyclo-oxygenase 2 (Cox2) and prostaglandin E2 (PGE2) [24]. These mechanisms have been described in a number of recent reviews [1, 25–27].
One of the main questions of MDSC biology is how MDSC accumulation is regulated. This question is critically important for the development of the effective therapeutic strategies to control these cells. In recent years data emerged linking several signaling pathways with MDSC expansion. They may become attractive targets for therapeutic intervention. Here we will discuss recent advances in this field.
Signal transducer and activator of transcription 3 (Stat3)
Stat3 plays a central role in many molecular events governing tumor cell proliferation, survival and invasion. At the same time, Stat3 is involved in inhibition of antitumor immune responses [28]. In myeloid cells, Stat3 signaling drives the expression of Bcl-xL, c-myc, cyclin D1, or survivin, which prevents cell apoptosis, promotes cell proliferation, and prevents differentiation to mature cell types [1, 28]. Previous studies have established a critical role for Stat3 in MDSC expansion in mice [29, 30] and for the contribution of MDSCs to angiogenesis [31]. Recently, an association was demonstrated between upregulated Stat3 activation and MDSC accumulation in melanoma patients [6]. As we discuss briefly in this review, Stat3 is also known to be critically important for MDSC suppressive activity. In that study, inhibition of Stat3 in vitro abolished the suppressive activity of MDSC. The important role of Stat3 in MDSC expansion is further supported by an in vivo study using the multitargeted tyrosine kinase inhibitor sunitinib[32]. Sunitinib blocked MDSC expansion in tumor-bearing mice via inhibition of Stat3 signaling in myeloid cells. Myeloid cell-specific overexpression of apoptosis inhibitor 6 (API6) caused activation of Stat3 in myeloid cells and systemic expansion of MDSCs. In the lung, it resulted in severe inflammation and massive tissue remodeling with an increased frequency of lung adenocarcinoma [33].
Several pathways downstream of Stat3 might be involved in the regulation of MDSC expansion and function. One such pathway involves the calcium-binding pro-inflammatory proteins S100A9 and S100A8 [34]. Activation of Stat3 in hematopoietic progenitor cells (HPC) upregulates S100A8 and S100A9; this, in turn, inhibits DC differentiation and promotes MDSC accumulation [35]. In the absence of S100A9, MDSC accumulation is inhibited in the spleen of tumor-bearing mice. At the same time, over-expression of S100A9 results in the accumulation of MDSCs in tumor-free mice [35]. The precise mechanism of this effect is not clear, but it was suggested that the S100A9 and S100A8 heterodimer might participate in the formation of the NADPH oxidase (Nox2) complex that is responsible for production of ROS in myeloid cells. Increased production of ROS contributes to inhibition of myeloid cell differentiation [35]. S100A9 also plays an important role in the migration of MDSCs to the tumor site. The effects of S100A9 and S100A8 proteins are mediated via carboxylated N-glycans, which are expressed on the receptor for advanced glycation end-products and other MDSC cell surface glycoprotein receptors. These receptors signal the effects of S100A9 and S100A8 through activation of the NF-κB pathway [36].
Another target of Stat3 in MDSCs is Nox2 itself. Up-regulation of ROS in MDSCs is dependent on increased expression of Nox2. Stat3 activation is directly responsible for upregulating transcription of the Nox2 components p47phox and gp91phox, increasing ROS production by MDSCs in tumor-bearing mice [37]. PKCβII, is required for DC differentiation and is downregulated by activated Stat3. Downregulation of PKCβII could prevent the differentiation of HPCs into mature cells and then support MDSC differentiation [38].
Recent work provide new insight into the signaling pathways downstream of Stat3 that are responsible for MDSC differentiation. The transcription factor CCAAT-enhancer-binding protein beta (C/EBPβ) was reported to play a crucial role in controlling differentiation of myeloid precursors to functional MDSCs [39]. C/EBPβ expression was previously associated with accumulation of CD11b+ Gr-1+ cells, in response to G-CSF or during infection [40]. The new work shows that the accumulation of MDSCs in the spleen of tumor-bearing mice is abrogated in C/EBPβ deficient mice. Moreover, when C/EBPβ is deleted in bone-marrow cells these cells lose the ability to differentiate in vitro into functional MDSCs. Interestingly, a direct link between Stat3 and C/EBPβ has been reported [41]. Stat3 controlled the G-CSF-responsive induction of C/EPBβ expression in myeloid progenitor cells. For instance, G-CSF driven Stat3 activation resulted in the induction of c-Myc expression by increasing the binding of C/EBPβ to Myc promoter [41]. It is conceivable that Stat3, at least partially, can induce MDSC expansion via up-regulation of C/EBPβ.
Stat3 could also play an indirect role in MDSC differentiation. Stat3 controls the expression of molecules such as acute-phase proteins, which might be responsible for MDSC mobilization, accumulation and survival [42]. In a model of polymicrobial sepsis, IL-6-activated Stat3 signaling in hepatocytes, through gp130 ligation, results in the expression of serum amyloid A and chemokine CXCL1, which cooperate in promoting the accumulation of MDSCs in the spleen [42].
It was recently demonstrated that heat-shock protein 72 (Hsp72), which is associated with tumor-derived exosomes, induces suppressive activity of the MDSCs via activation of Stat3. Hsp72 triggered Stat3 activation in MDSCs in a TLR2 and MyD88-dependent manner through autocrine production of IL-6 [43]. Similarly, it was shown that exosomes from tumor cells induce IL-6 release from MDSCs in a TLR2 and Stat3-dependent manner [44]. However, the effect of exosomes on MDSCs needs further clarification. The source of exosomes and the role of tumor stroma in the signaling pathways triggered by exosomes can impact the effect of exosomes on MDSC[ET1].
Thus, Stat3 utilizes various molecular mechanisms to regulate MDSC expansion and function. It is likely that the nature of these mechanisms depends on the type of tumor and specifics of the tumor microenvironment. Exactly how the tumor microenvironment can regulate Stat3-mediated effects on MDSC remains to be elucidated.
Stat1, Stat5, and Stat6
Stat1 is the main transcription factor activated by IFNγ or IL-1β signaling and is implicated in the regulation of inducible nitric oxide synthase (iNOS) and arginase activity. Stat1-deficient MDSCs are unable to inhibit T cell activation due to an inability to upregulate iNOS and arginase activity. Blocking IFNγ secretion by T cells also abrogates MDSC-mediated suppression mainly via the block of iNOS upregulation [45, 46]. A recent study demonstrated that STAT1 is particularly important for the function of M-MDSCs [47].
Stat5 has an important role in regulating MDSC survival. Sunitinib was found to prevent MDSC accumulation and restore normal T-cell function in the spleens of tumor-bearing mice [48]. Within tumors, however, MDSCs were resistant to sunitinib in both mice and humans. The resistance was conferred by GM-CSF, which inhibited Stat3 and promoted Stat5 activation. Thus the regionalized effect of sunitinib on MDSCs was suggested to result from compartment-specific GM-CSF exposure.
Stat6 activation in MDSCs occurs in response to binding of IL-4 or IL-13 to their receptor, CD124. This receptor is also described as one of the MDSC markers and is responsible for up-regulation of arginase activity [49–51] and increased TGF β production by MDSCs [22, 52]. Recently, the critical role of Stat6 in expansion of MDSCs after traumatic stress was demonstrated [53]. Thus, it appears that Stat1, 5, and 6 play an important role in MDSC activation and that these STATs mediate the immune suppressive function of MDSCs.
NF-κB transcription factor
In recent years, the critical role of NF-κB in MDSC accumulation and function has become apparent. In myeloid cells, the toll-like receptor (TLR) family plays a prominent role in NF-κB activation, primarily via the myeloid differentiation primary response gene 88 (MyD88). This is consistent with MDSC accumulation and activation during microbial and viral infections, as well as in trauma and sepsis. NF-κB, acting downstream of MyD88, is required for accumulation of MDSCs in a model of polymicrobial sepsis[54]. TLR4 was shown to be directly involved in MDSC function [55] and LPS, in combination with IFNγ, could promote MDSC expansion, probably by inhibiting differentiation of DC [56]. It was demonstrated that stimulation of myeloid cells with GM-CSF, IL-3 or via MyD88-dependent TLR ligands induced the expression of a triggering receptor expressed on myeloid cells 1 (TREM-1) [57]. During LPS-induced endotoxaemia, and in tumor-bearing mice, TREM-1 was also up-regulated on cells that resembled M-MDSCs. In another study, it was observed that CD11b+Ly-6CintLy-6G− M-MDSC accumulate in the ears of mice three days after inoculation of Mycobacterium bovis bacillus Calmette-Guérin (BCG) intradermal vaccine [58]. These cells differed from inflammatory monocytes and required MyD88-dependent BCG-specific signals to invade skin; whereas, neutrophil influx was MyD88 independent. Upon BCG phagocytosis, M-MDSC produced NO. These cells were unable to kill BCG or the nonpathogenic Mycobacterium smegmatis, but markedly impaired T cell priming in the draining lymph node. Mice pretreated with tumor exosomes had a significant acceleration of tumor metastasis in the lung associated with recruitment of MDSC [59]. These effects were blunted when Myd88 knockout (KO) mice were pretreated with tumor exosomes. Myd88 KO MDSC had substantially reduced ability to suppress T-cell activity and release cytokines than their wild-type counterparts, both in vitro and in vivo. In mice, exposure to a high LPS dose blunted house dust mite-induced airway eosinophilia and T-helper 2 (Th2) cytokine production. LPS promoted the development of lung-resident MDSC in a TLR4 and MyD88-dependent manner that suppressed lung DC-mediated reactivation of primed Th2 cells. LPS effects switched from suppressive to stimulatory in Myd88−/− mice. Suppression of Th2 effector function was reversed by IL-10 antibody or inhibition of arginase 1. Bone marrow progenitor cells could be induced by LPS to develop into MDSC cells, both in vivo and in vitro [60].
Stomach-specific expression of human IL-1β in transgenic mice leads to spontaneous gastric inflammation and cancer, which correlates with early recruitment of MDSCs to the stomach. IL-1β activates MDSCs in vitro and in vivo through the NF-kB pathway. The antagonism of IL-1 receptor signaling inhibited the development of gastric preneoplasia and suppressed MDSC mobilization [61]. The data accumulated so far suggest that although NF-κB can be involved in expansion of MDSC, its main role is to signal activation of these cells and acquisition of immune suppressive function. M-MDSC is the main population of MDSC where NF-κB pathway was shown to be active.
Cyclooxygenase 2 (Cox 2) and prostaglandin E2 (PGE2)
Prostaglandins, in particular PGE2, have been widely implicated in MDSC-mediated T-cell inhibition. In an early study, signaling through the PGE2 receptor E-prostanoid (EP) 4 in MDSC was found to induce arginase 1 Expression and activity [24]. Furthermore, blockade of arginase 1 expression with Cox-2 inhibitors elicited a lymphocyte-mediated antitumor response in 3LL lewis lung carcinoma tumor-bearing mice. These observations that suggest a regulatory role for Cox2 in MDSC function have been confirmed and expanded in more recent work. For example, accumulation of MDSC and Foxp3+ regulatory T cells was found in tumors formed by inoculation with Fas-overexpressing 3LL Lewis Lung carcinoma cells [62]. Fas ligation induced 3LL tumor cells to produce PGE2 by activating p38. PGE2, in turn, contributed to the Fas ligation-induced MDSC chemoattraction. Furthermore, in vivo administration of the Cox2 inhibitor significantly reduced MDSC accumulation in the Fas-overexpressing tumors in mice. The MDSC accumulation might result from the activity of exosomes isolated from these Fas-overexpressing tumors, which were found to switch the differentiation of myeloid cells to MDSC [63]. The exosomes induced the accumulation of MDSCs expressing Cox2, IL-6, VEGF, and arginase-1. MDSC-mediated promotion of tumor progression was dependent on PGE2 and TGF-β. Antibodies against PGE2 and TGF-beta blocked the activity of these exosomes on MDSC induction and, therefore, attenuated MDSC-mediated tumor-promoting ability. EP receptor agonists, including PGE2, induced the generation of MDSC from bone marrow stem cells; whereas, receptor antagonists blocked differentiation[64]. BALB/c Ptger2 (EP2) knockout mice, inoculated with the spontaneous metastatic 4T1 mammary carcinoma, had delayed tumor growth and reduced numbers of MDSC relative to wild-type mice, suggesting that PGE2 partially mediates MDSC induction through the EP2 receptor. Treatment of 4T1-tumor-bearing wild-type mice with the Cox2 inhibitor, SC58236, delayed primary tumor growth and reduced MDSC accumulation. In another study, the frequency of splenic MDSCs directly correlated with cox2, nos2, and arg1 message levels. Cox2 mRNA levels also directly correlated with the arg1 and nos2 transcript levels from tumor-infiltrating leukocytes, supporting the role of PGE2 as a regulator of arg1 and nos2 expression [65]. The tumor-mediated inhibition of antigen-presenting cell differentiation was associated with the up-regulated expression of Cox2 and the simultaneous repression of PGE2-catabolizing enzyme, 15-PGDH. Addition of COX2 inhibitor to the bone marrow cell cultures could prevent the tumor-induced skewing of myeloid cell differentiation, partially restoring cell phenotype and down-regulating the arginase expression in the myeloid cells [66]. The data accumulated so far strongly suggest that Cox2 and PGE2 represent a critical signaling pathway regulating both MDSC function and differentiation. The specific role of this pathway in different aspects of MDSC biology remains to be elucidated.
In vitro differentiation of MDSC
In patients with asthma, autoimmune diseases, conditions of graft versus host disease (GVHD), or organ transplantations there is a need for therapeutic intervention to inhibit antigen-specific immune responses. The powerful immune suppressive features of MDSC makes these cells attractive candidates for use in cell therapy of these diseases. The main challenge is to generate relatively stable immune suppressive MDSC ex vivo from cell populations obtained from tumor-free individuals. Different tumor-secreted factors, as well as bacterial products, have been shown to drive MDSC expansion and the ability of these factors to drive differentiation ex vivo has been tested in a number of recent studies. It was reported that early treatment with LPS and IFNγ can shift the differentiation of bone marrow progenitor cells (under GM-CSF condition) from DC to MDSC [56]. Another study reported the possibility of differentiating MDSC from hematopoietic stem cells by culturing them with a cocktail of cytokines. IL-3, IL-6, c-kit ligand, TPO, FLT3L, VEGF and M-CSF induced the differentiation of embryonic stem cells into functional MDSC, capable of inhibiting T cell activation in vitro and in vivo in a model of GVHD [67]. Surprisingly, the authors also reported that GM-CSF inhibited the expansion of MDSC in that specific model. In contrast, in other studies, the combination of GM-CSF with IL-6 or G-CSF has been sufficient to drive MDSC expansion [39, 68]. Incubation of peripheral blood mononuclear cells from a healthy donor, in the presence of GM-CSF alone or in combination with IL-1β, IL-6, TNFα, or VEGF, resulted in the generation of CD33+ MDSC-like cells [68]. These combinations of cytokines induced the up-regulation of iNOS, TGFβ, VEGF and Nox2 in these cells and the ability of these cells to suppress T-cell function. The combination of GM-CSF and IL-6, as well as the combination of GM-CSF and G-CSF, induced the differentiation of bone-marrow cells into MDSC in humans or mice [39]. GM-CSF and G-CSF, in combination with IL-13, induce preferential differentiation of M-MDSC which exerted their suppressive activity mainly through an arginase mechanism[69]. Taken together, these promising results demonstrated that generation of MDSC ex vivo is feasible. Their clinical utility, however, need to be established in future studies.
Concluding remarks: a two signal model for MDSC expansion and activation?
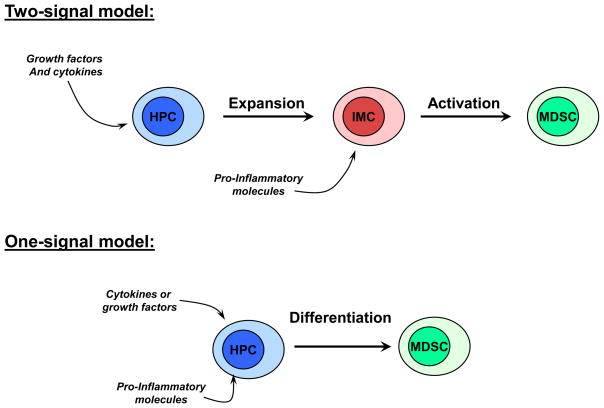
Despite progress made in recent years in understanding the molecular mechanisms governing MDSC accumulation, many specific details remain unclear. One of the major challenges to uncover the biology of MDSC is to understand whether the accumulation of these cells requires one or two signals. The “one-signal” model implies that one factor, for instance GM-CSF, can initiate the differentiation program of MDSC and activate these cells to convert them into immune suppressive cells (Fig. 1). We have discussed evidence suggesting that this can be the case. However, we are more in favor of the “two-signal” model. This model suggests that expansion of MDSC can be separated into two processes governed by different signal transduction pathways (Fig. 1). One pathway is predominantly responsible for MDSC expansion (Fig. 2) and the second one for driving MDSC activation (Fig. 3). The first process is induced by various cytokines and growth factors produced by tumors or bone marrow stroma in response to chronic stimulation. It involves such factors as GM-CSF, M-CSF, G-CSF, IL-6, VEGF, etc and signals primarily via STAT3 and STAT5. This signaling prevents differentiation of MDSC and promotes proliferation of immature myeloid cells. However, this signaling alone is not sufficient to generate accumulation of MDSC (Fig. 2). MDSC require a second activating signal, which manifests in up-regulation of arginase, NO, production of immune suppressive cytokines, etc. This type of signaling is provided by pro-inflammatory molecules such as IFNγ, IL-1β, IL-13, TLR ligands, etc. It utilizes the STAT1 and NF-κB transcription factors, and Cox2 upregulation (Fig. 3). This model may explain why, at steady state, activation of STAT3 and STAT5 in response to various growth factors that are required for normal hematopoiesis, does not result in accumulation of MDSC in the absence of strong signal from pro-inflammatory factors. It also can explain why acute inflammation, associated with the release of pro-inflammatory factors in the absence of sustained up-regulation of growth factors, also does not result in accumulation of MDSC. It is likely, as is always the case in biological systems, that there is an overlap in these signaling pathways. In steady state hematopoiesis different factors might contribute to drive differentiation towards either one or the other subset of MDSCs. Moreover, transcription factors can modulate the activity of each other, as was shown, for instance, for Stat3 and NF-κB [70] [71]. However, this model suggests that accumulation of MDSC is possible only if strong two signals are provided. The validity of this concept and its practical utility should be tested in future studies.
Figure 1. Potential models of MDSC accumulation.
Schematic description of the models of MDSC accumulation that require either one or two signals. HPC – hematopoietic progenitor cells. IMC – immature myeloid cells that have phenotype similar to MDSC but lacking immune suppressive activity
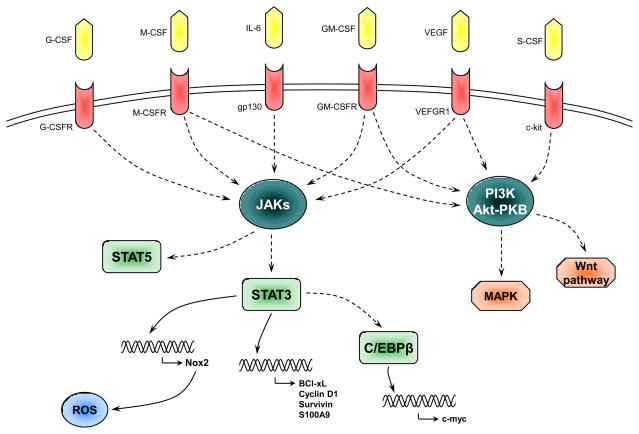
Figure 2. Schematics of possible signaling pathways involved in MDSC expansion.
Various cytokines produced by tumors or bone marrow stroma in response to chronic infections or inflammation activate several signal transduction pathways that result in activation of Stat3, Stat5. Stat3 regulates transcription of subunits of Nox2 that results in increased production of ROS, as well as up-regulation of a number of anti-apoptotic proteins and possibly CEBPβ that, in turn, up-regulate c-myc. All together these proteins contribute to proliferation and survival of immature myeloid cells and prevent their differentiation to mature cells. This manifests in expansion of cells with the phenotype of MDSC.
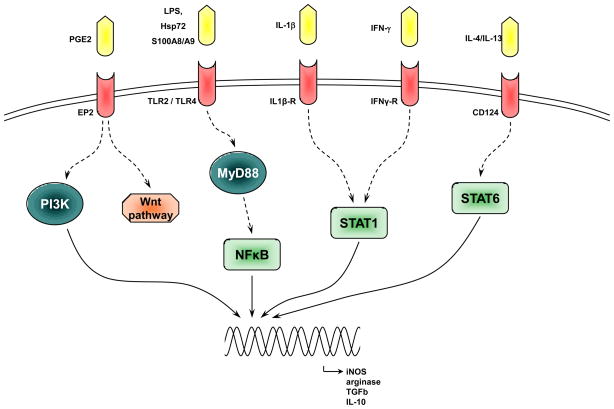
Figure 3. Schematics of possible signaling pathways involved in MDSC activation.
Bacterial and viral products, as well as cytokines released by activated T cells or myeloid cells induce activation of Stat1, Stat6 and TLR signaling. Stat1 and Stat6 act directly and TLR signaling via MyD88 and NF-κB to up-regulate proteins directly involved in the immune suppressive activity of MDSC.
Box 1. Arginase activity, NO and ROS production suppress T cell function.
These factors represent the main effectors of the suppressive function of MDSC. The L-arginine metabolism has been associated with MDSC suppression [14, 15]. Both Arginase 1 (Arg1) and iNOS (inducible NO synthase) use L-arginine as a substrate to produce urea and L-ornithine or NO respectively. Upregulation of Arg1 activity leads to the depletion of L-arginine from the microenvironment which induces the loss of the CD3ζ chain [72] and prevents the upregulation of cell cycle regulator in T cells [73] resulting in an inhibition of T cell proliferation. Upregulation of iNOS activity leads to increased NO production that has also been shown to suppress T cell function. NO is able to inhibit the downstream pathway of the IL2 receptor by blocking the phosphorylation of signaling proteins (like Jak3 or Stat5) [74] or to directly induce T cell apoptosis [75]. These mechanisms are mostly associated with M-MDSC. In contrast, reactive oxygen species (ROS), produced by the NADPH oxydase complex, is the main mediator responsible for G-MDSC suppression [3, 17]. The major effect is mediated by peroxynitrite which is the product of a chemical reaction between NO and superoxyde anion [76]. Peroxynitrite is capable of inhibiting CD8+ T cells by inducing the nitration of their T-cell receptors (TCR), resulting in an altered TCR/MHC-peptide recognition.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hestdal K, et al. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J Immunol. 1991;147:22–28. [PubMed] [Google Scholar]
- 3.Youn JI, et al. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almand B, et al. Increased production of immature myeloid cells in cancer patients. A mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 5.Filipazzi P, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 6.Poschke I, et al. Immature immunosuppressive CD14+HLA-DR-/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer Res. 2010;70:4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 7.Mandruzzato S, et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J Immunol. 2009;182:6562–6568. doi: 10.4049/jimmunol.0803831. [DOI] [PubMed] [Google Scholar]
- 8.Liu CY, et al. Population alterations of L: -arginase- and inducible nitric oxide synthase-expressed CD11b(+)/CD14 (−)/CD15 (+)/CD33 (+) myeloid-derived suppressor cells and CD8 (+) T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2009 doi: 10.1007/s00432-009-0634-0. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava MK, et al. Lung cancer patients' CD4(+) T cells are activated in vitro by MHC II cell-based vaccines despite the presence of myeloid-derived suppressor cells. Cancer Immunol Immunother. 2008;57:1493–1504. doi: 10.1007/s00262-008-0490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez PC, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peranzoni E, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol. 2010;22:238–244. doi: 10.1016/j.coi.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Ribechini E, et al. Subsets, expansion and activation of myeloid-derived suppressor cells. Med Microbiol Immunol. 2010;199:273–281. doi: 10.1007/s00430-010-0151-4. [DOI] [PubMed] [Google Scholar]
- 13.Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells in human cancer. Cancer J. 2010;16:348–353. doi: 10.1097/PPO.0b013e3181eb3358. [DOI] [PubMed] [Google Scholar]
- 14.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez PC, Ochoa AC. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol Rev. 2008;222:180–191. doi: 10.1111/j.1600-065X.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117:1155–1166. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Movahedi K, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 18.Li H, et al. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol. 2009;182:240–249. doi: 10.4049/jimmunol.182.1.240. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang B, et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006;66:1123–1131. doi: 10.1158/0008-5472.CAN-05-1299. [DOI] [PubMed] [Google Scholar]
- 21.Pan PY, et al. Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res. 2010;70:99–108. doi: 10.1158/0008-5472.CAN-09-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serafini P, et al. Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res. 2008;68:5439–5449. doi: 10.1158/0008-5472.CAN-07-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava MK, et al. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010;70:68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez PC, et al. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J Exp Med. 2005;202:931–939. doi: 10.1084/jem.20050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talmadge JE. Pathways mediating the expansion and immunosuppressive activity of myeloid-derived suppressor cells and their relevance to cancer therapy. Clin Cancer Res. 2007;13:5243–5248. doi: 10.1158/1078-0432.CCR-07-0182. [DOI] [PubMed] [Google Scholar]
- 26.Marigo I, et al. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- 27.Ostrand-Rosenberg S. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother. 2010;59:1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, et al. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nefedova Y, et al. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J Immunol. 2004;172:464–474. doi: 10.4049/jimmunol.172.1.464. [DOI] [PubMed] [Google Scholar]
- 30.Nefedova Y, et al. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 2005;65:9525–9535. doi: 10.1158/0008-5472.CAN-05-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kujawski M, et al. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin H, et al. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res. 2009;69:2506–2513. doi: 10.1158/0008-5472.CAN-08-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu P, et al. Myeloid-specific expression of Api6/AIM/Sp alpha induces systemic inflammation and adenocarcinoma in the lung. J Immunol. 2009;182:1648–1659. doi: 10.4049/jimmunol.182.3.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foell D, et al. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 35.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:3325–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha P, et al. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corzo CA, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farren MR, et al. Tumor-mediated inhibition of dendritic cell differentiation is mediated by down regulation of protein kinase C beta II expression. Immunol Res. 2010;46:165–176. doi: 10.1007/s12026-009-8118-5. [DOI] [PubMed] [Google Scholar]
- 39.Marigo I, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Hirai H, et al. C/EBPbeta is required for 'emergency' granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, et al. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010 doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander LE, et al. Hepatic acute-phase proteins control innate immune responses during infection by promoting myeloid-derived suppressor cell function. J Exp Med. 2010;207:1453–1464. doi: 10.1084/jem.20091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalmin F, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang X, et al. TLR2-Mediated Expansion of MDSCs Is Dependent on the Source of Tumor Exosomes. Am J Pathol. 2010 doi: 10.2353/ajpath.2010.100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallina G, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol. 2005;174:4880–4891. doi: 10.4049/jimmunol.174.8.4880. [DOI] [PubMed] [Google Scholar]
- 47.Movahedi K, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T-cell suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 48.Ko JS, et al. Direct and differential suppression of myeloid-derived suppressor cell subsets by sunitinib is compartmentally constrained. Cancer Res. 2010;70:3526–3536. doi: 10.1158/0008-5472.CAN-09-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha P, et al. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol. 2005;174:636–645. doi: 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- 50.Sinha P, et al. Interleukin-13-regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer Res. 2005;65:11743–11751. doi: 10.1158/0008-5472.CAN-05-0045. [DOI] [PubMed] [Google Scholar]
- 51.Bronte V, et al. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170:270–278. doi: 10.4049/jimmunol.170.1.270. [DOI] [PubMed] [Google Scholar]
- 52.Terabe M, et al. Transforming growth factor-beta production and myeloid cells are an effector mechanism through which CD1d-restricted T cells block cytotoxic T lymphocyte-mediated tumor immunosurveillance: abrogation prevents tumor recurrence. J Exp Med. 2003;198:1741–1752. doi: 10.1084/jem.20022227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munera V, et al. Stat 6-dependent induction of myeloid derived suppressor cells after physical injury regulates nitric oxide response to endotoxin. Ann Surg. 2010;251:120–126. doi: 10.1097/SLA.0b013e3181bfda1c. [DOI] [PubMed] [Google Scholar]
- 54.Delano MJ, et al. MyD88-dependent expansion of an immature GR-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J Exp Med. 2007;204:1463–1474. doi: 10.1084/jem.20062602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunt SK, et al. Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through Toll-like receptor 4. J Leukoc Biol. 2009;85:996–1004. doi: 10.1189/jlb.0708446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greifenberg V, et al. Myeloid-derived suppressor cell activation by combined LPS and IFN-gamma treatment impairs DC development. Eur J Immunol. 2009;39:2865–2876. doi: 10.1002/eji.200939486. [DOI] [PubMed] [Google Scholar]
- 57.Zanzinger K, et al. Regulation of triggering receptor expressed on myeloid cells 1 expression on mouse inflammatory monocytes. Immunology. 2009;128:185–195. doi: 10.1111/j.1365-2567.2009.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martino A, et al. Mycobacterium bovis bacillus Calmette-Guerin vaccination mobilizes innate myeloid-derived suppressor cells restraining in vivo T cell priming via IL-1R-dependent nitric oxide production. J Immunol. 2010;184:2038–2047. doi: 10.4049/jimmunol.0903348. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, et al. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–2499. doi: 10.2353/ajpath.2010.090777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arora M, et al. TLR4/MyD88-induced CD11b(+)Gr-1(int)F4/80(+) non-migratory myeloid cells suppress Th2 effector function in the lung. Mucosal Immunol. 2010 doi: 10.1038/mi.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu S, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Fas signal promotes lung cancer growth by recruiting myeloid-derived suppressor cells via cancer cell-derived PGE2. J Immunol. 2009;182:3801–3808. doi: 10.4049/jimmunol.0801548. [DOI] [PubMed] [Google Scholar]
- 63.Xiang X, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha P, et al. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007;67:4507–4513. doi: 10.1158/0008-5472.CAN-06-4174. [DOI] [PubMed] [Google Scholar]
- 65.Donkor MK, et al. Mammary tumor heterogeneity in the expansion of myeloid-derived suppressor cells. Int Immunopharmacol. 2009;9:937–948. doi: 10.1016/j.intimp.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 66.Eruslanov E, et al. Pivotal Advance: Tumor-mediated induction of myeloid-derived suppressor cells and M2-polarized macrophages by altering intracellular PGE2 catabolism in myeloid cells. J Leukoc Biol. 2010 doi: 10.1189/jlb.1209821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Z, et al. Development and function of myeloid-derived suppressor cells generated from mouse embryonic and hematopoietic stem cells. Stem Cells. 2010;28:620–632. doi: 10.1002/stem.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lechner MG, et al. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Highfill SL, et al. Bone marrow myeloid-derived suppressor cells (MDSC) inhibit graft-versus-host (GVHD) disease via an arginase-1 dependent mechanism that is upregulated by IL-13. Blood. 2010 doi: 10.1182/blood-2010-06-287839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nefedova Y, et al. Activation of dendritic cells via inhibition of Jak2/STAT3 signaling. J Immunol. 2005;175:4338–4346. doi: 10.4049/jimmunol.175.7.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez PC, et al. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–1573. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bingisser RM, et al. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J Immunol. 1998;160:5729–5734. [PubMed] [Google Scholar]
- 75.Mannick JB, et al. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 76.Nagaraj S, et al. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]