Abstract
Background
Many of the complications encountered during diabetes can be linked to the non-enzymatic glycation of proteins, including human serum albumin (HSA). However, there is little information regarding how the glycation pattern of HSA changes as the total extent of glycation is varied. The goal of this study was to identify and conduct a semi-quantitative comparison of the glycation products on HSA that are produced in the presence of various levels of glycation.
Methods
Three glycated HSA samples were prepared in vitro by incubating physiological concentrations of HSA with 15 mmol/l glucose for 2 or 5 weeks, or with 30 mmol/l glucose for 4 weeks. These samples were then digested and examined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) to identify the glycation products that were formed.
Results
It was found that the glycation pattern of HSA changed with its overall extent of total glycation. Many modifications including previously-reported primary glycation sites (e.g., K199, K281, and the N-terminus) were consistently found in the tested samples. Lysines 199 and 281, as well as arginine 428, contained the most consistently identified and abundant glycation products. Lysines 93, 276, 286, 414, 439, and 524/525, as well as the N-terminus and arginines 98, 197, and 521, were also found to be modified at various degrees of HSA glycation.
Conclusions
The glycation pattern of HSA was found to vary with different levels of total glycation and included modifications at the 2 major drug binding sites on this protein. This result suggests that different modified forms of HSA, both in terms of the total extent of glycation and glycation pattern, may be found at various stages of diabetes. The clinical implication of these results is that the binding of HSA to some drug may be altered at various stages of diabetes as the extent of glycation and types of modifications in this protein are varied.
Keywords: Non-enzymatic glycation, human serum albumin, diabetes, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, glycation-related modifications
1. Introduction
The glycation of various proteins in the body is accelerated during diabetes [1]. This process, known as the Maillard reaction, involves the addition of reducing sugars and/or their degradation products to free primary and secondary amine groups on proteins [2,3]. The first step of this reaction involves the nucleophilic addition of a reducing sugar to a primary amine group (e.g., as found on a lysine or at the N-terminus of a protein). In this stage a reversible Schiff base is formed which can undergo a slow irreversible rearrangement to form more stable Amadori products that accumulate over time. The total amount of accumulation of these products is known to be dependent on the type of sugar that is causing the glycation [4], the incubation time and sugar concentration [5], and the type protein that is being modified [6–8].
The Maillard reaction is fairly well-characterized for proteins with long lifetimes in the body (e.g., collagen [9–12] and lens crystalline [13–15]) and for some proteins with shorter half-lives (e.g., hemoglobin [16–18]). However, until recently there has been little information on this process for human serum albumin (HSA) [8], which is the major carrier agent for many drugs and small endogenous solutes in blood [2,19]. HSA has 2 major binding regions for drugs. These sites are known as Sudlow sites I and II and are located in subdomains IIA (within residues 150–292) and IIIA (within residues 384–489) of HSA, respectively [2,20]. Previous studies have found that glycation can alter the interactions of HSA with certain solutes [21–23] and may, under certain conditions, affect the overall structure of HSA [24,25]. It is expected from these observations that characterization of the modifications that occur on HSA would be valuable in helping determine why such changes in binding can occur during glycation.
Previous studies with glycated HSA prepared under specific conditions have indicated that the most significant sites for early glycation are the N-terminus and lysines 199, 281, 439, and 525 [2,26,27]. Similar work looking at modification of HSA with methylglyoxal has identified certain regions that are prone to advanced glycation end product (AGE) modification [28], including arginine 410 as a major region of modification and arginines 114, 186, 218, and 428 as regions with smaller amounts of modification. These findings are consistent with recent qualitative [29] and quantitative studies [30] that have been conducted by our group using minimally-glycated HSA. However, no past studies have looked at how these glycation patterns are altered when comparing samples of HSA with different levels of glycation.
In this study, HSA with various levels of glycation will be prepared in vitro under solution conditions and reaction times that are similar to those found for HSA in serum during diabetes [31,32]. The modifications that occur on the resulting samples of glycated HSA will be examined by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) [33]. A comparison will then be made between the results obtained for the HSA made with different extents of glycation and with previous results reported for minimally glycated HSA [29,30]. The modifications that are found will also be compared to the known locations of the major drug binding sites on HSA. This data should provide a more complete picture of how the structure of HSA and its function as a carrier agent for drugs are affected by various stages of glycation during diabetes.
2. Materials and methods
2.1. Materials
The HSA (99% pure, essentially fatty acid and globulin free), guanidine-HCl (99%), ammonium bicarbonate (99%), tris-HCl (99%), α-cyano-4-hydroxycinnamic acid (CHCA, 99%), 2,3-dihyroxybenzoic acid (DHB, 98%), dithiothreitol (99%), iodoacetamide (99%), D-glucose (99%), sodium azide (99%), trifluoroacetic acid (TFA, 98%), formic acid (96%), molecular biology grade water (DNase, RNase, and protease free), methanol (HPLC grade, 99.9%), acetonitrile (HPLC grade, 99.9%), and sequence-grade trypsin, Glu-C or Lys-C were all purchased from Sigma-Aldrich (St. Louis, MO). The enzymatic assay used to determine the overall degree of glycation for HSA was purchased from Diazyme Laboratories (Poway, CA).
2.2. Apparatus
Siliconized low retention microcentrifuge tubes (0.6 ml and 1.5 ml volume), Slide-A-Lyzer dialysis cassettes (7 kDa cut off, 0.1–0.5 ml and 0.5–3 ml sample volumes), μ-C18 Zip-tip pipette tips (0.2 μL bed volume), 0.20 μm nylon filters, and 10 DG size exclusion columns (10 ml volume) were obtained from Fisher Scientific (Pittsburgh, PA). The Speedvac used to concentrate protein samples was from Savant Instruments Inc (Farmingdale, NY). Mass spectra were acquired on a Voyager 6148 MALDI-TOF MS system (Applied/Perspective Biosystems, CA). A positive ion delayed extraction mode was used on this system with an accelerating voltage of 25 kV, a grid wire voltage which was 0.008% of the accelerating voltage, a grid voltage which was 77% of the accelerating voltage, and a delay time of 100 ns. Matlab 2009a, including the bioinformatics toolbox, was purchased from Mathworks (Natick, MA). Mascot wizard was obtained from the Matrix Science website [34].
2.3. Preparation of glycated HSA
Glycation was initiated by incubating a physiological concentration of HSA (42 g/l) with a 15 mmol/l glucose solution. The glucose solution was prepared in pH 7.4, 200 mmol/l phosphate buffer which also contained 1 mmol/l sodium azide (Note: sodium azide was added to prohibit bacterial growth; this agent is toxic, and care must be used to avoid contact of this compound with skin [35–37]). HSA samples containing 3 different levels of total glycation were examined. To make these samples, the starting HSA/glucose solution was divided into several portions and incubated at 37 °C for 2–5 weeks [38], which gave the “HSA-2” and “HSA-5” samples used in this report. A control sample was prepared under identical conditions by using a solution of 42 g/l HSA dissolved in the same pH 7.4 buffer with sodium azide (but without the addition of glucose) and that was incubated for five weeks at 37 °C. A third glycated HSA sample was prepared under these conditions but with a total added glucose concentration of 30.7 mmol/l (i.e., representing hyperglycemic conditions [37]) and an incubation time of 4 weeks; these conditions provided the “HSA-4” sample used in this report. A second control sample of HSA was also prepared under these latter conditions in the absence of any glucose.
Each sample of glycated HSA was isolated from its reaction mixture by using size-exclusion chromatography and equilibrium dialysis. First, a 10 DG size exclusion column was washed with 20 ml of water, followed by the application of 3 ml of the desired glycated HSA solution. A 5 ml portion of water was passed through the column to elute the glycated HSA and this fraction was collected. A 2.5 ml aliquot of this fraction was transferred to a 0.5–3 ml dialysis cassette and dialyzed extensively against two 1000 ml portions of water for 2 h at room temperature. An additional dialysis step against 1000 ml water was performed overnight at 4 °C. The glycated HSA solution was then removed from the dialysis cassette, lyophilized, and stored at 4 ºC until later use. A modified commercial fructosamine assay kit was used to determine the level of HSA glycation [39]. In this assay, glycated proteins were cleaved into protein fragments using proteinase K, and the fructosamine residues were specifically targeted by fructosaminase to produce amino acids and H2O2. The resulting H2O2 was then detected by using a colorimetric peroxidase-linked reaction [40,41]. None of the AGEs examined in this report have been reported by the manufacturer of the kit to create interferences in this method.
2.4. Pretreatment and digestion of HSA
The glycated HSA samples and control samples were pretreated and digested using the same procedure, equipment and reagents to minimize any differences in digestion patterns due to changes in experimental conditions. Prior to pretreatment of the HSA, molecular biology-grade water was used to prepare a pH 8.5, 100 mmol/l ammonium bicarbonate buffer. Solutions containing 6 mol/l guanidine-HCl or 1 mol/l dithiothreitol reagent were prepared using this ammonium bicarbonate buffer. A solution containing 1 mol/l iodoacetamide and 1 mol/l sodium hydroxide was prepared in water. A 1 ml solution of 3 mg/ml HSA was prepared in the guanidine-HCl solution and was gently mixed. A 15 μl aliquot of DTT reagent was then added to the HSA solution. This mixture was incubated at 37 °C for 1 h, after which 36 μl of the iodoacetamide/sodium hydroxide reagent was added to the HSA solution. The resulting solution was incubated in the dark at room temperature for 30 min. Following this incubation, an additional 150 μl of the DTT reagent was added to the mixture. The HSA solution was then divided into 0.5 ml portions that were placed into 0.1–0.5 ml dialysis cassettes. Dialysis was performed on the contents of each cassette against three 1000 ml portions of water, where each dialysis cycle was performed for 4 h at room temperature. The dialyzed HSA samples were divided into 50 μl aliquots, each containing about 100 μg HSA, and placed into low-retention microcentrifuge tubes. These aliquots were reduced to dryness and stored at −80 °C.
Stock enzyme solutions containing 1 mg/ml of trypsin, Lys-C, or Glu-C were prepared in water. A pH 7.8, 100 mmol/l ammonium bicarbonate buffer and a pH 9.0, 75 mmol/l Tris-HCl buffer were also prepared. For the trypsin digest, 200 μL of ammonium bicarbonate buffer was added to vials containing pretreated HSA/glycated HSA. Each vial of the reconstituted HSA was mixed for 3–5 min, with a 3.3 μl portion of the trypsin stock solution then being added to each reconstituted HSA sample, giving a substrate-to-enzyme ratio of 30:1. These solutions were incubated at 37 °C for 18 h; a 10 μl aliquot of concentrated formic acid was added to each HSA solution and the resulting HSA digests were stored at −80 °C. For the Glu-C digest, 200 μL of a pH 7.8, 100 mmol/l ammonium bicarbonate buffer was added to vials containing pretreated HSA/glycated HSA, and this mixture was mixed for 3–5 min. A 10 μl portion of the Glu-C stock solution was added to each HSA sample and the solutions were incubated at 37 °C for 8 h. Another 5 μl portion of the Glu-C stock solution was added to each HSA sample, giving a final substrate-to-enzyme ratio of 7:1, and the resulting solutions were incubated for an additional 18 h. A 10 μl aliquot of concentrated formic acid was added to each digests and the resulting solutions were stored at −80 °C. For the Lys-C digest, 200 μL of pH 9.0, 75 mmol/l tris-HCl buffer was added to vials containing pretreated HSA/glycated HSA, and these vials were mixed for 3–5 min. A 5 μl portion of the Lys-C stock solution was added to each vial of HSA and the resulting mixtures were incubated at 37 °C for 18 h. A 10 μl aliquot of concentrated formic acid was added to the digests, and the final solutions were stored at −80 °C.
2.5 Collection and analysis of mass spectra
A MALDI matrix that contained both CHCA and DHB was prepared as described previously [30]. A stock calibration mixture containing 32.5 pmol/l glufibrinopeptide (1570.6774 Da), 25 pmol/l des-Arg-bradykinin (904.4681 Da), and 32.5 pmol/l angiotensin I (1296.6853 Da) was prepared in 100% acetonitrile. The MALDI matrix was spiked by adding 4 μL of the stock calibration mixture to 96 μl of the matrix. This spiked matrix solution was placed on every other row of a 96-well MALDI-TOF MS plate in positions that were adjacent to those used for the HSA samples. μ-C18 Zip-tips were used to fractionate the peptide digests [33]. A 1 μl aliquot of the fractionated peptides was mixed with 1 μL of the spiked matrix solution and 0.80 μl of the resulting mixture was spotted onto a 96-well MALDI-TOF MS plate. All of the mass spectra acquired in this study were first externally calibrated using the calibration spots on the adjacent rows to the samples. These spectra were then internally calibrated by using the spiked calibrant masses found in the mass spectra for the HSA samples.
An evaluation of unspiked HSA samples indicated that there was no overlap in the m/z values for the calibrants and the detected HSA-derived peptides. A maximum allowable difference (or error) of 25 ppm in the measured masses, representing 2 times the precision of the experimentally-measured mass deviation for unmodified peptides, was found to be sufficient for matching the actual and predicted peptide masses when this technique was used [42]. Programs written in Matlab were used to assist in the data analysis (Note: Copies of these programs are available upon request). Raw mass values from each mass spectrum were extracted using Mascot Wizard and these values were compared to an expanded modification list (see Table 1) that was generated by using a program written in Matlab. Only unique peptides that were found in HSA-2, HSA-4, or HSA-5 samples were matched to this search list. To determine which peptides were unique, a separate Matlab program was used to compare the extracted mass list for the glycated HSA samples versus the control HSA samples and to match those masses within 50 ppm. The peaks in the glycated sample that did not match were selected for further analysis. During this process, any modified peptides with masses that matched predicted values within 25 ppm were considered to potentially contain glycation-related adducts.
Table 1.
List of advanced glycation end products and early stage glycation products considered in this study.
| Glycation adducts involving lysine | Abbreviation | Δm (Da) |
|---|---|---|
| Fructosyl-lysine | FL | 162.05 |
| Fructosyl-lysine-1H2O | FL-1H2O | 144.04 |
| Fructosyl-lysine-2H2O | FL-2H2O | 126.03 |
| Nε-Carboxyethyl-lysine | CEL | 72.02 |
| Nε-Carboxymethyl-lysine | CML | 58.01 |
| Pyrraline | Pyr | 108.02 |
| Glycation adducts involving arginine | Abbreviation | Δm (Da) |
|---|---|---|
| Nε-[5-(2,3,4-Trihydroxybutyl)-5-hydro-4-imidazolon-2-yl]ornithine | 3-DG-H1 | 144.04 |
| Tetrahydropyrimidine | THP | 144.04 |
| Imadazolone B | IB | 142.03 |
| Argpyrimidine | ArgP | 80.03 |
| Nε-(5-Hydro-5-methyl-4-imidazolon-2-yl)ornithine | MG-H1 | 54.01 |
| Nε-(5-Hydro-4-imidazolon-2-yl)ornithine | G-H1 | 39.99 |
| Glycation adducts involving both lysine and arginine | Abbreviation | Δm (Da) |
|---|---|---|
| 1-Alkyl-2-formyl-3,4-glycosyl-pyrrole | AFGP | 270.07 |
3. Results and discussion
2.5. Control of extent of glycation and determination of sequence coverage
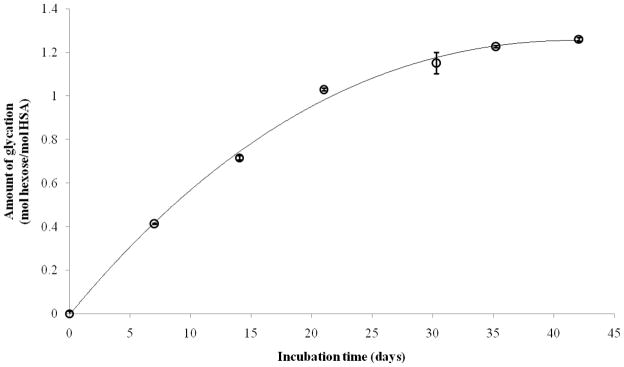
Figure 1 shows how the extent of glycation of HSA, as measured by a fructosamine assay, changed over time during the in vitro preparation of this modified protein. These results were obtained for the reaction of 42 g/l HSA with 15 mmol/l glucose at pH 7.4 and at 37 °C over six weeks. Most of the increase in glycation occurred during the first three weeks, with the rate of glycation slowing down between three and six weeks. Similar trends have been noted in previous work examining the incubation of HSA [43] or bovine serum albumin with glucose [37]. Two of the samples selected for more detailed analysis were prepared under these conditions using two or five weeks of incubation (i.e., the samples referred to as “HSA-2” and “HSA-5” in this report). These conditions were used to obtain glycated HSA that was similar in its glycation levels to that found in serum during pre-diabetes or typical diabetes. A third sample was prepared by incubating 42 g/l HSA with 30 mmol/l glucose for four weeks. This sample was used to represent higher levels of glycation, as might occur in advanced or uncontrolled diabetes. An assay of these samples gave the following measured levels of glycation: HSA-2, 0.72 (± 0.1) mol hexose/mol HSA; HSA-5, 1.23 (± 0.1) mol hexose/mol HSA; and HSA-4, 3.35 (± 0.1) mol hexose/mol HSA. These levels of modification were similar to the range of HSA glycation that is typically found in vivo during diabetes (i.e., 1–3 mol hexose/mol HSA) [44]. A similar assay of the original HSA and control sample that was used to prepare these glycated samples was found to have no measurable level of fructosamine (i.e., less than 0.02 mol hexose/mol HSA).
Figure 1.
Measured level of glycation for HSA, as measured by a fructosamine assay and obtained as a function of time for the reaction of 42 g/l HSA with 15 mmol/l glucose at pH 7.4 and 37 °C. The error bars represent a range of ± 1 S.D.
Details on the sequence coverage that was obtained for the HSA and glycated HSA samples are provided in the Supplemental Information. For the control HSA sample, the tryptic, Lys-C and Glu-C digests covered 78%, 47% and 56% of the primary sequence of HSA, respectively. Similarly, digestion of the HSA-2 sample with the same enzymes gave sequence coverages of 63%, 49% and 44%; the HSA-5 sample gave 54%, 44% and 49% sequence coverages; and the HSA-4 sample had sequence coverages of 69%, 45%, and 55%. When the results of these digests were combined, the total sequence coverages for the control HSA, HSA-2, HSA-4, and HSA-5 samples were 90%, 81%, 84%, and 82%, respectively. A decreased coverage for the glycated HSA samples versus the control HSA was expected because glycation would reduce the amount of unmodified peptides and decrease the signal for some of these peptide below the selected signal-to-noise ration of detection (i.e., S/N = 5, in this case). The coverage for the HSA-2, HSA-4 and HSA-5 samples all increased to approximately 85% when assigned glycated adducts were factored into the sequence coverage calculation.
For the control sample and the glycated samples, all of the arginine residues in HSA were included in this sequence coverage. For the control sample, 83% of the lysine residues were included, with the exception of lysines 4, 233, 240, 317, 534, 536, 538, 541, 545, and 560. Similarly, 81% of the lysines in the HSA-2 sample were included, with the exception of lysines 136, 233, 240, 313, 317, 323, 359, 534, 536, 538, and 560. Lysines 4, 541, and 545, which were not covered in the control HSA sample, were covered in the HSA-2 sample. For the HSA-5 sample, 76% of the lysines were part of the sequence coverage, with the exception of lysines 233, 240, 262, 313, 323, 317, 351, 359, 444, 536, 538, 541, 545, and 560. Lysines 4 and 534, which were not covered in the control HSA sample, were covered in the HSA-5 sample. For the HSA-4 sample, 85% of the lysines were covered, excluding lysines 233, 240, 323, 359, 534, 536, 538, 541, and 545. Lysines 4, 317, and 560, which were not covered in the control HSA sample, were covered in the HSA-4 sample. The lack of coverage for these regions of HSA was of limited consequence in this study because none of these residues have been found in earlier work to have major amounts of modification on glycated HSA [26,28] (Note: minor modifications have been noted previously for lysines 136, 233, 323, 351 or 359, and 536 or 538 [29]).
2.6. Identification of glycation products
The glycation products formed on HSA were identified by first determining the peptide ions with mass values that were unique to the glycated HSA samples. The modifications in these ions were then determined by comparing these mass with a theoretical list of modified peptides that might result from a glycated HSA digest. When this approach was used, several early and late-stage glycation adducts were assigned to the glycated HSA samples (see Table 2). For the HSA-2 sample, 8 peptides were found to be modified to form a mixture of early glycation products and AGEs. Similarly, 8peptides were found to be modified to produce glycation products in both the HSA-4 and HSA-5 samples.
Table 2.
Suspected modification sites found on various samples of in vitro glycated HSA
| Digest | Residues | Relevant Amino Acid | Suspected Modification Sitesa | |
|---|---|---|---|---|
| HSA-2 | Trypsin | 1–10 | N+, K4, R10 | N+ (FL-1H2O) + K4 (AFGP) |
| Trypsin | 275–286 | K276, K281, K286 | K276 & K281 (FL & FL-1H2O) | |
| Trypsin | 539–557 or 542–560 | K541, K545, K557 K545, K557, K560 |
K545 & K557 (FL-2H2O & FL-2H2O) or (FL-1H2O & Pyr) K545 & K557 (FL-2H2O & Pyr) or (FL + CEL) |
|
| Glu-C | 278–292 | K281, K286 | K281 & K286 (FL & CEL) or (FL-2H2O & Pyr) | |
| Glu-C | 83–100 | K93, R98 | K93 (FL-2H2O) or K93 & R98 (CEL & MG-H1) | |
| Glu-C | 566–585 or 168–188 | K573, K574 K174, K181, R186 |
K573 and K574 (AFGP*2) K174 & R186 (FL & ArgP) |
|
| Lys-C | 196–205 | K199, K205, R197 | K199 & R197 (FL & 3-DG-H1/THP) | |
| Lys-C | 414–432 | K414, K432, R428 | K414 & R428 (FL-1H2O & AFGP) or K414 & R428 (AFGP and 3-DG-H1/THP) | |
| HSA-5 | Trypsin | 520–525 | K524, K525, R251 | R521 & K524 (Pyr & G-H1) |
| Trypsin | 275–286 | K276, K281, K286 | K276 & K281 (FL & FL-1H2O) | |
| Glu-C | 281–294 | K281, K286 | K281 & K286 (FL-2H2O & CEL) | |
| Glu-C | 426–442 | K432, K436, K439, R428 | R428 & K439 (G-H1 & Pyr) and (ArgP & CEL) | |
| Glu-C | 189–208 | K190, K195, K199, K205, R197 | K199 & R197 (Pyr & IB) | |
| Lys-C | 5–20 | K12, K20, R10 | K12 (FL) | |
| Lys-C | 414–432 | K414, K432, R428 | K414 & R428 (FL-1H2O & AFGP) or (AFGP & 3-DG-H1/THP) | |
| Lys-C | 525–534 | K525, K534 | K525 (FL-1H2O) | |
| HSA-4 | Trypsin | 1–12 | N+, K4, K12, R10 | N+ & K4 (CEL & FL-2H2O) or N+ & R10 (FL-1H2O & MG-H1) |
| Trypsin | or 200–212 or 403–413 |
K205, K212, R209 K413, R410 |
K205 & R209 (CML & ArgP) K413 or R410 (AFGP) |
|
| Trypsin | 198–209 or 182–195 or 206–218 |
K199, K205, R209 K190, K195, R186 K212, R209, R218 |
K199 & K205 AFGP, (FL & Pyr), or (FL-1H2O+FL-2H2O) R186 & K190 (ArgP & Pyr) R209 & K218 (FL-1H2O & MG-H1) |
|
| Trypsin | 258–274 | K262, K274 | K262 (FL-1H2O) | |
| Glu-C | 312–321 | K313, K317 | K313 & K317 (AFGP & AFGP) | |
| Glu-C | 557–571 | K557, K560, K564 | K560 & K564 (FL & CML) | |
| Glu-C | 426–442 | K432, K436, K439, R428 | K439 (FL) or R428 & K439 (Pyr+MG-H1) | |
| Glu-C | 83–100 | K93, R98 | K93 (FL-2H2O) or K93 (CEL) & R98 (MG-H1) | |
| Glu-C | 189–208 | K190, K195, K199, K205, R197 | K199 & R197 (Pyr & IB) | |
The most likely modification sites, which were identified based on computational and structural data, are given in bold. The abbreviations for the types of glycation are defined in Table 1.
One feature that was used for glycation site assignment was the fact that modifications could not occur on the same amino acids that were acting as cleavage sites for trypsin or Lys-C [29,45]. This fact allowed for the unambiguous assignment of glycation products to three peptides in the HSA-2 sample. These three peptides encompassed regions 1–10, 196–205, and 275–286 on HSA and corresponded to changes in mass (Δm) of 414, 306, or 306 Da respectively. The peptide encompassing residues 1–10 was found to be modified to form dehydrated fructosyl-lysine (FL-1H2O), which is specific for lysines, and 1-alkyl-2-formyl-3,4-glycosyl-pyrrole (AFGP), which can be formed on either lysine or arginine residues. This peptide contained the N-terminus (N+), K4 and R10 on HSA. Thus, the observed modifications were occurring on N+ or K4 because the C-terminal end of R10 was being cleaved by trypsin. Modifications by AFGP plus Nε-[5-(2,3,4-trihydroxybutyl)-5-hydro-4-imidazolon-2-yl]ornithine (3-DG-H1) or tetrahydropyrimidine (THP) would also give a Δm value of 414 Da. However, 3-DG-H1 and THP are arginine-specific modifications, making them less likely to have been present.
The peptide containing 196–205 in the HSA-2 sample was found to be modified by FL plus 3-DG-H1 or THP. A modification by THP or 3-DG-H1 would result in the same change in mass and could not be differentiated based solely on the Δm values. This peptide contained K199, R197, and K205, but the C-terminal end of K205 was cleaved by trypsin. This indicated that the most likely sites of modification on this peptide were at R197 and K199, which formed THP/3-DG-H1 and FL, respectively. A Δm value of 306 Da could also have been produced by the presence of both FL and FL-1H2O; however, the presence of these combined changes was unlikely because it would require that both K199 and K205 (which was cleaved by Lys-C) were being modified. Similarly, for residues 275–286 in the HSA-2 sample both K276 (FL or FL-1H2O) and K281 (FL or FL-1H2O) were assigned as modification sites.
There were several residues in both the HSA-4 and the HSA-5 samples that could be assigned with a high level of certainty because of the pattern of missed cleavage points that was observed. For instance, residues 520–525, 275–286, 5–20, and 525–534 were found to be modified to form pyrraline (Pyr) plus Nε-(5-hydro-4-imidazolon-2-yl)ornithine (G-H1) (Δm = 148 Da), FL + FL-1H2O (Δm = 306 Da), FL (Δm = 162 Da), and FL-1H2O (Δm = 144 Da), respectively, in the HSA-5 sample. Residues 275–286 were found to have identical modifications (i.e., forming FL + FL-1H2O) in both the HSA-2 and HSA-5 samples, which indicated that this region had a high propensity for the creation of such adducts. Modifications were assigned to K524 (Pyr) and R521 (G-H1), K12 (FL), and K525 (FL-1H2O) within these peptides based on the expected pattern of missed cleavage points for Lys-C and trypsin. The same approach was used to assign modifications in the HSA-4 sample for residues 258-274, 312–321, and 83–100, which were found to form FL-1H2O (Δm = 144 Da) at K262, AFGP + AFGP(Δm = 540 Da) at K313 and K317, and FL-2H2O or Nε-carboxyethyl-lysine (CEL) at K93 plus MG-H1 (Δm = 126 Da) at MG-H1, respectively.
2.7. Computational analysis of glycation sites
The pKa of a given amino acid residue, the fractional accessible surface area of this residue, and the location of the residue relative to local catalytic sites (i.e., histidines or lysines) were all used to help compare the expected reactivity of lysines and/or arginines during glycation. For this analysis, lysines that were within 15 Å of other positively-charged groups in the crystal structure of HSA were considered to have a higher amount of reactivity versus other lysines on HSA [26]. In this previous study, the proximity of lysine residues to histidine residues on albumin, particularly in regions close to disulfide bonds, was found to influence the level of total glycation at these sites. This observation was the rational for using a distance-based approximation based on the known crystal structure of HSA and Visual Molecular Dynamics [46,47], along with the information provided by MALDI-TOF MS, as an initial computational approach to help identify the most reactive regions of HSA during glycation. Although some small differences may be present between the crystal structure of HSA and the structure of this protein’s soluble form when doing these calculations, the information this approach provided was still found to be useful as an initial tool in comparing the expected reactivities of various residues to glycation.
Arginines and lysines that had low pKa values versus other lysines/arginines on HSA, as estimated using PROPKA [48], were also used with the MALDI-TOF MS data as a second computational approach to help identify sites on HSA that were susceptible to glycation-related modifications. When information about local catalytic sites or pKa values did not provide insight as to the likelihood of modification at a given residue, computational results using the fractional accessible surface area (FAS) of the residues were also used as indicators of reactivity. This overall approach has been shown in previous work with MALDI-TOF MS, quantitative proteomics and minimally-glycated HSA to be valuable in helping identify possible glycation sites [30]. A more detailed discussion of these computational techniques is given in the Supplemental Information. These computational approaches were employed in this study in cases where multiple lysines or arginine residues were present on the same peptide or when several modified peptides had mass-to-charge ratios within the allowed mass accuracy that could be associated with a given peak in the mass spectra for glycated HSA.
The HSA-4 digest contained four peptides for which computational methods were used to determine the most likely modification sites. The majority of these peptides were from the Glu-C digest, which had no cleavage specificity for lysines or arginines. In the HSA-4 Glu-C digest, residues 557–571, 426–442, and 189–208 were found to form FL + CML (Δm = 220 Da), FL or Pyr + MG-H1 (Δm = 162 Da), and Pyr + IB (Δm = 250 Da), respectively. Residues 557–571, which was modified to form two lysine-specific adducts, contains K557, K560, and K564. K560 and K564 are located near a disulfide bridge (C558 and C567), which places these two residues within 11 Å of each other. K557 is not located within 25 Å of any basic amino acids, which indicated that the formation of any glycation products at this residue would not have been enhanced by local acid-base catalysis. The proximity of K560 and K564 to each other suggested that these residues were more likely to form FL and CML. These residues also had large FAS values of 0.82 and 0.72, respectively, versus a FAS value of 0.68 for K557. These results all indicated that K560 and K564 were the most likely modification sites within residues 557–571.
Lysine 439 was tentatively determined to be the most reactive site within residues 426–442 in both the HSA-4 and HSA-5 samples, as based on the location of this residue relative to H440 (3.83 Å) in a loop region of HSA. This result indicated that K439 was being modified to form FL or Pyr in the HSA-4 sample and Pyr or CEL in the HSA-5 sample. The assignment of modifications to K414 (FL-1H2O or AFGP) and R428 (AFGP and 3DG-H1/THP) was made without the use of computational data, with these modifications being found in both the HSA-2 and HSA-5 samples. This information suggested that the region spanning residues 414–442 of HSA had a strong tendency to be modified during glycation. R428 was found to be modified in residues 414–432 and 426–442, indicating this arginine was particularly prone to modification, in agreement with a previous study examining the formation of HSA adducts with methylglyoxal [28]. Residues 189–208 were found to be modified to form Pyr + IB in both the HSA-4 and HSA-5 samples. In this case the assignment of IB to R197 could be made without the use of computational data, and K199 was found to be modified by Pyr, as determined based on the proximity of K199 to H247 (15.19 Å) and its relatively low pKa value of 7.47. This latter result agrees with previous work examining glycation-related modifications that occur at K199 [26].
In situations where a particular modification site could not be identified based on computational data, a comparison with the previous results in the literature was conducted [25–27]. For instance, an ion with a mass of 1622 Da that was identified in the HSA-4 digest could have been produced by modifications occurring on residues 1–12 (CEL + FL-2H2O or FL-1H2O + MG-H1, Δm = 198 Da), 200–212 (CML + ArgP, Δm = 138 Da), or 403–413 (AFGP, Δm = 270 Da). The N-terminus and K4 in residues 1–10 were determined to be the most likely sites of modification in this case because the N-terminus has previously been identified as a glycation site on HSA [2,27], and K4 was located beside H3 (i.e., which would have enhanced the rate of glycation products at this residue). The corresponding modifications at these residues were CEL and FL-2H2O. Similarly, modifications occurring at K199 (to form AFGP, FL, or Pyr) and K205 (to form AFGP, FL, or Pyr) in the HSA-4 sample were determined to be the most likely source of the observed ion with a mass of 1707 Da because these lysines were located close to H247 and H242, respectively, which would have enhanced the formation of glycation products [26].
Two modified peptide ions with masses of 2409 and 2547 Da were identified in the HSA-2 sample. The first of these two peptides represented modifications on either residues 539–557 (FL-2H2O + FL-2H2O, FL-1H2O + Pyr, Δm = 252 Da) or 542–560 (FL-2H2O + Pyr, FL + CEL, Δm = 234 Da) and the second was due to modifications on residues 566–585 (AFGP + AFGP, Δm = 540 Da) or 168–188 (FL + ArgP, Δm = 242 Da). The assignment of modifications on these particular peptides was difficult because there was no clear indicator of the amino acid reactivity from either computational analysis or the literature to help in this assignment.
2.8. Semi-quantitative analysis of glycation sites
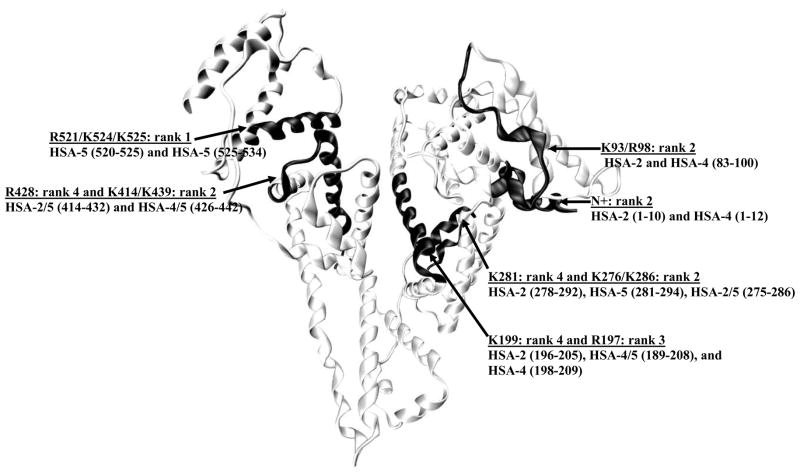
There were several cases in this study in which a given lysine or arginine residue was found to be modified in multiple samples, such as in different digests of a particular glycated HSA sample or in HSA samples representing different stages of glycation. In these cases, the number of times that a given residue was found to be modified was used as a semi-quantitative indicator of the reactivity of that particular residue with regards to glycation. A structure of HSA that includes these semi-quantitative assignments is given in Figure 2. In this structure, 6 regions of HSA were found to be modified to form at least 2 glycation products.
Figure 2.
Structure of HSA showing the most likely sites for glycation-related modifications, as based on a semi-quantitative comparison of the data obtained in this study. The regions that were found to have the highest levels of modification are highlighted. The amino acids that contained the highest amounts of modification within these regions are also shown.
The highest amount of modification was found in regions 196–209, 278–294, and 414–442 of HSA. Four peptides were modified within region 196–209 when all stages of glycation were considered: one modified peptide (196–205) in the HSA-2 sample, 2 modified peptides in the HSA-4 sample (189–208 and 198–209), and one modified peptide (189–208) in the HSA-5 sample. All of these peptides had a modification assigned to K199. This result indicated that K199 was prone to glycation for in vitro glycated HSA, in agreement with previous studies that have found this residue to be a major glycation site on HSA [26,29]. It was also found that K199 was modified by FL in the early stages of glycation (i.e., in HSA-2), while Pyr, and FL-1H2O or FL-2H2O appeared to be the dominant modifications at this location during later stages of glycation (i.e., in HSA-4 and HSA-5). This result was not surprising considering the increased amount of AGEs that would occur as a protein like HSA is subjected to glycation over long periods of time [5,49]. A large number of modifications were also assigned in this study to R197. For this site, the predominant adduct that was formed was either 3-DG-H1 or THP in the early stages of glycation (i.e., HSA-2 sample) or IB in later stages of glycation (i.e., the HSA-4 and HSA-5 samples). An identical modification of R197 to form 3-DG-H1 or THP has been identified in a previous study using a commercial preparation of minimally-glycated HSA [29].
Four modified peptides from region 278–294 of HSA were found when all stages of glycation were considered. In this case, residues 278–292 were modified in the HSA-2 sample, residues 281–294 were modified in the HSA–5 sample, and residues 275–286 were modified in both the HSA-2 and HSA-5 samples. K281 was modified in all four peptides to form FL-related modifications, CEL, and/or Pyr. The identification of K281 as a major modification site is consistent with data from other reports using both in vitro and in vivo glycated HSA [26,29]. It was found that there were also minor contributions from modifications at K276 and K286 within residues 278–294 to form FL or FL-1H2O and FL, FL-2H2O, CEL, or Pyr, respectively.
Residues 414–442 on HSA were found to contain at least four modified peptides, as noted in all of the glycated HSA samples. R428 was identified as a modification site in all four of these peptides and was involved in the formation of a number of AGEs (AFGP or 3-DG-H1/THP, ArgP or G-H1, and MG-H1), depending on the level of total glycation. There were also minor modifications at K414 (FL-1H2O or AFGP) and K439 (Pyr or CEL, and FL or CEL) within region 414–442 of HSA. These findings are again consistent with previous reports, in which R428 [28] and K439 [26,29] have been identified as prominent modification sites on HSA. Several modifications were also found at the N-terminus (FL-1H2O, FL-2H2O, AFGP, and/or CEL) and K525 (FL-1H2O), both regions of which have been identified as having high levels of modification in other studies [2,26,27,29].
The glycation sites noted in this report were compared to modification regions that have been previously identified using commercial preparations of minimally glycated HSA [29,30]. These comparisons were made based on 1) qualitative data using shifts in peptide masses [29] and 2) data based on quantitative proteomics and 16O/18O-labeling [30]. The results obtained from this analysis are summarized in Table 3. Seven of the modified peptides that were found in this current study to have high levels of modification were also found to be modified in previous studies [29,30]. Three of these modified peptides contained K525, K439, or K199, which are also the residues expected from prior work to have high levels of modification in glycated HSA [2]. In addition, K12 R81, R197, R428, K205, K286, and K524 were all identified as modification sites in both this report and in previous studies using glycated HSA [29,30].
Table 3.
Comparison of modification sites identified in this study with modification sites found in previous work with minimally glycated HSA
| HSA Sample | Modified Region 1a | Modified Region 2b | Modified Region 3c | Overlapping Residues | Modification |
|---|---|---|---|---|---|
| HSA-5 | 520–534 | 520–525 | 525–534 | K521, K524, & K525 | FL-1H2O |
| HSA-2, HSA-5 | 414–432 | 426–442 | 415–439 | K439/R428 | FL-1H2O or AFGP |
| HSA-4, HSA-5 | 426–442 | 426–442 | 415–439 | K439 | FL, FL-H2O, or Pyr |
| HSA-2, HSA-4 | 196–205, 198–209 | 196–209, 191–205 | n/a | K199, K205 | FL, FL-H2O, or Pyr |
| HSA-5 | 281–294 | 286–294 | n/a | K286 | FL-2H2O or CEL |
| HSA-2, HSA-4, HSA-5 | 1–10, 1–12, 5–12 | 7–17 | n/a | K12 | FL, FL-H2O, or CEL |
| HSA-2, HSA-4 | 83–100 | n/a | 71–84 | R81 | MG-H1 |
Modified region 1 represents the region of HSA that was found to contain modified sites in this current study. The abbreviations used to represent these modifications are defined in Table 1.
Modified region 2 represents the region of HSA that was found in Ref. [29] to be modified to form early and late stage glycation products when using a commercial preparation of minimally-glycated HSA.
Modified region 3 represents the region of a commercial preparation of minimally-glycated HSA that was previously found in Ref. [30] to have significant levels of glycation, as measured by using 16O/18O-labeling and mass spectrometry.
3.5 Physiological relevance of results
The physiological and pathological relevance of the changes in glycation patterns that were seen in this study can be demonstrated by comparing these results with recent solute binding studies that have used similar samples of glycated HSA. In one such study, the association equilibrium constant for L-tryptophan, an essential amino acid that binds to Sudlow site II on HSA, had up to a 5.8-fold increase in affinity in going from normal HSA to in vitro glycated HSA that was prepared in the same manner as used in this study [39]. Using the same preparations of glycated HSA as in Ref. [39], the sulfonylurea drug tolbutamide (i.e., which has been used in the treatment of type II diabetes) was found to have up to a 1.3- to 1.4-fold increase in its association equilibrium constants at Sudlow sites I and II in going from normal to glycated HSA [50]. These changes in binding strength with the level of glycation clearly indicate that such modifications can alter interactions of drugs and other solutes with HSA, which in turn could change the effective activity and transport of these agents in the circulation [39,50,51].
The need to consider how changes in glycation patterns may vary with the extent of glycation is demonstrated by a similar binding study for the sulfonylurea drug acetohexamide [51]. This binding study used the same preparations of in vitro glycated HSA [39,50] and similar preparations to those utilized in this report. Acetohexamide gave an overall increase in binding affinity to HSA as the level of glycation increased but different local changes in binding strength at Sudlow sites I and II. At Sudlow site I, there was a 40% increase in the association equilibrium constant when going from normal HSA to mildly glycated HSA (i.e., a sample similar to HSA-2). This change was followed by a decrease in affinity for HSA with higher amounts of glycation and that was identical to the HSA-4 sample examined in this current report. In the case of Sudlow site II, the binding of acetohexamide to the same samples of glycated HSA showed a 40% decrease in affinity at low levels of modification, followed by an increase in affinity at higher levels of glycation. This trend fits the observations made in this current report in that the modifications at Sudlow site I for residues 278–292 (FL, CEL, and/or Pyr), and 196–205 (FL, 3-DG-H1) appeared to transition to dehydrated FL and CEL (281–294), and Pyr and IB (196–205) in the more highly glycated sample (HSA-5).
In this report, residues 414–432 at Sudlow site II were found to be modified by combinations of FL-1H2O and AFGP or AFGP and 3-DG-H1/THP in the HSA-2 and HSA-5 samples. Residues 426–442 were only found to be modified in the HSA-5 sample, giving mixtures of G-H1 and Pyr or ArgP and CEL. The HSA-4 sample, which contained the highest amount of glycation in this current study and in the previous binding studies with acetohexamide and tolbutamide [50,51], appeared to have a smaller number of modifications at Sudlow site II, with only a single modified peptide (i.e., residues 426–432 containing FL or Pyr+MG-H1) being detected at this site and in this sample. These changes in the number and types of modifications as the extent of glycation was varied suggest that 1) the changes in binding affinity seen for acetohexamide and tolbutamide during glycation [50,51] are related to the relative rates of formation and accumulation of the various possible modifications, and 2) different modifications may affect this binding to different degrees. Further studies based on quantitative proteomics and mass spectrometry are now being conducted to further characterize these changes in structure. However, the correlation already been noted between the structural and functional information indicates that the glycation level of HSA can affect its binding to pharmaceutical agents, including drugs such as acetohexamide and tolbutamide that are used to treat diabetes.
3. Conclusion
In this report, regions 189–209, 275–294, and 414–442 of HSA all appeared to be particularly prone to modification as a result of glycation. Two of these regions (189–209 and 275–294) are located within Sudlow site I, which is a major drug binding site on HSA [2]. Lysine 199, which has repeatedly been identified in the past as a modification site within this highly glycated region of HSA, is also known to be directly involved in the interactions of many drugs and fatty acids with HSA [2,52]. Lysine 281 was also found to have high levels of modification during glycation; this residue is located in the structure of HSA at a location that is only 13 Å away from a known ligand binding residue within Sudlow site I [46]. Residues R428, K414, and K439, which were all found to have high levels of modifications in the glycated HSA samples, are located within Sudlow site I, the second major drug binding site of HSA [46].
The close vicinity of these modification sites to known drug binding sites on HSA helps explain why glycation may alter the interaction of many drugs and solutes that bind to Sudlow site I on HSA. For instance, the binding of bilirubin at Sudlow site I, has been shown to be altered when comparing normal HSA to in vivo glycated HSA [25]. In addition, binding by drugs such as phenytoin and valproate, which can involve multiple sites on HSA) [53] has been shown to be affected by glycation in diabetes [54]. It has also been shown previously that the binding of several solutes at Sudlow site II can be altered when using glycated HSA [21,28,38].
A semi-quantitative analytes of glycation adducts that were found in this report indicated that the N-terminus, lysines 93, 199, 276, 281, 286, 414, 439, 524, and 525, as well as arginines 98, 197, and 428, can all contain significant amounts of modification. All of the previously identified glycation sites on HSA (e.g.., the N-terminus, K199, K281, K439, K525 [26,27,29]) were found to be modified in the glycated HSA samples that were examined in this study. In addition, a known site for advanced glycation end products in HSA (i.e., R428) [45] was also found to have significant amounts of modification in this study. This information indicated that the results from this study gave good agreement with previous reports examining the glycation of HSA. Quantitative studies using 16O/18O-labeling and mass spectrometry are currently being conducted to more accurately determine the extent of these modifications.
A unique feature of this study was that a semi-quantitative analysis indicated that the modification pattern of HSA changed with the total extent of glycation. For example, K199 and R197 were found to be modified in all three stages of glycation, with the predominant modifications on these residues shifting from FL (K199) and 3-DG-H1 or THP (R197) in the HSA-2 sample to Pyr (K199) and IB (R197) in the HSA-4 and HSA-5 samples. Similarly, modifications at R428 were found in all three stages of glycation, with this residue forming AFGP, 3-DG-H1, or THP in the HSA-2 sample, these same modifications plus G-H1 or Arg-P in the HSA-5 sample, and MG-H1 in the HSA-4 sample. K199 and R197 are both located in Sudlow site I on HSA and R428 is located in Sudlow site II, which indicates that drug-protein interaction at these two regions may, in turn, also depend on the level of glycation.
Comparison of the data obtained in this report with previous binding studies was used to demonstrate the physiological and pathological implications of these results [39,50,51]. This comparison suggested that the changes in binding affinity seen for drugs such as acetohexamide with in vitro glycated HSA can be affected by both the level of glycation and the glycation pattern of HSA. The fact that the glycation pattern and types of modification changed at different levels of glycation indicates that these changes may lead to changes in drug interactions with HSA during diabetes and with the extent of control for this disease. By using this work with in vitro glycated HSA as an initial reference point, further work using 16O/18O-labeling in mass spectrometry and/or work with in vivo glycated HSA is now being conducted to more fully characterize the nature and extent of these changes in structure and drug interactions. Preliminary results from these ongoing studies indicate that R98, R197, K199, K276, K281, K286, and R428 are all modified within in vivo glycated HSA from patients with moderate to advanced diabetes, which is consistent with the semi-quantitative results provided in this report. In addition, many modifications found in these in vivo glycated HSA samples are identical to the modifications noted here for the HSA-4 sample. Tandem mass spectrometry is also being used to verify the presence of some of the modifications detected in this report for glycated HSA (e.g., this approach has already verified the presence of CEL at K281, FL-2H2O at K286, Pyr at K205, and variants of FL at K557, K560, and K564, as determined in this study by using mass spectrometry and computational methods). The data provided in this current report and these future studies should provide valuable information that will lead to a better understanding of how drug transport and binding in blood can change during diabetes. This information, in turn, is expected to eventually lead to the development of improved strategies based on personalized medicine for the selection of drugs and drugs dosages for the treatment of diabetes.
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Health (NIH) under grant R01 DK069629. Support for the remodeled facilities used to perform these experiments was provided by NIH grant RR015468-001. Part of this work was carried out in the Nebraska Center for Mass Spectrometry, as supported by the National Cancer Institute under grant P30 CA36727, the NIH under grants P20 RR15635 and RR015468, and the Nebraska Research Initiative.
List of Abbreviations
- AFGP
1-alkyl-2-formyl-3,4-glycosyl-pyrrole
- AGE
advanced glycation end product
- ArgP
argpyrimidine
- CEL
Nå-carboxyethyl-lysine
- CHCA
α-cyano-4-hydroxycinnamic acid
- CML
Nå-carboxymethyl-lysine
- 3-DG-H1
Nå-[5-(2,3,4-trihydroxybutyl)-5-hydro-4-imidazolon-2-yl]ornithine
- DHB
2,5-dihydroxybenzoic acid; fractional accessible surface area (FAS)
- FAS
fractional accessible surface area
- FL
fructosyl-lysine
- G-H1
Nå-(5-hydro-4-imidazolon-2-yl)ornithine
- HSA
human serum albumin
- IB
imadazolone B
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MG-H1
Nå-(5-hydro-5-methyl-4-imidazolon-2-yl)ornithine
- Pyr
pyrraline
- TFA
trifluoroacetic acid
- THP
tetrahydropyrimidine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li W, Ota K, Nakamura J, Naruse K, Nakashima E, Oiso Y, et al. Antiglycation effect of glyclazide on in vitro AGE formation from glucose and methylglyoxal. Exp Biol Med. 2008;233:176–9. doi: 10.3181/0705-BC-131. [DOI] [PubMed] [Google Scholar]
- 2.Peters T. All about Albumin: Biochemistry, Genetics, and Medical Applications. San Diego: Academic Press; 1996. [Google Scholar]
- 3.Makita Z, Vlassara H, Cerami A, Bucala R. Immunochemical detection of advanced glycosylation end products in vivo. J Biol Chem. 1992;267:5133–8. [PubMed] [Google Scholar]
- 4.Syrovy I. Glycation of albumin: Reaction with glucose, fructose, galactose, ribose or glyceraldehyde measured using four methods. J Biochem Biophys Meth. 1994;28:115–21. doi: 10.1016/0165-022x(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 5.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem J. 1999;344:109–16. [PMC free article] [PubMed] [Google Scholar]
- 6.Magalhaes PM, Appell HJ, Duarte JA. Involvement of advanced glycation end products in the pathogenesis of diabetic complications: The protective role of regular physical activity. Eur Rev Aging Phys Act. 2008;5:17–29. [Google Scholar]
- 7.Hayashi T, Mase S, Namiki M. Formation of three-carbon sugar fragment at an early stage of the browning reaction of sugar with amines or amino acids. Agric Biol Chem. 1986;50:1959–64. [Google Scholar]
- 8.Zhang Q, Ames JM, Smith RD, Baynes JW, Metz TO. A perspective on the maillard reaction and the analysis of protein glycation by mass spectrometry: Probing the pathogenesis of chronic disease. J Proteome Res. 2009;8:754–69. doi: 10.1021/pr800858h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito M, Marumo K. Collagen cross-links as a determinant of bone quality: A possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus. Osteoporos Int. 2010;21:195–214. doi: 10.1007/s00198-009-1066-z. [DOI] [PubMed] [Google Scholar]
- 10.Sell DR, Nemet I, Monnier VM. Partial characterization of the molecular nature of collagen-linked flourescence: Role of diabetes and end-stage renal disease. Arch Biochem Biophys. 2010;493:192–206. doi: 10.1016/j.abb.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bank RA, Baylis MT, Lafeber FP, Maroudas A, Tekoppele JM. Ageing and zonal variation in post-translational modification of collagen in normal human articular cartilage. the age-related increase in non-enzymatic glycation affects iomechanical properties of cartilage. Biochem J. 1998;330:345–51. doi: 10.1042/bj3300345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monnier VM, Kohn RR, Cerami A. Accelerated age related browning of human collagen in diabetes mellitus. Proc Natl Acad Sci. 1984;81:583–7. doi: 10.1073/pnas.81.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansirani, Anathanaryanan PH. A comparitive study of lens protein glycation in various forms of cataracht. Indian J Clin Biochem. 2004;19:110–2. doi: 10.1007/BF02872403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BS, Harding JJ. The effects of aminoguanidine on the glycation (non-enzymatic glycosylation) of lens proteins. Exp Eye Res. 1990;50:463–7. doi: 10.1016/0014-4835(90)90033-q. [DOI] [PubMed] [Google Scholar]
- 15.Turk Z, Misur I, Turk N. Temporal association between lens protein glycation and cataract development in diabetic rats. Acta Diabetol. 1997;34:49–54. doi: 10.1007/s005920050066. [DOI] [PubMed] [Google Scholar]
- 16.Biroccio A, Urbani A, Massoud R, di Ilio C, Sacchetta P, Bernardini S, et al. A quantitative method for the analysis of glycated and glutathionylated hemoglobin by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Anal Biochem. 2005;336:279–88. doi: 10.1016/j.ab.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi T, Miyazaki A, Kishikawa M, Yasuda M. Quantification of glycated hemoglobin by electrospray ionization mass spectrometry. J Mass Spectrom. 1997;32:773–8. doi: 10.1002/(SICI)1096-9888(199707)32:7<773::AID-JMS533>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Chandalia HB, Krishnaswamy PR. Glycated hemoglobin. Curr Sci. 2002;83:1522–32. [Google Scholar]
- 19.Rodkey FL. Direct spectrophotometric determination of albumin in human serum. Clin Chem. 1965;11:478–87. [PubMed] [Google Scholar]
- 20.Ghuman J, Zunszain PA, Petitpas I, Bhattachara AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 21.Koyama H, Sugioka N, Uno A, Mori S, Nakajima K. Effects of glycosylation of hypoglycemic drug binding to serum albumin. Biopharm Drug Dispos. 1997;18:791–801. doi: 10.1002/(sici)1099-081x(199712)18:9<791::aid-bdd66>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Vorum H, Fisker K, Otagiri M, Pedersen AO, Hansen UK. Calcium ion binding to clinically relevant chemical modifications of human serum albumin. Clin Chem. 1995;41:1654–61. [PubMed] [Google Scholar]
- 23.Chiou YJ, Tomer KB, Smith PC. Effect of nonenzymatic glycation of albumin and superoxide dismutase by glucuronic acid and suprofen acyl glucuronide on their functions in vitro. Chem Biol Interact. 1999;121:141–59. doi: 10.1016/s0009-2797(99)00098-8. [DOI] [PubMed] [Google Scholar]
- 24.Nakajou K, Watanabe H, Kragh-Hansen U, Maruyama T, Otagiri M. The effect of glycation on the structure, function and biological fate of human serum albumin as revealed by recombinant mutants. Biochim Biophys Acta. 2003;1623:88–97. doi: 10.1016/j.bbagen.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Shaklai N, Garlick RL, Bunn HF. Nonenzymatic glycosylation of human serum albumin alters its confirmation and function. J Biol Chem. 1984;259:3812–7. [PubMed] [Google Scholar]
- 26.Iberg N, Fluckiger R. Nonenzymatic glycosylation of albumin in vivo. J Biol Chem. 1986;261:13542–5. [PubMed] [Google Scholar]
- 27.Robb DA, Olufemi SO, Williams DA, Midgley JM. Identification of glycation at the N-terminus of albumin by gas chromatography-mass spectrometry. Biochem J. 1989;261:871–8. doi: 10.1042/bj2610871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed N, Dobler D, Dean M, Thornalley PJ. Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J Biol Chem. 2005;280:5724–32. doi: 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- 29.Wa C, Cerny RL, Clarke WA, Hage DS. Characterization of glycation adducts on human serum albumin by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2007;385:48–60. doi: 10.1016/j.cca.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnaby OS, Wa C, Cerny RL, Clarke W, Hage DS. Quantitative analysis of glycation sites on human serum labeling using 16O/18O labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2010;411:1102–10. doi: 10.1016/j.cca.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;25:S5–S20. doi: 10.2337/diacare.26.2007.s5. [DOI] [PubMed] [Google Scholar]
- 32.Turner RC, Cull CA, Frighi V, Holman RR. Glycemic control with diet, sulfonylurea, metformin, or insulin in patients with type 2 diabetes mellitus. J Am Med Assoc. 1999;281:2005–12. doi: 10.1001/jama.281.21.2005. [DOI] [PubMed] [Google Scholar]
- 33.Wa C, Cerny R, Hage DS. Obtaining high sequence coverage in matrix-assisted laser desorption time-of-flight mass spectrometry for studies of protein modification: Analysis of human serum albumin as a model. Anal Biochem. 2006;349:229–41. doi: 10.1016/j.ab.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Lapolla A, Fedele D, Reitano R, Aricò NC, Seraglia R, Traldi P, et al. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J Am Soc Mass Spectrom. 2004;15:496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Ney KA, Karen J, Colley KJ, Pizzob SV. The standardization of the thiobarbituric acid assay for nonenzymatic glucosylation of human serum albumin. Anal Biochem. 1981;118:294–300. doi: 10.1016/0003-2697(81)90585-6. [DOI] [PubMed] [Google Scholar]
- 37.Voziyan PA, Khalifah RG, Thibaudeau C, Yildiz A, Jacob J, Serianni AS, et al. Modification of proteins in vitro by physiological levels of glucose. J Biol Chem. 2003;278:46616–24. doi: 10.1074/jbc.M307155200. [DOI] [PubMed] [Google Scholar]
- 38.Barzegar A, Moosavi-Movahedi AA, Sattarahmady N. Spectroscopic studies of the effects of glycation of human serum albumin on L-trp binding. Protein Pept Lett. 2007;14:13–8. doi: 10.2174/092986607779117191. [DOI] [PubMed] [Google Scholar]
- 39.Joseph KS, Hage DS. The effects of glycation on the binding of human serum albumin to warfarin and l-tryptophan. J Pharm Biomed Anal. 2010;53:811–8. doi: 10.1016/j.jpba.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouzuma T, Usami T, Yamakoshi M, Takahashi M, Imamura S. An enzymatic method for the measurement of glycated albumin in biological samples. Clin Chim Acta. 2002;324:61–71. doi: 10.1016/s0009-8981(02)00207-3. [DOI] [PubMed] [Google Scholar]
- 41.Armbuster DA. Fructosaminase: Structure, analysis and clinical usefulness. Clin Chem. 1987;33:2153–63. [PubMed] [Google Scholar]
- 42.Zubarev R, Manns M. On the proper use of mass accuracy in proteomics. Mol Cell Proteomics. 2007;6:377–81. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Syrovy I. Glycation of albumin: Reaction with glucose, fructose, galactose, ribose or glyceraldehyde measured using four methods. J Biochem Biophys Meth. 1994;28:115–21. doi: 10.1016/0165-022x(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 44.Lapolla A, Fedele D, Seraglia R, Catinella S, Baldo L, Aronica R, et al. A new effective method for the evaluation of glycated intact plasma proteins in diabetic subjects. Diabetologia. 1995;38:1076–81. doi: 10.1007/BF00402178. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Cocklin RR, Bidasee KR, Wang M. Rapid determination of advanced glycation end products using MALDI-TOF-MS and PERL script peptide searching algorithm. J Biomol Tech. 2003;14:224–30. [PMC free article] [PubMed] [Google Scholar]
- 46.Sugio S, Kashima A, Mochizuki S, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–21. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 49.Thornalley PJ. Dicarbonyl intermediates in the maillard reaction. Ann N Y Acad Sci. 2005;1043:111–7. doi: 10.1196/annals.1333.014. [DOI] [PubMed] [Google Scholar]
- 50.Joseph KS, Anguizola J, Hage DS. Binding of tolbutamide to glycated human serum albumin. J Pharm Biomed Anal. doi: 10.1016/j.jpba.2010.09.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joseph KS, Anguizola J, Jackson AJ, Hage DS. Chromatographic analysis of acetohexamide binding to glycated human serum albumin. J Chromatogr B. 2010;878:2775–81. doi: 10.1016/j.jchromb.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol. 1998;5:827–35. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 53.Mandula H, Parepally JMR, Feng R, Smith QR. Role of site-specific binding to plasma albumin in drug availability to brain. J Pharmacol Exp Therapeut. 2006;317:667–75. doi: 10.1124/jpet.105.097402. [DOI] [PubMed] [Google Scholar]
- 54.Doucet J, Fresel J, Hue G, Moore N. Protein binding of digitoxin, valproate and phenytoin in sera from diabetics. Eur J Clin Pharmacol. 1993;45:577–9. doi: 10.1007/BF00315318. [DOI] [PubMed] [Google Scholar]
- 55.Willard L, Ranjan A, Zhang H, et al. VADAR: a web server for quantitative evolution of protein structure quality. Nucleic Acids Res. 2003;31:3316–9. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.