Abstract
Vasa is a broadly conserved DEAD-box RNA helicase associated with germ line development and is expressed in multipotent cells in many animals. During embryonic development of the sea urchin Strongylocentrotus purpuratus, Vasa protein is enriched in the small micromeres despite a uniform distribution of vasa transcript. Here we show that the Vasa coding region is sufficient for its selective enrichment and find that gustavus, the B30.2/SPRY and SOCS box domain gene, contributes to this phenomenon. In vitro binding analyses show that Gustavus binds the N-terminal and DEAD-box portions of Vasa protein independently. A knockdown of Gustavus protein reduces both Vasa protein abundance and its propensity for accumulation in the small micromeres, whereas overexpression of the Vasa-interacting domain of Gustavus (GusΔSOCS) results in Vasa protein accumulation throughout the embryo. We propose that Gustavus has a conserved, positive regulatory role in Vasa protein accumulation during embryonic development.
Keywords: Vasa, Gustavus, multipotent cells, embryogenesis, E3 ligase
Introduction
Animals specify primordial germ cells (PGCs) with a variety of different developmental strategies, yet they employ a shared set of genes (Extavour and Akam, 2003; Seydoux and Braun, 2006). The most extensively studied of these is vasa, a highly conserved DEAD box RNA helicase, which has proven a reliable germ line marker in many animal species (e.g. Gustafson and Wessel, 2010; Raz, 2000). Originally identified in Drosophila, vasa is essential for oocyte development, posterior patterning, and PGC specification in this embryo (Lasko and Ashburner, 1988; Schupbach and Wieschaus, 1986; Styhler et al., 1998; Tomancak et al., 1998). These germ line requirements of vasa are also evident in the mouse and in Caenorhabditis elegans (Kuznicki et al., 2000; Spike et al., 2008; Tanaka et al., 2000).
Although vasa is not essential for normal physiology of every cell type within a developing or adult animal, its requirement for fecundity in the adult is supported by loss of function mutation analyses in Drosophila, Xenopus, mouse, the budding tunicate Polyandrocarpa misakiensis, the crustacean Parhyale hawaiensis, the monogenean Neobenedenia girellae and the nematode Caenorhabditis elegans; each analysis substantiates similar roles for vasa homologs in germ cell development (Hay et al., 1990; Ikenishi and Tanaka, 1997; Kuznicki et al., 2000; Lasko and Ashburner, 1988; Liang et al., 1994; Ohashi et al., 2007; Ozhan-Kizil et al., 2009; Schupbach and Wieschaus, 1986; Spike et al., 2008; Styhler et al., 1998; Sunanaga et al., 2007; Tanaka et al., 2000). In chicken embryonic stem cells, ectopic expression of vasa is sufficient to induce expression of specific germ line and meiotic genes e.g. the chicken vasa homolog (Cvh) (Lavial et al., 2009). However, to date, these in vitro cell culture properties of vasa have not been observed in an intact embryo. Moreover, data from polychaetes, Hydra, sea urchins and flatworms suggest that vasa can have broader stem cell functions (Gustafson and Wessel, 2010; Kuznicki et al., 2000; Mochizuki et al., 2001; Noce et al., 2001; Pfister et al., 2008; Raz, 2000; Rebscher et al., 2007; Shibata et al., 1999; Tanaka et al., 2000).
Localized Vasa protein expression within the embryo at some point during development is a common feature in all animals studied and it appears that animals accomplish this by utilizing several independent regulatory mechanisms, both transcriptional and posttranscriptional (Knaut et al., 2000; Knaut et al., 2002; Liu et al., 2003; Sugimoto et al., 2009; Van Doren et al., 1998; Voronina et al., 2008; Wolke et al., 2002; Yoon et al., 1997). Taken together, these data underscore the fact that proper expression and localization of Vasa protein is crucial for its roles in development. During the course of evolution, multiple layers of regulation may have compounded to accommodate a variety of increasingly diverse developmental strategies while maintaining cell type-specific Vasa expression. Evidence of this regulatory complexity can be seen in the larval ascidian Ciona intestinalis, the colonial ascidian Botryllus primigenus and the sea urchin S. purpuratus where removal of Vasa-positive cells induces de novo Vasa expression (Sunanaga et al., 2006; Takamura et al., 2002; Voronina et al., 2008).
Although several animals localize Vasa protein within a developing embryo through transcriptional and translational regulation, this alone does not always account for select Vasa protein accumulation. A growing body of evidence suggests that selective Vasa proteolysis is a contributing factor. Despite early localization of vasa mRNA in zebrafish embryos, Vasa protein is distributed uniformly and is localized to PGCs only later in development (Braat et al., 2000; Knaut et al., 2000; Yoon et al., 1997), due in part to soma-specific degradation of Vasa protein (Wolke et al., 2002). In the developing Drosophila oocyte, Vasa protein accumulates at the posterior pole despite a uniform distribution of vasa mRNA (Hay et al., 1988; Lasko and Ashburner, 1990). A regulatory balance between addition and removal of ubiquitin modifications appears to contribute to this phenotype. The deubiquitinating enzyme Fat facets physically interacts with Vasa protein, promotes its pole plasm accumulation and Fat facets mutants show increased levels of ubiquitylated Vasa (Liu et al., 2003). Recent data in Drosophila identified two paralogous E3 ubiquitin ligase specificity receptors, fsn and gustavus, involved in this regulatory balance of Vasa ubiquitylation controlling pole plasm accumulation (Kugler et al., 2010; Styhler et al., 2002). Drosophila Gustavus protein contains a B30.2/SPRY sequence comprising a single structural domain that biochemically interacts with Vasa in vitro (Woo et al., 2006a; Woo et al., 2006b). Sequence analysis of fsn predicts a similar B30.2/SPRY domain and both Fsn and Gustavus bind Vasa protein in vivo. Gustavus contains a SOCS-box that interacts with ElonginB/C-Cullin 5 complex, whereas fsn contains an F-box that interacts with a Cullin 1 scaffolding complex (Kugler et al., 2010; Woo et al., 2006a). These complexes can link target proteins to the ubiquitin-proteasome machinery for degradation or other fate modifications (Piessevaux et al., 2008).
In the sea urchin S. purpuratus, vasa transcripts are present uniformly during early embryogenesis, but Vasa protein is enriched in the 16-cell stage micromeres and subsequent small micromeres (Juliano et al., 2006; Voronina et al., 2008). One possible explanation for this difference in appearance is that vasa mRNA is translated in the small micromeres and is translationally repressed in all other cells. This is a well-documented phenomenon and data from several different animals demonstrate how translational repression allows localized protein production from a ubiquitous transcript during embryonic development (reviewed by (Kuersten and Goodwin, 2003). In many cases translation is controlled by cis-regulatory sequences within the 5′ and 3′ untranslated regions (UTRs) of the transcript (reviewed by (Chatterjee and Pal, 2009). Alternatively, the vasa transcript may be translated throughout this embryo and selective Vasa protein enrichment results from Vasa proteolysis in all cells except the small micromeres. This study provides insight into the regulatory mechanisms governing the small micromere accumulation of Vasa protein in the presence of uniform vasa transcript and supports the model that the E3 ubiquitin ligase specificity receptor gustavus has conserved functions regulating Vasa protein accumulation.
Materials and Methods
Animals and Embryos
S. purpuratus husbandry and embryo culturing was carried out as described previously (Gustafson and Wessel, 2008).
Sp-gustavus Cloning, Mutagenesis, Plasmid Construction and Phylogenetic Analysis
Sp-gustavus was identified from computational gene sequence predictions in the S. purpuratus genome (http://www.hgsc.bcm.tmc.edu/projects/seaurchin/) using a BLAST analysis against the D. melanogaster gustavus protein sequence. ClustalW analysis was used to align amino acid sequences (http://www.ch.embnet.org/software/ClustalW.html). SPRY and SOCS box domains were identified using the pfam program (http://pfam.janelia.org/; (Finn et al., 2006) and the B30.2/SPRY domains were identified by comparative analysis to the B30.2/SPRY domain in D. melanogaster gustavus as defined previously (Woo et al., 2006a). Domain sequence identities in gustavus orthologs were analyzed using the EMBOSS Needle Pairwise Alignment Algorithm (http://www.ebi.ac.uk/Tools/emboss/align). Template cDNA was prepared using the TaqMan® Reverse Transcription Reagents kit (Applied Biosystems, Foster City, CA) from purified S. purpuratus mesenchyme blastula-stage embryo RNA as described previously (Bruskin et al., 1981). Full-length Sp-gustavus was PCR amplified from mesenchyme blastula cDNA, cloned into pGEM-T Easy (Promega, Madison, WI) and verified by sequencing (Table 2). This wildtype construct was used as a template to generate a gustavus plasmid encoding a R146W mutation by PCR stitch mutagenesis (Table 2) and GusΔSOCS constructs made with template plasmids encoding either Sp-gustavus wt or Sp-gustavus R146W. GST-GusΔSOCS wt and R146W constructs (Table 2) were generated with the pGEX-5X-3 expression vector (GE Healthcare Lifesciences, Piscataway, NJ). GusΔSOCS wt constructs containing the Xenopus β-globin 5′ and 3′ UTRs as well as an Sp6 promoter and transcription start site (pSp6 β-globin UTRs) were also generated (a generous gift from Dr. Kimberly Mowry, Brown University).
Table 2.
GFP and Vasa-GFP fusion protein localization summary
| RNA construct | Small micromere localization sufficiency |
Perinuclear granule protein localization |
Nuclear enrichment |
|---|---|---|---|
| Vasa UTRs | No | No | No |
| Vasa1F-GFP | Yes | Yes | No |
| Vasa2F-GFP | No | Yes | No |
| Vasa4F-GFP | No | No | No |
| Vasa6F-GFP | No | No | Yes |
| Vasa3R-GFP | No | No | No |
| Vasa4R-GFP | No | No | No |
| Vasa5R-GFP | No | No | Yes |
Sp-vasa Plasmid Construction and Mutagenesis
Various N-terminal and C-terminal deletions to the Sp-vasa coding region were generated by PCR-mediated mutagenesis (see primers in Table 2) and subcloned into the pSp6 β-globin UTRs plasmid between the Xenopus β-globin 5′ and 3′UTRs. A GFP variant EGFP (Clontech, Mountain View, CA) coding region was added in frame to the C-terminus of each Sp-vasa deletion construct (Table 2). Sp-vasa regions encoding N-terminal (N-term), middle (DEAD) and C-terminal (C-term) fragments were generated by PCR (Table 2) and subcloned into pT7-MAT-Flag-2 (Sigma-Aldrich, St. Louis, MO).
MG132 Treatment and Analysis
S. purpuratus eggs were fertilized in ASW containing 1 mM 3-aminotriazole (3-AT; Sigma-Aldrich, St Louis, MO). Embryos were cultured in ASW at 16°C to early blastula-stage (~10 hours). The embryos were then cultured in 10 μM, 25 μM or 50 μM MG132 (Z-Leu-Leu-Leu-CHO; Enzo Life Sciences International, Inc., Plymouth Meeting, PA) ASW containing 0.5% Dimethyl sulfoxide (DMSO), ASW alone, or ASW with 0.5% DMSO as negative controls at 16°C for 12 hours. Protein samples from each culture were prepared for immunoblot analysis by pelleting the embryos, resuspending in 2X sample buffer (100 mM Tris-HCl pH 6.8, 4% SDS, 20% glycerol, 15 mM DTT, 0.2% bromophenol blue). The samples were analyzed by western blot probing with either affinity-purified rabbit anti-Vasa (1:1000) (Voronina et al., 2008) or rabbit anti-Actin (1:2000) (Sigma, St. Louis, MO) as described previously (Voronina et al., 2003). Using Cy3-conjugated goat anti-rabbit IgGs (1:5000) (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA) as secondary antibodies, Actin and Vasa were imaged on the membranes with a Typhoon™ 9410 Variable Mode Imager (GE Healthcare Life Sciences, Piscataway, NJ) and were quantified using MultiGauge Imaging Software (Fujifilm, Tokyo, Japan). Remaining embryos from each culture were fixed for whole-mount immunofluorescent analysis as described previously (Laidlaw and Wessel, 1994). Anti-Vasa antibodies (1:500) and Cy3-conjugated goat anti-rabbit IgG (1:300) (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA) were used for immunofluorescence analysis and images were acquired with a Zeiss Axiovert 200 M microscope using a 40X water immersion objective (Carl Zeiss Incorporated, Thornwood, NY). Fluorescent images were quantified using Metamorph imaging software (Universal imaging Corporation, Downington, PA).
Recombinant Sp-Gustavus and Sp-Vasa Protein Production and Purification
Recombinant fusion proteins were expressed in E. coli BL21 cells and cell pellets were resuspended in 10 ml of MAT Binding Buffer (25 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100 and Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN) containing 0.75% Sarkosyl. GST-GusΔSOCS proteins were prepared with MAT Binding Buffer containing 1 mM EDTA. After lysing the cells with 20 mg of lysozyme for 30 minutes at 25°C while rotating, and freeze-thawing at −80°C and 37°C 3 times, the lysates were cooled on ice for 15 minutes and sonicated 2 cycles: 1 minute each at 100% duty cycle and 1.5 power using a Branson 3000 Sonifier. Insoluble material was removed by centrifugation and the soluble protein lysate was collected and stored at −80°C. GST-GusΔSOCS wt and GST-GusΔSOCS R146W fusion proteins were affinity purified with glutathione-coupled agarose beads (Sigma-Aldrich, St. Louis, MO) by incubating 1 ml of swollen beads with 10 ml of lysate at 4°C for 3 hours. The beads were washed 6 times with 10 ml of MAT Binding Buffer. Purification of the recombinant proteins was confirmed by SDS-PAGE followed by Western blotting with anti-GST antibodies (1:5000) and Coomassie Brilliant Blue staining.
Recombinant Gustavus-Vasa Binding Assay
MAT-Vasa-Flag N-term, DEAD and C-term cell extracts were thawed and 1 ml of each was precleared with 50 μl of a GST protein glutathione-coupled agarose bead 50% slurry in MAT Binding Buffer for 1 hour at room temperature under rotation. The beads were pelleted and the supernatant collected 2 times. Each cell extract was divided into 3 equal volumes and added to glutathione-coupled agarose beads containing either purified GST, GST-GusΔSOCS wt or GST-GusΔSOCS R146W. MAT Binding Buffer was added to a final volume of 1.2 ml for each binding reaction. Following a 3-hour incubation at room temperature under rotation, each binding reaction was washed in 1 ml of MAT Binding Buffer 6 times. Protein samples were prepared by adding 200 μl of 2X Sample Buffer containing 15 mM DTT to the beads, boiled for 5 minutes, centrifuged for 30 seconds at 18,000 x (g) and the denatured protein supernatants were collected. The samples (20 μl) were resolved on a 4-20% gradient Tris-glycine polyacrylamide gel (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes for immunoblotting with mouse monoclonal anti-Flag primary antibodies (1:5000) (Sigma-Aldrich, St. Louis, MO) and peroxidase-conjugated rabbit anti-mouse secondary antibodies (1:10000) (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA). The MAT-Vasa-Flag proteins were detected by incubation in a chemiluminescence solution (1.25 mM luminol, 68 μM coumeric acid, 0.0093% hydrogen peroxide and 0.1 M Tris pH 8.6) for 1 minute, exposed to film and developed.
Whole-mount in situ RNA Hybridization and Immunohistochemistry
A digoxigenin (DIG) RNA Labeling Kit (Roche, Indianapolis, IN) and plasmid templates (see above) were used to generate DIG-labeled antisense RNA probes. DIG-labeled RNA probes generated from a pSPT 18 vector were used as nonspecific negative controls. Whole-mount in situ RNA hybridizations (WMISH) were carried out as described previously (Arenas-Mena et al., 2000). Images were acquired with a Zeiss Axiovert 200 M microscope using a 40X oil immersion objective (Carl Zeiss Incorporated, Thornwood, NY, USA). Sp-gustavus WMISH embryos and were blocked for 1 hour in MOPS-Tween20 buffer containing 10% sheep serum, incubated with 1 ml of MOPS-Tween20 buffer with 10% sheep serum and anti-Vasa primary antibodies (1:500) overnight at room temperature, Cy3-conjugated goat anti-rabbit secondary IgGs (1:300) and counterstained with 0.2 μg/ml Hoechst 33258 (Invitrogen, Carlsbad, CA). The embryos were imaged on a TCS SP2 AOBS confocal scanning microscope (Leica Microsystems; Bannockburn, IL) and the Sp-gustavus in situ staining was pseudocolored green using Photoshop®. Embryos mock-injected with Dextran or microinjected with synthetic β-globin UTRs GusΔSOCS wt RNA were fixed in paraformaldehyde and blocked as detailed above. These embryos were then incubated with affinity-purified anti-Vasa primary antibodies (1:500) as above, and Alexa488-conjugated goat anti-rabbit secondary IgGs (1:300) and imaged under the same detector settings on a Zeiss Axioplan microscope or LSM510 META laser confocal microscope.
Sp-Gustavus-specific Antibody Production, Purification and Immunofluorescence
Polyclonal antibodies were generated using purified recombinant full-length Sp-Gustavus protein. An amino-terminal biotin-conjugated synthetic peptide containing the Sp-Gustavus VSGGMKQVAREPNFKSLHRELTYSGDLQ sequence (GenScript, Piscataway, NJ) was immobilized on streptavidin-agarose beads (Sigma-Aldrich, St. Louis, MO) and used for affinity-purification of the anti-Gustavus antiserum. S. purpuratus eggs and embryos were fixed for whole-mount immunofluorescence in 4% paraformaldehyde and washed with MOPS-Tween20, as described above, incubated with the affinity-purified anti-Gustavus primary antibodies (1:50), Cy3-conjugated goat anti-rabbit secondary IgGs (1:300), counterstained with 0.2 μg/ml Hoechst 33258 and imaged.
Synthetic RNA and Morpholino Antisense Oligonucleotide Injection and Analysis
Capped sense RNA was synthesized using the mMessage mMachine® Sp6 Kit (Ambion, Austin, TX). Synthetic RNA transcripts were purified with RNeasy Micro Kit (Qiagen, Valencia, CA) yielding RNA concentrations between 1.5-3 μg/μl. Each RNA was mixed with synthetic β-globin UTRs mCherry RNA to a final concentration of approximately 1 μg/μl and 0.5 μg/μl respectively. Approximately 2 pl of each RNA mixture was injected into each fertilized egg. Injected embryos were cultured in artificial seawater (ASW) at 16°C and live-imaged at various developmental stages with either a Zeiss Axioplan microscope or LSM510 META laser confocal microscope (Carl Zeiss Incorporated, Thornwood, NY). A morpholino antisense oligonucleotide (MASO) complementary to the Sp-gustavus transcript 5′UTR was generated to block translation (GeneTools, Philomath, OR). Using 500 μM of Gustavus MASO, microinjections were performed as previously described (Juliano et al., 2010). Fluorescent images were quantified as described above. Cy3 fluorescent Actin and Vasa immunoblots were imaged with a Typhoon™ 9410 Variable Mode Imager and Gustavus chemiluminescent immunoblot images were collected on film as described above.
Results
The Vasa coding region directs small micromere enrichment
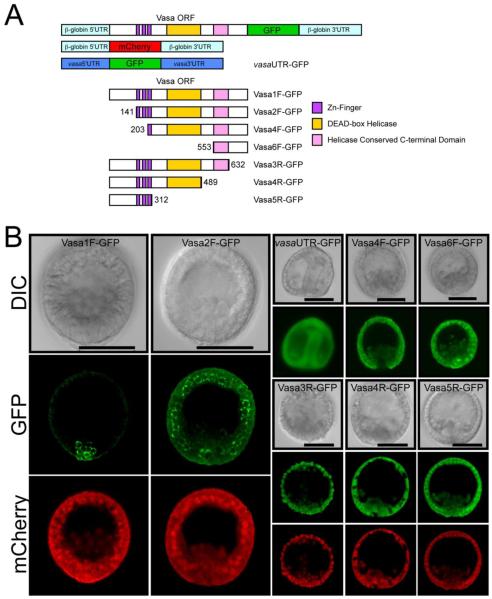
Analysis of the endogenous vasa transcript and protein localization during S. purpuratus embryo development was indicative of a post-transcriptional regulatory mechanism (Voronina et al., 2008). Despite a uniform vasa transcript distribution during early development, Vasa protein is detectable only in the small micromeres. To test if vasa mRNA is translated only in the small micromeres, we asked if the vasa UTRs impart sufficient regulatory information for small micromere-specific protein enrichment. A reporter was constructed which contains the GFP open reading frame (ORF) flanked by vasa 5′ and 3′UTRs (Figure 1A). Synthetic vasaUTR-GFP RNA was transcribed in vitro, injected into fertilized S. purpuratus eggs, and accumulation of GFP in embryos was monitored. We found that GFP fluorescence was present uniformly in the embryos suggesting that vasa UTRs do not contain sufficient regulatory information to account for selective Vasa protein accumulation in small micromeres (Table 2; Figure 1B).
Figure 1. The entire S. purpuratus Vasa open reading frame is sufficient for small micromere enrichment of Vasa protein.
(A) A schematic depiction for each synthetic RNA construct analyzed. Domain locations are representative of its position within each protein and were identified using the pfam program (http://pfam.janelia.org/) (Finn et al., 2006). (B) Synthetic mCherry RNA co-injected with RNA generated from the indicated Vasa-GFP constructs in (A). GFP (green) and mCherry (red) fluorescence assayed in mesenchyme blastula or gastrula-stage embryos following microinjection of synthetic RNA along with corresponding DIC images. See Table 2 for summary of results. Scale bar = 50 μm.
We next tested whether the Vasa ORF is sufficient for selective protein accumulation in the small micromeres. The entire Vasa coding region was fused to GFP and flanked by Xenopus β-globin UTRs to direct constitutive translation throughout the embryo (Vasa 1F-GFP; Figure 1A). Synthetic Vasa ORF-GFP RNA was co-injected into fertilized eggs with mCherry RNA, also flanked by the β-globin UTRs, as a control. While mCherry fluorescence was robustly detected in all cells, GFP fluorescence was enriched only in the small micromeres. Further, the Vasa 1F-GFP protein was localized to perinuclear structures (Voronina et al., 2008; Table 2; Figure 1B).
To test which portion of the Vasa coding region is sufficient for its small micromere enrichment, a series of N-terminal and C-terminal Vasa deletion constructs were generated, fused to GFP and flanked by β-globin UTRs (Figure 1A). The truncation sites in the Vasa ORF were made outside known and predicted tertiary structural domains based on crystallographic data and computational predications in order to minimize any unfavorable folding conditions. Although Vasa 2F-GFP only lacks residues 1-140 of the Vasa ORF, it accumulates in a punctate perinuclear location, but in all cells of the embryo (Table 2; Figure 1A, 1B). Removal of residues 1-202 (Vasa 4F-GFP), which includes the first 2 predicted CCHC Zn-knuckles, resulted in uniform GFP fluorescence in all cells and lacked any punctate perinuclear localization (Vasa 4F-GFP; Table 2; Figure 1A-B). Taken together, these data demonstrate that the first 140 residues are required for enrichment of Vasa protein in the small micromeres and the first two CCHC Zn-knuckles are crucial for the perinuclear localization of Vasa protein. However, Vasa 5R-GFP, which contains only the N-terminal 312 residues and includes all 3 CCHC Zn-knuckles, is not sufficient for either small micromere protein accumulation or perinuclear localization (Table 2; Figure 1A-B). In addition, deletion of just the 150 C-terminal residues results in loss of selective GFP accumulation (Vasa 3R-GFP; Table 2; Figure 1). Thus the Vasa ORF contains multiple regions required for both its subcellular localization and for its enrichment in the multipotent small micromere lineage of the sea urchin embryo.
Vasa protein is subject to proteasome-mediated degradation
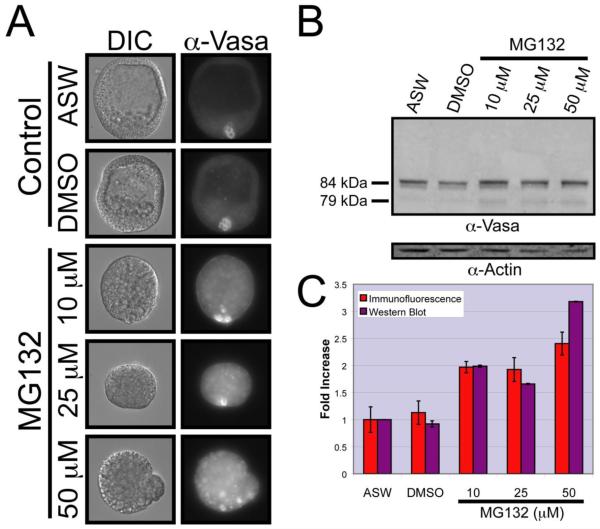
The Vasa ORF is sufficient to drive selective accumulation in the small micromere lineage. One mechanism that could explain this observation is that protein turnover may regulate selective Vasa protein accumulation. To test this possibility, embryos were treated with the reversible proteasome inhibitor MG132 and endogenous Vasa protein levels were assayed by quantitative immunofluorescence and western blotting. While control embryos had normal Vasa expression patterns, MG132-treated embryos showed an increase in the Vasa immunofluorescence signal throughout the embryo (Figure 2A). Despite apparent morphological defects in the embryos, likely due to complications arising from abnormal cell cycle regulation, Vasa protein enrichment was still evident in small micromeres following MG132 treatment (Figure 2A). Quantitative analyses of Vasa protein levels, from both immunoblotting and immunofluorescence, show a dose dependent increase of approximately 2 to 3-fold upon MG132 treatment (Figure 2B-C). These data argue that Vasa protein is subject to proteasome-dependent degradation in non-small micromeres, which is consistent with the sufficiency of the Vasa coding region to direct its small micromere-specific accumulation. At present it is unclear why Vasa protein continually accumulates to highest levels in the small micromeres, even under a variety of perturbations. It is not a general phenomenon, since mCherry and other GFP reporters do not show such enrichment in the small micromeres.
Figure 2. Vasa protein accumulates in all cells of the embryo following proteasome inhibition.
(A) Immunofluorescence localization of endogenous Vasa protein in embryos cultured in artificial seawater (ASW) alone, ASW containing 0.5% dimethyl sulfoxide (DMSO), or ASW containing 10 μM, 25 μM or 50 μM MG132. Images of Vasa protein staining are shown in grayscale next to their corresponding DIC images. (B) Quantitative immunoblot analysis of Vasa levels normalized to actin levels using protein extracts from each culture. (C) Quantitative analysis of Vasa fluorescence per embryo in (A, purple) and immunoblotting (B, red). Error bars correspond to the fluorescence intensity standard deviation within the individual embryos assayed (ASW, n = 26; DMSO, n = 29; 10 μM MG132 n = 30; 25 μM MG132 n = 27; 50 μM MG132 n = 31) in immunofluorescence quantification and the standard deviation from 3 separate immunoblots in Western blotting quantification.
Identification and analysis of the sea urchin gustavus ortholog
The canonical mechanism for directing proteins to the proteasome involves target protein acquisition of a polyubiquitin chain following its recognition by an E3 ubiquitin ligase (Voges et al., 1999). Recent work in Drosophila suggests the E3 ubiquitin ligase substrate specificity receptor Fsn functions as a negative regulator of Vasa protein stability in the embryo by directing ubiquitylation of Vasa and its subsequent degradation by proteosomes (Kugler et al., 2010). Although Drosophila Vasa is ubiquitylated in vivo, we were unable to detect a ubiquitin-modified form of sea urchin Vasa (Liu et al., 2003); data not shown).
Our efforts turned to another E3 ubiquitin ligase specificity receptor, gustavus, since Drosophila Gustavus binds to Vasa in vitro and in vivo through its B30.2/SPRY domain (Kugler et al., 2010; Woo et al., 2006a; Woo et al., 2006b). However, despite the ability to interact with both Vasa and E2 ubiquitin-conjugating machinery Elongin B/C-Cullin5, Drosophila Gustavus appears to function as a positive regulator of Vasa protein stability (Kugler et al., 2010). To examine whether this functional relationship exists in sea urchins, a S. purpuratus gustavus ortholog was identified and characterized. BLAST analysis of the S. purpuratus genome (http://www.hgsc.bcm.tmc.edu/projects/seaurchin), using the Drosophila Gustavus protein sequence, identified a single gene. This was deduced to be the sea urchin gustavus ortholog based on its sequence similarity as well as the presence and organization of B30.2/SPRY and SOCS box domains (Supplementary Figure 1A-B). Genes with the same domain architecture exist in a variety of animals and show the closest sequence identity to the S. purpuratus gustavus gene within their B30.2/SPRY domain (Supplementary Figure 1A-B). Structural data of Drosophila Gustavus protein reveals that several loops within two β-sheets form a prominent pocket that directly interacts with a Vasa peptide. The Vasa-interacting residues within these loops are completely conserved in all other identified gustavus orthologs including S. purpuratus gustavus (Supplementary Figure 1A; Woo et al., 2006b).
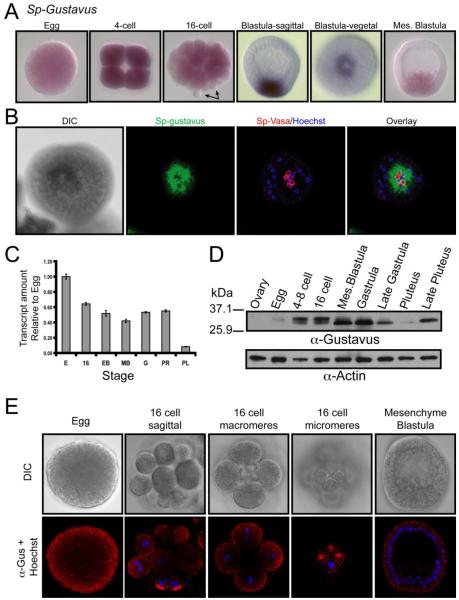
Gustavus transcripts were analyzed by quantitative PCR and in situ RNA hybridization during embryonic development (Figure 3A and 4C). Of particular note is a uniform and strong signal in eggs, which remains uniform during early cleavage divisions. By the 16-cell stage, however, the transcripts decrease in the micromeres while remaining uniform throughout the rest of the embryo. Gustavus transcripts in blastula-stage embryos are restricted to a group of cells forming a ring around the vegetal pole. In mesenchyme blastula, the transcript remains at the vegetal pole, but is also present in the ingressing primary mesenchyme cells. Consistent with the qPCR analysis, the gustavus mRNA in situ signal progressively declines from gastrula to larval pluteus stages (Figure 3A, 3C and data not shown). Co-labeling blastula embryos for gustavus mRNA and Vasa proteins demonstrates that gustavus transcripts are present in a ring of adjacent vegetal blastomeres, but are mostly excluded from the Vasa-positive small micromeres (Figure 3B). The predicted size of S. purpuratus Gustavus protein is ~31 kDa and anti-Gustavus antibodies detect a doublet migrating at this molecular weight (Figure 3D). Despite the presence of Gustavus transcript in unfertilized eggs, Gustavus protein is not abundant in ovary or egg extracts by western blot (Figure 3D). However, both Gustavus bands are detected in 4-cell to 16-cell embryos, while the smaller band persists into gastrula, decreases in late gastrula and 3-day larvae, and increases again in 12-day larvae (Figure 3D). Both Gustavus bands in 16-cell embryo protein extracts persisted following phosphatase treatment, suggesting that phosphorylation is not responsible for the two Gustavus antibody-reactive protein species (data not shown). Immunofluorescence demonstrates that Gustavus protein accumulation is punctate throughout the egg and early embryo and then becomes enriched in the vegetal cortex in micromeres (Figure 3E). Gustavus appears in all cells in mesenchyme blastula embryos and we found no embryos specifically lacking Gustavus protein in the small micromeres despite a depletion of gustavus transcripts in these cells.
Figure 3. S. purpuratus gustavus transcripts, but not protein, are excluded from the micromere lineage.
(A) Spatial expression of gustavus mRNA using WMISH. Arrows in 16-cell embryo indicate micromeres devoid of gustavus transcript. (B) Blastula-stage embryos co-labeled for gustavus mRNA by WMISH (pseudocolored green) and Vasa protein by immunofluorescence (red). (C) Quantitative RT-PCR (qPCR) measurement of relative gustavus mRNA levels. All values are normalized to Sp-ubiquitin RNA and are represented as a fold difference to egg RNA levels. Error bars indicate the standard deviation from triplicate qPCR reactions for each template. (D) Immunoblot detection of Gustavus and Actin proteins in cell extracts from various developmental stages. (E) Immunofluorescence analysis of endogenous Gustavus protein localization (red) and DNA (blue) at indicated developmental stages. Independent exposure and detector imaging conditions were used for each developmental stage.
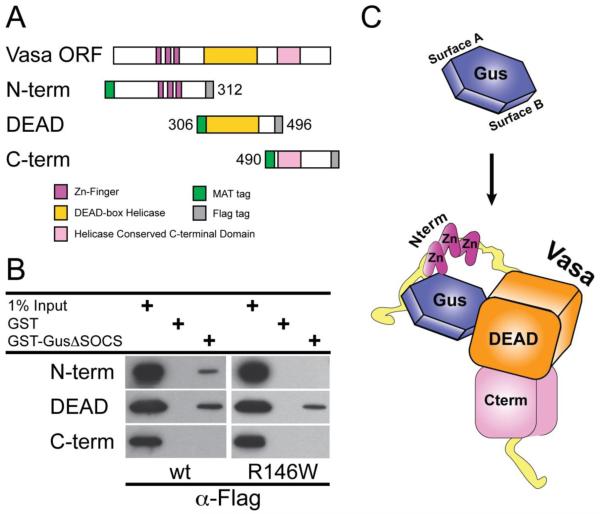
Figure 4. GusΔSOCS-Vasa interacts with discrete segments of Vasa protein.
(A) A graphic representation of each Vasa construct containing an N-terminal Metal Affinity Tag (MAT), C-terminal Flag epitope, corresponding domains and DEAD-box sequence motifs. (B) Gustavus binding domains in Vasa. Bacterial extracts containing N-term, DEAD and C-term recombinant Vasa proteins were incubated with immobilized GST, GST-GusΔSOCS wt or GST-GusΔSOCS R146W and bound proteins were analyzed by immunoblotting with anti-Flag antibodies. (C) A bipartite Gustavus-Vasa binding model in S. purpuratus. The gustavus B30.2/SPRY domain is shown in blue with binding surface A and binding surface B indicated as predicted previously (Woo et al., 2006a). A schematic representation of the S. purpuratus vasa DEAD box domain in orange, C-terminus domain in pink, N-terminus CCHC zinc knuckles in purple and unstructured glycine-rich flexible sequence in yellow.
The Gustavus B30.2/SPRY domain interacts with multiple portions of Vasa
The Vasa binding properties of the Gustavus B30.2/SPRY domain are well characterized in Drosophila, where biochemical and structural data identified a minimal DINNN Gustavus-interacting sequence in the Vasa protein N-terminus with respect to the DEAD-box domain (Kugler et al., 2010; Woo et al., 2006a; Woo et al., 2006b). Mutational analyses of the Gustavus B30.2/SPRY domain identified 4 phylogenetically conserved residues (Y133A, G149Y, R150W and W221L) that are essential for its binding to Vasa and are all located on the same binding surface (Woo et al., 2006a). Although the S. purpuratus Gustavus B30.2/SPRY domain is well conserved and includes the 4 corresponding residues essential for Vasa binding (Supplemental Figure 1A and 1B), the S. purpuratus Vasa N-terminus lacks any similarity to the minimal Gus-interacting sequence in Drosophila. Therefore, we tested if the Gustavus B30.2/SPRY domain can interact with different portions of the S. purpuratus Vasa protein using a GST-pull down assay. Recombinant GST-GusΔSOCS wt and R146W (corresponding to the conserved R150 in Drosophila Gustavus) fusion proteins were first constructed, expressed in E. coli and purified with glutathione-agarose beads. Flag-tagged proteins containing Vasa residues 1-312 (N-term), 306-496 (DEAD) and 490-732 (C-term) were expressed in E. coli (Figure 4A). These soluble cell extracts were incubated with immobilized GST-GusΔSOCS wt protein, GST-GusΔSOCS R146W mutant protein or GST alone and bound proteins were analyzed by immunoblotting with anti-Flag antibodies. GST alone was incapable of binding any of the Vasa protein fragments whereas GST-GusΔSOCS wt bound both the Vasa N-term and DEAD proteins, but not the C-term protein fragment. GST-GusΔSOCS R146W also bound the DEAD protein, but was unable to bind the N-term or C-term Vasa proteins (Figure 4B). These data indicate that Gustavus has at least 2 interaction sites on the Vasa protein: one within the N-terminal portion and another within the DEAD box domain. Since a Gustavus R146W mutation does not disrupt its DEAD protein interaction and abolishes its interaction with the N-term protein, Gustavus may interact with the N-term and DEAD residues on Vasa protein independently through two separate binding surfaces (Figure 4C). The DEAD-box domain of interaction is consistent with gustavus regulating the selective accumulation of this ancient protein in many organisms throughout the animal kingdom.
Overexpression of full-length gustavus and a gustavus mutant lacking its SOCS-box causes ectopic Vasa protein accumulation
Gustavus may function as a negative regulator of Vasa protein stability by targeting it for polyubiquitylation and degradation by the proteasome. Such a polyubiquitylation event would depend on the ability of Gustavus to directly interact with Vasa through its B30.2/SPRY domain and link it to the ubiquitin conjugating machinery through the interaction of its SOCS-box with the Elongin B/C complex (Figure 5A). Alternatively, Gustavus may function as a positive regulator of Vasa protein stability by competing for Vasa binding with other proteins that target Vasa for proteolysis. In this scenario, Gustavus may indeed link Vasa to the ubiquitin conjugating machinery, but such an interaction may not result in Vasa degradation (Figure 5A).
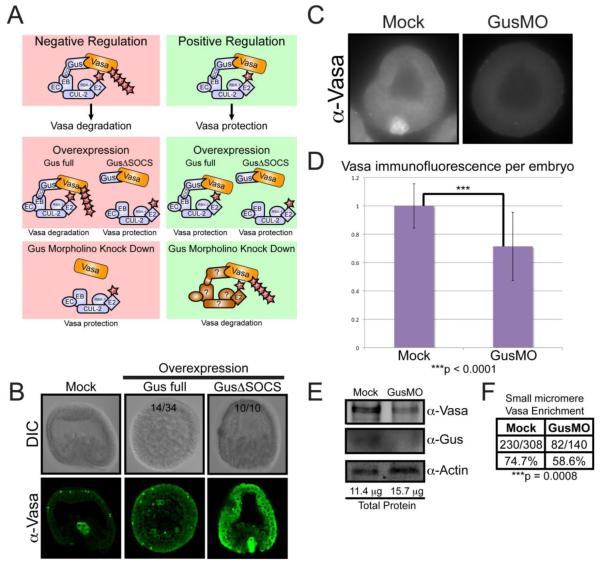
Figure 5. Gustavus is required for normal Vasa abundance and small micromere enrichment.
(A) Two hypothesis for how Gustavus function regulates Vasa protein stability. Negative Regulation: Gustavus targets Vasa for proteolysis through direct binding and ubiquitin conjugation. Overexpression of full-length Gustavus protein throughout the embryo would promote global Vasa proteolysis including the small micromeres, whereas overexpression of Gustavus lacking its SOCS-box would compete with endogenous Gustavus for Vasa binding, thereby protecting and elevating global Vasa protein levels. Morpholino knock down of endogenous Gustavus would remove any proteolysis targeting of Vasa resulting in elevated Vasa protein throughout the embryo. Positive Regulation: Gustavus interacts with Vasa and competes with other proteins that can target Vasa for proteolysis. Overexpression of full-length Gustavus or Gustavus lacking its SOCS-box throughout the embryo would both bind and protect Vasa, resulting in elevated Vasa levels throughout the embryo. Morpholino knock down of endogenous Gustavus would leave Vasa protein susceptible to proteolytic targeting by other Vasa-binding E3 ligase adapter proteins throughout the embryo resulting in a global reduction in Vasa protein. Gus: Gustavus, SOCS: SOCS-box, EB: Elongin B, EC: Elongin C Cul-2: Cullin 2, RBX1: RING Box Protein-1, E2: E2 ubiquitin conjugating enzyme, Ub: Ubiquitin (B) Immunofluorescence localization of endogenous Vasa in early gastrula embryos cultured in ASW following mock injection or injection with synthetic RNA containing a GusΔSOCS coding region (depicted in Figure 4A) or full-length Gus coding region. Numbers shown reflect representation within the resultant population, and Gus-full length was markedly more variable in the vasa phenotype. (C) Immunofluorescence of endogenous Vasa protein in 20 hour-old embryos either mock injected or treated with 500 μM GusMO. (D) Quantitative analysis of Vasa fluorescence per embryo. Error bars correspond to the fluorescence intensity standard deviation within the individual embryos assayed. The Vasa immunofluorescence difference between Mock (n = 76) and GusMO-injected embryos (n = 61), as calculated by a paired t-test, was statistically significant based on a two-tailed P value less than 0.0001. (E) Immunoblot analysis of protein extracts from mock and GusMO-injected embryos. A Bradford Assay calculated the total protein amount used for each sample. (F) The number of mock and GusMO-injected embryos displaying Vasa protein enrichment in the small micromeres, as observed by immunofluorescence. The difference in small micromere Vasa protein enrichment frequency between mock and GusMO embryos, as calculated using a Fisher’s Exact Test, was statistically significant (p < 0.0008) based on a two-tailed test.
In order to test these two hypotheses, we first investigated the effects of Gustavus overexpression on endogenous Vasa protein accumulation by injecting RNA encoding either the full-length Gustavus or GusΔSOCS flanked by the β-globin UTRs in the early embryo. If Gustavus acts as a negative regulator, we predict overexpression of a full-length Gustavus protein throughout the embryo would promote global Vasa proteolysis including the small micromeres. However, overexpression of Gustavus lacking its SOCS-box would compete with endogenous Gustavus for Vasa binding, yet fail to direct Vasa turnover resulting in elevated Vasa protein levels throughout the embryo (Figure 5A). While Vasa protein accumulated selectively in the small micromeres of mock-injected embryos, embryos overexpressing wildtype full-length Gustavus did not gastrulate, arrested development after ~24 hours and exhibited morphological abnormalities including an expanded ingression of primary mesenchyme cells (PMCs) into the blastocoel (Figure 5B). While some cells had a noticeable Vasa enrichment, they were dispersed among PMCs throughout the blastocoel instead of the small micromere location at the vegetal pole (Figure 5B). It is likely that Gustavus binds and regulates the stability of other proteins besides Vasa so these abnormalities may reflect aberrant regulation of Gustavus targets stemming from its multiplicity of target proteins. Such pleiotropic effects make it difficult to distinguish between positive and negative regulatory Gustavus functions with regard to Vasa protein stability.
However, embryos overexpressing GusΔSOCS displayed a dramatic increase in Vasa protein in all cells (Figure 5B). Unlike the MG132-treated embryos, these embryos exhibited completely normal development and morphology. Despite this increase throughout the embryo, Vasa protein was still more abundant in the small micromeres (Figure 5B). These results are consistent with both positive and negative Gustavus regulator hypotheses.
Sp-gustavus is required for normal Vasa protein abundance and small micromere enrichment
To further test a functional relationship between gustavus and vasa in S. purpuratus, gustavus translation was impaired by microinjection of a gustavus-specific morpholino antisense oligonucleotide (GusMO) and endogenous Vasa protein accumulation was assayed in 20 hour-old embryos. Immunoblot analysis confirmed that GusMO treatment effectively decreased Gustavus protein compared to mock-injected embryos (Figure 5E). While typical abundance of Vasa protein was observed in mock-injected embryos, we find a statistically significant Vasa protein decrease in GusMO-injected embryos (Figure 5C-E). Furthermore, GusMO-injected embryos showed a significant reduction in the frequency of small micromere Vasa protein enrichment compared to control embryos (Figure 5C and 5F). These data are consistent with a positive regulatory role for Gustavus in both Vasa protein abundance and small micromere enrichment during embryonic development.
Discussion
The results shown here suggest that regulation of Vasa protein accumulation by proteolysis is a conserved mechanism (Kugler et al., 2010; Liu et al., 2003; Wolke et al., 2002). In addition, we provide the first evidence that gustavus has a conserved regulatory role in embryonic Vasa expression, as it appears to have analogous functions to promote Vasa protein stability in both S. purpuratus and Drosophila (Styhler et al., 2002; Kugler et al., 2010). Four pieces of evidence provide functional insight into the posttranslational regulation of Vasa during S. purpuratus embryonic development: 1) Vasa protein accumulates throughout the embryo following proteasome inhibition; 2) Knockdown of endogenous Gustavus protein reduces endogenous Vasa protein and reduces the frequency of Vasa protein enrichment in the small micromeres; 3) Overexpression of the Vasa-interacting domain of Gustavus (Gus B30.2/SPRY ΔSOCS) results in ectopic accumulation of Vasa protein; and 4) micromere removal abrogates this regulation and promotes the ectopic accumulation of Vasa protein throughout the embryo (Voronina et al., 2008).
Vasa enrichment in the small micromeres
While the results presented here suggest Gustavus generally functions as a positive regulator of Vasa protein, additional regulation likely contributes to Vasa enrichment in the small micromeres. For example, the enhanced enrichment of Vasa protein in the small micromeres following proteasome inhibition with MG132 indicates that Vasa is also subject to degradation in the small micromeres. A similar increase in Vasa protein following GusΔSOCS overexpression, along with the decreased frequency of small micromere enrichment following Gustavus knockdown, is consistent with Gustavus functioning as a positive regulator in small micromeres to promote Vasa enrichment. Moreover, the accumulation of Gustavus in the vegetal cortex of the forming micromere suggests that Gustavus presence is not sufficient to degrade Vasa. Finally, it is also possible that the role of Gustavus in Vasa protein regulation may change during development or between different cell types. Western blot analysis shows two distinct bands detected by Gustavus-specific antibodies from cleavage through 16-cell embryo (Figure 3D). However, further work is required to determine whether these represent two functionally distinct Gustavus protein species and how their regulation may change upon removal of the micromeres when Vasa accumulates throughout the embryo.
Conservation of a bipartite Gustavus-Vasa interaction
Extensive structural and biochemical studies of Drosophila show Gustavus is an E3 ligase substrate specificity receptor. Gustavus can form a complex with Elongins B and C through an interaction with the SOCS box in vitro while maintaining its Vasa binding properties (Woo et al., 2006a). In vitro binding analysis identified a 30 amino acid stretch of Drosophila Vasa sufficient to bind Gustavus with an affinity similar to full length Vasa. Deletion of these residues abolished the Vasa-Gustavus interaction in a yeast two-hybrid analysis (Styhler et al., 2002). Comparison to other B30.2/SPRY domain proteins predicted two binding surfaces on Gustavus and a mutational analysis identified 4 residues on the B30.2/SPRY domain essential for binding the 30 amino acid Vasa peptide, thus demonstrating that it interacts with surface A on Gustavus (Woo et al., 2006a; Figure 4C). A subsequent atomic structure and biochemical study narrowed the minimal peptide sufficient for interaction with Gustavus to a DINNN sequence in a 20 amino acid Vasa peptide. This binds as a linear motif to a rigid preformed binding pocket on surface A of the B30.2/SPRY domain and argues against an induced-fit binding mechanism. The Asn residues at positions 3-5 in the essential residues are completely buried in the Gustavus binding pocket and contribute most to the high binding affinity (Woo et al., 2006b). However, DINNN peptides did not efficiently compete for the Vasa-Gustavus interaction in co-immunoprecipitation assays, suggesting that Drosophila Gustavus makes additional contacts with Vasa outside of the DINNN motif (Kugler et al., 2010).
A direct Vasa-Gustavus protein interaction is conserved in sea urchins, but several features of this interaction are different than in Drosophila. Despite a complete conservation of Gustavus residues shown to directly contact the minimal Drosophila Vasa DINNN sequence, no corresponding Vasa sequence exists in S. purpuratus. Further, vasa from other organisms also lacks a DINNN motif (see Gustafson and Wessel, 2010), even though gustavus has strong sequence conservation. Our data suggest Vasa interacts with Gustavus through both its N-terminal region and its DEAD box domain independently while displaying no interaction in its C-terminal region (Figure 4C). S. purpuratus Gustavus Arg 146 resides on binding surface A, as described for Drosophila Gustavus, and an R146W mutant abolishes its interaction with the Vasa N-terminal region while maintaining its interaction with the DEAD box domain. This suggests that Gustavus may bind a sequence functionally analogous to DINNN in the Vasa N-terminal region through surface A whereas Gustavus may interact with the DEAD-box through a separate binding surface (Figure 4C). The glycine-rich composition of S. purpuratus Vasa N-terminal region is indicative of a flexible structure, which potentially could wrap around the B30.2/SPRY domain and accommodate a multi-contact binding model (Figure 4C).
Our data also suggests this bipartite Gustavus-Vasa binding mechanism contributes to Vasa protein stability in vivo. Deletion analysis of Vasa (Figure 1) shows that both of the Gustavus-interacting portions of the Vasa open reading frame (N-term and DEAD) are required for its small micromere enrichment. While the Vasa C-terminal portion is required for its small micromere enrichment, in vitro binding analyses indicate it does not interact with Gustavus (Figures 1 and 4). The Vasa C-terminal portion may be a target of a Gustavus independent regulation or a potential site of ubiquitylation.
Subcellular localization of Vasa may require zinc-fingers
The perinuclear granule localization of Vasa is reminiscent of the germ line-associated ribonucleoprotein (RNP) rich structures nuage, the mitochondrial cloud, polar granules, P-granules, chromatoid bodies and somatic cell miRNA processing bodies (P-bodies) found in other animals (Eddy, 1975). While the various names of these RNP rich structures correspond to differences in morphology, composition and animals in which they were first identified, it is believed they are functionally related entities. Indeed, several of the proteins identified in nuage structures function in mRNA regulation (Seydoux and Braun, 2006). Moreover, Vasa homologs localize to such perinuclear structures in all animals studied to date including examples both in germ line and somatic cells (Parvinen, 2005; Seydoux and Braun, 2006). Ectopic perinuclear localization of the Vasa 2F construct in non-small micromere cells shows that separate mechanisms are responsible for Vasa small micromere enrichment and Vasa subcellular localization (Figure 1B). These results imply that this portion of Vasa can ectopically nucleate a nuage-like structure. Further analyses help address the nature of these structures and Vasa’s role in their assembly. These results also suggest that, while the portion of the Vasa ORF N-terminal to the CCHC Zn-knuckles is dispensable for is nuage localization, it is required for its small micromere accumulation. Together, these results show there are several posttranslational aspects of Vasa regulation in S. purpuratus embryonic development.
Supplementary Material
Supplementary Figure 1. Identification and phylogenetic analysis of the Strongylocentrotus purpuratus gustavus homolog reveals high conservation across animal taxa. (A) Amino acid sequence alignment of several putative gustavus homologs identified by BLAST analysis of D. melanogaster gustavus (NP_724402.1). Sequences from vertebrates Mus musculus SSB-1 (NP_083311), SSB-4 (NP_660116), Homo sapiens SSB-1 (NP_079382), SSB-4 (NP_543138), Danio rerio SSB-1 (NP_001020631) and SSB-4 (XP_686153); a cephalochordate Branchiostoma floridae (EEN66863.1); cnidarians Nematostella vectensis (XP_001633549.1) and Hydra magnipapillata (XP_002160204.1); a tunicate Ciona intestinalis (NP_001027614.1); a crustacean Caligus clemensi (ACO15582.1) and a nematode Caenorhabditis elegans (NP_497320.2) are represented. Blue and purple bands correspond to B30.2/SPRY and SOCS box domains respectively. Drosophila gustavus residues that form a rigid binding pocket and directly interact with the DmVasa peptide are indicated by black diamonds. Orange-highlighted Sp-gustavus sequence corresponds to the peptide used for affinity purification of the anti-Gustavus antibodies. (B) Domain structure of S. purpuratus gustavus and comparative domain sequence identity to gustavus orthologs.
Table 1.
Plasmid Construction Primers
| Construct | Forward Primer* | Reverse Primer* | Plasmid |
|---|---|---|---|
| Vasa1F-GFP | GGGCCCATGTCAGAAGACTGGGGG | ACTAGTATCCCATGATTCATCATCAGCAGCTCC | pSP6 β-globin UTRs |
| Vasa2F-GFP | GGGCCCATGTCAGGAGATAGATCCTGTTATAAC | ACTAGTATCCCATGATTCATCATCAGCAGCTCC | pSP6 β-globin UTRs |
| Vasa4F-GFP | GGGCCCATGTCAGGTGGAAAGTGCTTTAGATGTC | ACTAGTATTCCCATGATTCATCATCAGCAGCTCC | pSP6 β-globin UTRs |
| Vasa6F-GFP | GGGCCCATGTCACTTCTCAGTCAATCTGAATTCC | ACTAGTATCCCATGATTCATCATCAGCAGCTCC | pSP6 β-globin UTRs |
| Vasa3R-GFP | GGGCCCATGTCAGAAGACTGGGGG | ACTAGTGTTTCCGACACGTCCGGTACG | pSP6 β-globin UTRs |
| Vasa4R-GF | GGGCCCATGTCAGAAGACTGGGGG | ACTAGTCCCTGCCTTCTCCTGGACCTCATTG | pSP6 β-globin UTRs |
| Vasa5R-GFP | GGGCCCATGTCAGAAGACTGGGGG | ACTAGTATCTGGATTGGGGCATTCCTTGGC | pSP6 β-globin UTRs |
|
| |||
| Nterm | AAGCTTATGTCAGAAGACTGGGGGAC | CTCGAGTACCATACTTCTGCACAGGTGTTG | pT7-MAT-Flag-2 |
| DEAD | AAGCTTACACCTGTGCAGAAGTATGGTATGC | CTCGAGTCAGGAAGAGGTAGTCGTTGAG | pT7-MAT-Flag-2 |
| Cterm | AAGCTTCTCAACGACTACCTCTTCCTGACG | CTCGAGAACTAGTATCCCATGATTCATCATCAGC | pT7-MAT-Flag-2 |
|
| |||
| EGFP | ACTAGTATGGTGAGCAAGGGCGAGGAG | ACTAGTTACTTGTACAGCTCGTCCATGCCG | pSP6 β-globin UTRs |
|
| |||
| Gus Full-length | ATGGGGCAGAAAGTGTCCGG | TCACTGAAAGAGGAGGTATTTTTTCACC | pGEM-T Easy |
|
| |||
| Gus R146W | ATGGGGCAGAAAGTGTCCGG CGTGGGGCTGGGACTTGGGATGGAAGAAGCTCTTCCACGACTC |
GAGTCGTGGAAGAGCTTCTTCCATCCCAAGTCCCAGCCCCACG TCACTGAAAGAGGAGGTATTTTTTCACC |
pGEM-T Easy |
|
| |||
| GusΔSOCS GusDSOCS |
GGATCCTTATGGGGCAGAAAGTGTCC GGGCCCATGGGGCAGAAAGTGTCC |
GAATTCTCATGAAGGTTCTAGGCCTCCAACATATTTG ACTAGTTCACTGAAAGAGGAGGTATTTTTTCACC |
pGEX-5x-3 pSP6 β-globin UTRs |
All primers are shown in the 5′ to 3′ direction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–43. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
- Braat AK, van de Water S, Goos H, Bogerd J, Zivkovic D. Vasa protein expression and localization in the zebrafish. Mech Dev. 2000;95:271–4. doi: 10.1016/s0925-4773(00)00344-0. [DOI] [PubMed] [Google Scholar]
- Bruskin AM, Tyner AL, Wells DE, Showman RM, Klein WH. Accumulation in embryogenesis of five mRNAs enriched in the ectoderm of the sea urchin pluteus. Dev Biol. 1981;87:308–18. doi: 10.1016/0012-1606(81)90154-8. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–62. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–84. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Polycomb group gene expression in the sea urchin. Dev Dyn. 2008;237:1851–61. doi: 10.1002/dvdy.21582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays. 2010;32:626–37. doi: 10.1002/bies.201000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–87. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109:425–33. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- Ikenishi K, Tanaka TS. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ. 1997;39:625–33. doi: 10.1046/j.1440-169x.1997.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–15. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Yajima M, Wessel GM. Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev Biol. 2010;337:220–32. doi: 10.1016/j.ydbio.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–88. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C. An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol. 2002;12:454–66. doi: 10.1016/s0960-9822(02)00723-6. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–37. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Kugler JM, Woo JS, Oh BH, Lasko P. Regulation of Drosophila Vasa in vivo through paralogous Cullin-RING E3 ligase specificity receptors. Mol Cell Biol. 2010 doi: 10.1128/MCB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000;127:2907–16. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- Laidlaw M, Wessel GM. Cortical granule biogenesis is active throughout oogenesis in sea urchins. Development. 1994;120:1325–33. doi: 10.1242/dev.120.5.1325. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–7. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4:905–21. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Lavial F, Acloque H, Bachelard E, Nieto MA, Samarut J, Pain B. Ectopic expression of Cvh (Chicken Vasa homologue) mediates the reprogramming of chicken embryonic stem cells to a germ cell fate. Dev Biol. 2009;330:73–82. doi: 10.1016/j.ydbio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–11. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Liu N, Dansereau DA, Lasko P. Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation. Curr Biol. 2003;13:1905–9. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Noce T, Okamoto-Ito S, Tsunekawa N. Vasa homolog genes in mammalian germ cell development. Cell Struct Funct. 2001;26:131–6. doi: 10.1247/csf.26.131. [DOI] [PubMed] [Google Scholar]
- Ohashi H, Umeda N, Hirazawa N, Ozaki Y, Miura C, Miura T. Expression of vasa (vas)-related genes in germ cells and specific interference with gene functions by double-stranded RNA in the monogenean, Neobenedenia girellae. Int J Parasitol. 2007;37:515–23. doi: 10.1016/j.ijpara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Ozhan-Kizil G, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2009;327:230–9. doi: 10.1016/j.ydbio.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319:146–59. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Piessevaux J, Lavens D, Peelman F, Tavernier J. The many faces of the SOCS box. Cytokine Growth Factor Rev. 2008;19:371–81. doi: 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-3-reviews1017. REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306:599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–8. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, Strome S. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics. 2008;178:1973–87. doi: 10.1534/genetics.107.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–76. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–78. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Koh E, Sin HS, Maeda Y, Narimoto K, Izumi K, Kobori Y, Kitamura E, Nagase H, Yoshida A, Namiki M. Tissue-specific differentially methylated regions of the human VASA gene are potentially associated with maturation arrest phenotype in the testis. J Hum Genet. 2009;54:450–6. doi: 10.1038/jhg.2009.59. [DOI] [PubMed] [Google Scholar]
- Sunanaga T, Saito Y, Kawamura K. Postembryonic epigenesis of Vasa-positive germ cells from aggregated hemoblasts in the colonial ascidian, Botryllus primigenus. Dev Growth Differ. 2006;48:87–100. doi: 10.1111/j.1440-169X.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- Sunanaga T, Watanabe A, Kawamura K. Involvement of vasa homolog in germline recruitment from coelomic stem cells in budding tunicates. Dev Genes Evol. 2007;217:1–11. doi: 10.1007/s00427-006-0112-5. [DOI] [PubMed] [Google Scholar]
- Takamura K, Fujimura M, Yamaguchi Y. Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev Genes Evol. 2002;212:11–8. doi: 10.1007/s00427-001-0204-1. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125:1723–32. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–6. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–86. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Marzluff WF, Wessel GM. Cyclin B synthesis is required for sea urchin oocyte maturation. Dev Biol. 2003;256:258–75. doi: 10.1016/s0012-1606(02)00134-3. [DOI] [PubMed] [Google Scholar]
- Wolke U, Weidinger G, Koprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289–94. doi: 10.1016/s0960-9822(02)00679-6. [DOI] [PubMed] [Google Scholar]
- Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 2006a;25:1353–63. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006b;24:967–76. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–65. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Identification and phylogenetic analysis of the Strongylocentrotus purpuratus gustavus homolog reveals high conservation across animal taxa. (A) Amino acid sequence alignment of several putative gustavus homologs identified by BLAST analysis of D. melanogaster gustavus (NP_724402.1). Sequences from vertebrates Mus musculus SSB-1 (NP_083311), SSB-4 (NP_660116), Homo sapiens SSB-1 (NP_079382), SSB-4 (NP_543138), Danio rerio SSB-1 (NP_001020631) and SSB-4 (XP_686153); a cephalochordate Branchiostoma floridae (EEN66863.1); cnidarians Nematostella vectensis (XP_001633549.1) and Hydra magnipapillata (XP_002160204.1); a tunicate Ciona intestinalis (NP_001027614.1); a crustacean Caligus clemensi (ACO15582.1) and a nematode Caenorhabditis elegans (NP_497320.2) are represented. Blue and purple bands correspond to B30.2/SPRY and SOCS box domains respectively. Drosophila gustavus residues that form a rigid binding pocket and directly interact with the DmVasa peptide are indicated by black diamonds. Orange-highlighted Sp-gustavus sequence corresponds to the peptide used for affinity purification of the anti-Gustavus antibodies. (B) Domain structure of S. purpuratus gustavus and comparative domain sequence identity to gustavus orthologs.