SUMMARY
The Cip/Kip CDK inhibitor (CKI) p21Cip1/WAF1 has a critical role in the nucleus to limit cell proliferation by inhibiting CDK-cyclin complexes. In contrast, cytoplasmic p21 regulates cell survival and the actin cytoskeleton. These divergent functions for p21 in different cellular compartments suggest the necessity for complex regulation. In this study, we identify the CRL2LRR-1 ubiquitin ligase as a conserved regulator of Cip/Kip CKIs that promotes the degradation of C. elegans CKI-1 and human p21. The nematode CRL2LRR-1 complex negatively regulates nuclear CKI-1 levels to ensure G1-phase cell cycle progression in germ cells. In contrast, human CRL2LRR1 targets cytoplasmic p21, acting as a critical regulator of cell motility that promotes a non-motile, stationary cell state by preventing p21 from inhibiting the Rho/ROCK/LIMK pathway. Inactivation of human CRL2LRR1 leads to the activation of the actin-depolymerizing protein cofilin, dramatic reorganization of the actin cytoskeleton, and increased cell motility.
Keywords: cell motility, actin cytoskeleton, ubiquitin ligase, cullin, CDK inhibitor, cell cycle
INTRODUCTION
Cell migration underpins fundamental processes in animal development and homeostasis. Defects in cell movement are associated with pathological processes such as vascular disease and tumor metastasis (Jaffe and Hall, 2005; Narumiya et al., 2009; Ridley et al., 2003; Shimokawa and Rashid, 2007). Cell migration involves the remodeling of the actin cytoskeleton, which occurs through the coordination of two opposing activities: actin depolymerization mediated by ADF/cofilin proteins, and actin polymerization catalyzed by actin nucleating and polymerization-promoting factors, such as Arp2/3 and formins (Jaffe and Hall, 2005; Ridley, 2006).
Small GTPases of the Rho family, Rho, Rac, and Cdc42, are key regulators of the actin cytoskeleton (Jaffe and Hall, 2005; Ridley et al., 2003). Cdc42 establishes cell polarity and is responsible for the formation of filopodia. Rac is required for the formation of lamellipodia and promotes cell spreading. RhoA, B, and C, collectively referred to here as Rho, induce the formation of actin stress fibers and focal adhesions, in part through the activation of formins (Jaffe and Hall, 2005; Ridley, 2006). In non-migrating cells, Rho maintains cell shape and the attachment to the substrate (Nobes and Hall, 1999). Rho regulates the actin cytoskeleton in part by activating the Rho-associated kinases ROCK I and II (referred to here as ROCK) (Narumiya et al., 2009), which in turn affects a number of ROCK effectors, of which a central effector is LIM kinase (LIMK) (Schmandke and Strittmatter, 2007). LIMK is activated by ROCK and inhibits the actin-depolymerizing protein cofilin via phosphorylation of cofilin on its serine 3 residue (Schmandke and Strittmatter, 2007). Cofilin plays a key role in cytoskeleton dynamics and cell migration by stimulating the severing and depolymerization of actin filaments, and under certain conditions can nucleate actin filaments (Bernstein and Bamburg, 2010). Cofilin’s activity is defined by its phosphorylation status, and the increased activity of the unphosphorylated cofilin is associated with enhanced cell movement and cancer metastasis (Bernstein and Bamburg, 2010; Ono, 2007; Wang et al., 2007).
High Rho activity promotes strong adhesion to the substrate and inhibits cell migration (Nobes and Hall, 1999; Pellegrin and Mellor, 2007). Cell movement is associated with the gradient-wise downregulation of Rho and the activation of Rac at the leading edge, while Rho activity is required for tail retraction in the migrating cell (Ridley et al., 2003). The Rho-family GTPases thus require tight spatial and temporal regulation for proper actin remodeling as cells transition between stationary and motile states.
Members of the mammalian Cip/Kip family of cyclin-dependent kinase (CDK)-inhibitors (CKIs), p21Cip1, p27Kip1, and p57Kip2, have the well-known function to regulate the cell cycle by inhibiting a broad range of CDK-cyclin complexes (Sherr and Roberts, 1999). Cip/Kip CKIs bind CDK-cyclin complexes in the nucleus, and elevated nuclear CKI levels inhibit cell division (Abukhdeir and Park, 2008; Bornstein et al., 2003; Gu et al., 1993; Harper et al., 1993). The CKIs function as tumor suppressors, with their inactivation increasing CDK-cyclin activity that drives cell proliferation (Abukhdeir and Park, 2008; Besson et al., 2004a). However, elevated levels of cytoplasmic p21 and p27 have been observed in a number of human cancers and are associated with high tumor grade and poor prognosis (Abukhdeir and Park, 2008; Besson et al., 2004a; Besson et al., 2008; Blagosklonny, 2002). This contradiction between the tumor suppressor function of the CKIs and their elevated cytoplasmic levels in aggressive cancers suggested that the CKIs have an alternate function in the cytoplasm. Two major functions have been assigned to cytoplasmic CKIs: anti-apoptotic activity and the regulation of the actin cytoskeleton (Besson et al., 2008). In particular, p21, p27, and p57 each negatively regulates the Rho/ROCK/LIMK pathway in the cytoplasm. p27 inhibits Rho, p21 inhibits ROCK, and p57 inactivates LIMK (Besson et al., 2004b; Lee and Helfman, 2004; McAllister et al., 2003; Tanaka et al., 2002; Yokoo et al., 2003).
The accumulation of cytoplasmic p21 and p27 is linked to the oncogenic transformation of cells by activated Ras, HER2/neu, or ErbB2 (Lee and Helfman, 2004; Liu et al., 2000; Xia et al., 2004). Cytoplasmic p21 levels are also elevated during developmental events, including the differentiation of monocytes and fibroblasts, as well as axon regeneration and neurite extension (Asada et al., 1999; Lee and Helfman, 2004; Manapov et al., 2005; Tanaka et al., 2002). These findings suggest that the cytoplasmic localization of CKIs is under complex regulation, however, the mechanisms of such regulation are poorly understood.
In this study, we identify a ubiquitin ligase (E3) that specifically regulates cytoplasmic p21 levels in human cells: the cullin 2-RING ubiquitin ligase (CRL2) complex. CRL2 complexes are multisubunit E3s that contain five components: the scaffold protein CUL2; a RING H2 protein (Rbx1/Roc1) that binds the ubiquitin conjugating enzyme; the adaptor Elongin C that is in complex with Elongin B; and a variable substrate-recognition subunit (SRS) that links to the complex through interaction with Elongin C (Bosu and Kipreos, 2008). The SRS determines the substrate specificity of the CRL complex, and thereby its associated cellular functions.
Here we show that the leucine-rich repeat protein LRR-1 functions as the SRS for a CRL2LRR-1 complex that targets the degradation of the Cip/Kip CDK-inhibitor CKI-1 in C. elegans to promote cell cycle progression in germ cells. We further show that the orthologous human CRL2LRR1 complex has a conserved function in human cells, where it mediates the degradation of the CDK-inhibitor p21Cip1. However, in human cells, the degradation of p21 by CRL2LRR1 does not appreciably affect cell cycle progression. Rather, human CRL2LRR1 targets the degradation of p21 in the cytoplasm to prevent the inhibition of the Rho/ROCK/LIMK pathway. Inactivation of LRR1 results in the activation of cofilin, the remodeling of the actin cytoskeleton, and increased cell motility in both normal and cancer cells. The human CRL2LRR1 complex is therefore a central regulator of actin-based cell movement.
RESULTS
C. elegans LRR-1 is the substrate-recognition subunit for a CRL2 complex
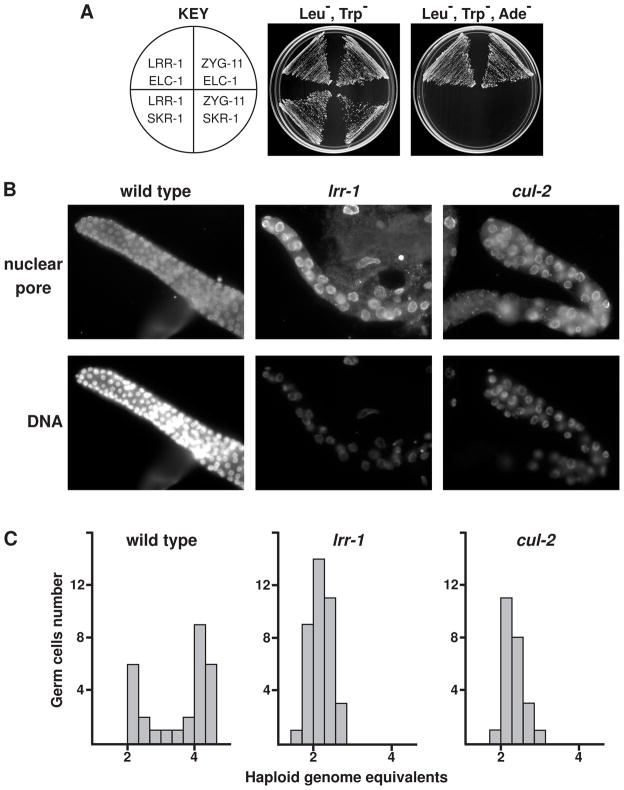
In order to identify proteins that interact with CUL-2, we performed affinity purification of CUL-2-FLAG protein expressed in C. elegans, followed by mass spectrometry. This analysis identified the core CRL2 components Rbx1 (RBX-1), Elongin C (ELC-1), and Elongin B (ELB-1), as well as three SRSs that we had previously identified: FEM-1, ZYG-11, and ZER-1 (Starostina et al., 2007; Vasudevan et al., 2007). The leucine-rich repeat protein 1 (LRR-1) also co-purified with CUL-2-FLAG (see Supplemental Information for details). To test whether LRR-1 functions as an SRS for a C. elegans CRL2 complex, we analyzed two-hybrid interactions between LRR-1 and the CRL2 adaptor ELC-1, which binds SRSs to the complex. LRR-1 interacted with ELC-1 to the same extent as the known SRS ZYG-11, but neither LRR-1 nor ZYG-11 interacted with an adaptor for the SCF complex, as expected (Fig. 1A). Our results are in agreement with the finding that the mammalian ortholog of LRR-1, LRR1, interacts with CUL2 as a putative SRS, although no function(s) have been reported for this complex (Kamura et al., 2004).
Figure 1.
C. elegans LRR-1 interacts with the CRL2 adaptor ELC-1, and lrr-1 mutants share a germ cell mutant phenotype with cul-2 mutants. (A) Yeast two-hybrid analysis revealed that LRR-1 binds to the CRL2 adaptor protein ELC-1, similar to the known CRL2 SRS ZYG-11, whereas neither LRR-1 nor ZYG-11 binds the SCF adaptor SKR-1. (B) Gonads dissected from young adult lrr-1 mutant, cul-2 mutant, or wild-type hermaphrodites, stained with Hoechst and anti-nuclear-pore antibody. (C) Histogram of DNA content in germ cells with 2C and 4C DNA content labeled on the x-axis.
To explore LRR-1 function, we analyzed the recessive lrr-1(tm3543) deletion null allele, which is predicted to generate a truncated LRR-1 protein that lacks 66% of the C-terminal residues. lrr-1 is an essential gene (Piano et al., 2002), and the lrr-1(tm3543) allele cannot be maintained as a homozygous strain. lrr-1(tm3543) heterozygotes appear overtly wild type, while homozygous lrr-1 mutant progeny from lrr-1 heterozygote parents develop to become sterile adults. These animals have a protruding vulva defect derived from a failure to produce the full complement of vulva cells [15.0 ± 0.6 (SEM) vulva cells in lrr-1 mutants versus 22.0 ± 0.0 in lrr-1 heterozygotes or wild type animals (n=10 for each)]. In contrast to the deficit of vulval cells, lrr-1 mutants have the normal number of vulval muscle cells [4.0 ± 0.2 (SD) per lateral side for lrr-1 mutants, n=38, and 4.0 ± 0.0 for both lrr-1 heterozygotes and wild type, n=24], and exhibit a modest increase in the number of epidermal seam cells [17.6 ± 0.5 (SEM) in lrr-1 mutants vs. 16.0 ± 0.0 in wild type, n=20]. The few extra seam cells in lrr-1 mutants often have less DNA than normal seam cells, suggesting a cell cycle defect (data not shown).
The sterility in lrr-1(tm3543) mutants or lrr-1(RNAi) animals is linked to a striking germline defect that is similar to that of cul-2 mutants: a reduced number and enlarged size of germ cells (Fig. 1B). Germ cell DNA content was analyzed to determine the cell cycle stage of the arrested cells. Wild-type germ cells demonstrate a bimodal distribution with peaks at 2C and 4C DNA content, corresponding to G1 and G2/M cell cycle phases, respectively, while lrr-1 and cul-2 mutant germ cells have only a single peak at 2C (Fig. 1C). This indicates that LRR-1, like CUL-2, is required for the G1-to-S-phase transition in C. elegans germ cells.
C. elegans CRL2LRR-1 negatively regulates CKI-1 levels in germ cells
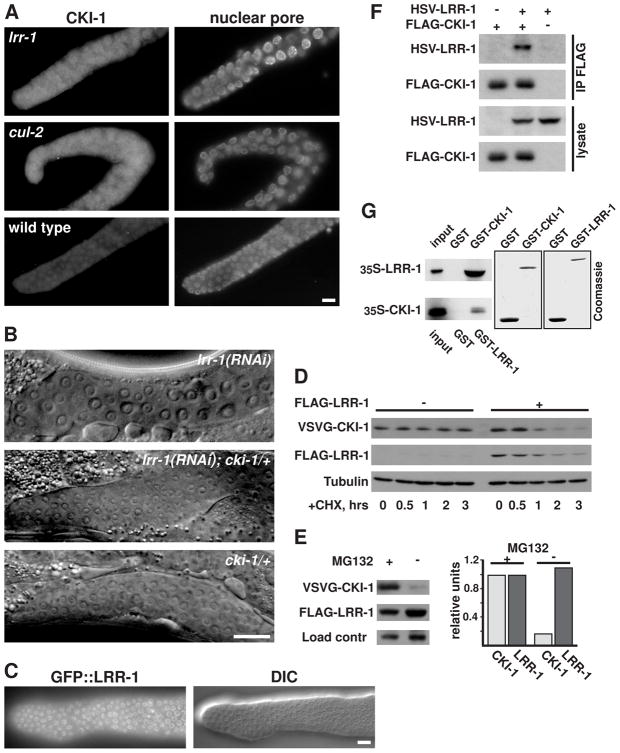
Next, we attempted to identify the critical substrate for the CRL2LRR-1 complex. Our previous work pointed to CKI-1 as the downstream effector for CUL-2-regulated G1-phase progression in germ cells (Feng et al., 1999). CKI-1 is a CDK-inhibitor of the Cip/Kip family, which negatively regulates cell cycle progression in C. elegans (Feng et al., 1999; Hong et al., 1998). We had previously found that G1-phase arrest in cul-2 mutant germ cells correlates with an accumulation of CKI-1 protein in the distal mitotic region of the gonad (Feng et al., 1999). We observed a similar accumulation of CKI-1 in the distal nuclei of lrr-1 mutant germ cells (Fig. 2A). The CKI-1 signal per germ cell nucleus was 13 times higher in lrr-1 mutants than in wild type (13.1 ± 4.2, n=71 vs. 1.0 ± 0.2, n=41, arbitrary units), and similar to the level observed in cul-2 mutants (11.6 ± 2.0 a.u., n=29). We observed that GFP::LRR-1 localized to nuclei in germ cells, which is consistent with a role in regulating nuclear-localized CKI-1 (Fig. 2C).
Figure 2.
C. elegans LRR-1 negatively regulates CKI-1 levels in germ cells and stimulates the proteasome-mediated degradation of CKI-1 when expressed in human cells. (A) Gonads dissected from young adult lrr-1 mutant, cul-2 mutant, or wild-type hermaphrodites, stained with anti-CKI-1 and anti-nuclear pore antibodies. (B) Genetic interaction between lrr-1 and cki-1. Differential-interference contrast (DIC) images of the distal gonad of wild type or cki-1/+ young adults treated with lrr-1 RNAi, and cki-1/+ young adults with no RNAi treatment (the latter is indistinguishable from that of wild type). The distal end of the gonad is to the left. In this experiment (out of three experiments with similar results), 16/16 (100%) of lrr-1(RNAi); cki-1/+ animals examined had germ cells of normal size, while 16/16 lrr-1(RNAi) animals had fewer, enlarged germ cells, and 7/7 cki-1/+ animals had germ cells of normal size. (C) Fluorescence and DIC micrographs of the distal tip of a dissected gonad from an adult that expresses GFP::LRR-1 translational fusion under the control of the germline-specific pie-1 promoter. Scale bars in A-C are 10 μm. (D) Pulse-chase experiment showing increased turnover of CKI-1 protein in the presence of LRR-1. VSVG-CKI-1 was expressed in HEK293T cells, in the presence or absence of FLAG-LRR-1, and in the presence of MG132 proteasome inhibitor. At time 0, MG132 was removed, and protein synthesis was blocked by cycloheximide (CHX). Cells were harvested at the indicated time points and analyzed by western blot with the indicated antibodies. (E) Proteasome inactivation blocks the LRR-1-mediated destabilization of CKI-1. VSVG-CKI-1 and FLAG-LRR-1 were co-expressed in HEK293T cells in the presence or absence of MG132, and cell lysates were analyzed by western blotting with the indicated antibodies. A cross-reacting band served as a loading control. (F) HSV-LRR-1 co-precipitates with FLAG-CKI-1 when co-expressed in HEK293T cells. Immunoprecipitated proteins were eluted from the beads with 0.4 mg/ml FLAG peptide and then analyzed by western blot with the indicated antibodies. (G) LRR-1 and CKI-1 bind each other in vitro. 35S-labeled in vitro translated LRR-1 or CKI-1 proteins were incubated with GST, GST-CKI-1, or GST-LRR-1 purified on glutathione Sepharose beads, the beads were washed and analyzed by SDS-PAGE/autoradiography. Input is 10% of the in vitro translated protein used in the binding reactions. The two right panels are Coomassie-stained gels showing the relative amounts of GST and GST-fusion proteins used in the reactions.
We previously showed that the G1 phase arrest of cul-2 mutant germ cells is partially suppressed by the deletion of one chromosomal copy of the cki-1 gene, suggesting that the arrest results from the increased levels of CKI-1 (Feng et al., 1999). We observed a similar effect of reducing CKI-1 levels on the number of germ cells in lrr-1 deficient animals. While wild-type animals treated with lrr-1 RNAi had germ cells that were larger and fewer in number, the germ cells of cki-1(gk132) heterozygotes exposed to lrr-1 RNAi were similar to those of wild-type animals (Fig. 2B and Fig. 2 legend). This suppression of the germ cell arrest by removal of one copy of CKI-1 suggests that the accumulation of CKI-1 mediates the G1 arrest observed in lrr-1 mutants. This conclusion was supported by the observation that cki-1 feeding-RNAi treatment can rescue lrr-1 mutant germ cells numbers from 76.2 ± 9.4 with no RNAi treatment to 220.8 ± 20.1 with the cki-1 RNAi, n=10 for each. The cki-1 feeding-RNAi is only modestly effective at inactivating CKI-1, as determined by a lack of hyperplasia in somatic tissues of wild type animals. Interestingly, we observed that more fully inactivating CKI-1 by injecting cki-1 dsRNA into lrr-1 heterozygote parents produced a genetic enhancement in which lrr-1 mutants had reduced numbers of post-embryonically dividing somatic cells, including seam cells and vulval cells (data not shown). CRLs generally have multiple cellular functions, and this observation suggests that the complete absence of CKI-1 is incompatible with the loss of a distinct CRL2LRR-1 function.
Because CKI-1 accumulates in germ cells of lrr-1 mutants, we tested whether CKI-1 is a direct target of the CRL2LRR-1 complex. We observed that CKI-1 protein had increased turnover when co-expressed with the LRR-1 protein in HEK293T cells, as would be expected for an E3 substrate (Fig. 2D). The level of CKI-1 was restored by treatment with the proteasome inhibitor MG132, indicating that the degradation required the proteasome (Fig. 2E). Moreover, LRR-1 co-precipitated with CKI-1 in human cells, and the proteins bound each other in vitro, demonstrating that the two proteins physically interact (Fig. 2F,G). Collectively, our data indicate that C. elegans LRR-1 functions as the SRS for a CUL-2-based complex that mediates the degradation of CKI-1 in germ cells to allow progression through G1 phase.
Human CRL2LRR1 targets p21Cip1 for ubiquitylation and degradation
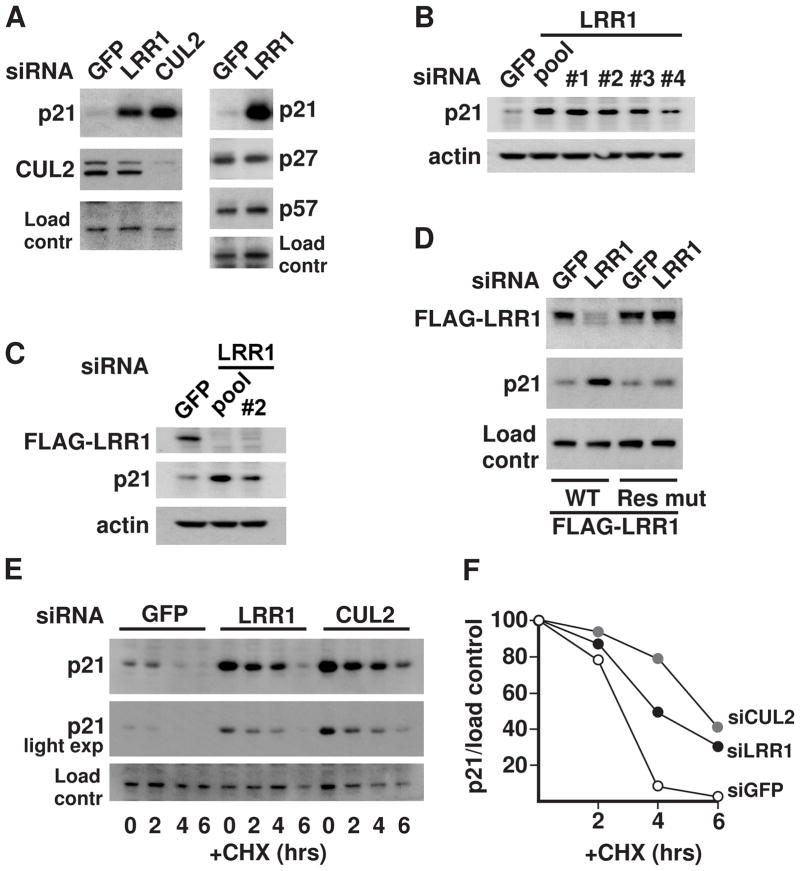
In mammals, there are three Cip/Kip family paralogs, p21Cip1, p27Kip1, and p57Kip2, that each inhibit multiple CDK-cyclin complexes (Sherr and Roberts, 1999). To determine whether LRR1 negatively regulates CDK-inhibitors in human cells, we analyzed the levels of the three Cip/Kip CKIs upon LRR1 knockdown in HeLa cells. The level of p21 increased significantly upon LRR1 knockdown, but there was no measurable effect on p27 or p57 protein levels (Fig. 3A). CUL2 knockdown also increased p21 levels to an extent comparable to that of LRR1 knockdown (Fig. 3A). While our initial experiments employed pools of LRR1 siRNAs, individual LRR1 siRNAs also produced an accumulation of p21 (Fig. 3B). The LRR1 siRNAs effectively knocked down the expression of exogenous 3xFLAG-LRR1 while producing an accumulation of endogenous p21 (Fig. 3C). Expression of an LRR1 rescue construct that is resistant to LRR1 siRNA prevented the increase in endogenous p21 level upon LRR1 siRNA treatment (Fig. 3D). The half-life of endogenous p21 protein was markedly extended in LRR1 or CUL2 knockdown cells, indicating that LRR1 and CUL2 increase the post-translational turnover of p21 (Fig. 3E,F).
Figure 3.
Endogenous p21 is negatively regulated by LRR1 and CUL2 in human cells. (A) HeLa cells were transfected with siRNA for GFP (control), LRR1, or CUL2, and the levels of the indicated proteins in whole cell lysates were assessed by western blot. (B) The efficiency of individual LRR1 siRNAs (#1 - #4) in increasing p21 levels is compared to a pool of all four individual LRR1 siRNAs (pool). (C) HeLa cells stably expressing 3xFLAG-LRR1 were transfected with pooled LRR1 siRNAs or LRR1 siRNA #2, and the levels of 3xFLAG-LRR1 were detected with anti-FLAG antibody. (D) Expression of an siRNA-resistant LRR1 rescue construct prevents the increase of p21 levels in cells treated with LRR1 siRNA. HeLa cells were co-transfected with LRR1 siRNA #1 or GFP siRNA, and with an expression construct encoding either FLAG-tagged wild-type LRR1 or a silent mutant LRR1 with 5 mismatches in the region targeted by the siRNA. (E) p21 protein turnover in LRR1 or CUL2 knockdown cells. HeLa cells were transfected with the indicated siRNAs in the presence of MG132. At time 0, MG132 was removed, and further protein synthesis was blocked by cycloheximide (CHX). Cells were collected at the indicated time points and analyzed by western blot. (F) Graph of p21 levels (standardized relative to the loading control) for the turnover rate experiment in (E). The values for each siRNA treatment were set at 100% for the time 0 values.
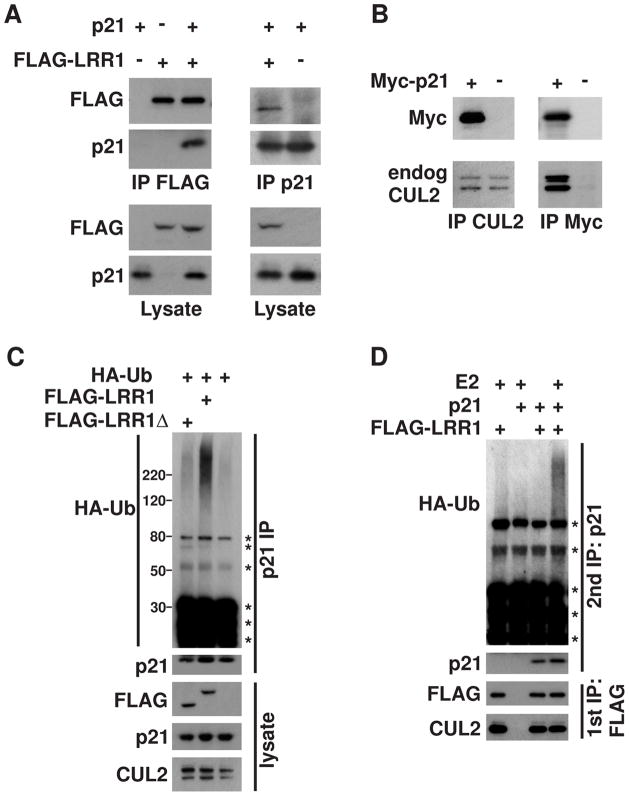
We sought to determine if the CRL2LRR1 complex directly targets p21 for degradation by testing for physical interaction between p21 and LRR1. We found that exogenous p21 and FLAG-LRR1 reciprocally co-immunoprecipitated each other in HEK293T cells (Fig. 4A). Additionally, both untagged p21 and Myc-tagged p21 were able to reciprocally bind endogenous CUL2 in HEK293T cells, providing further evidence that p21 binds to the CRL2LRR-1 complex (Fig. 4B; data not shown).
Figure 4.
LRR1 physically associates with p21 and stimulates its ubiquitylation. (A) p21 physically interacts with LRR1. p21 and FLAG-LRR1 were co-expressed in HEK293T cells as noted by (+) symbols above the lanes. Anti-p21 or anti-FLAG immunoprecipitations (IP) and lysates were analyzed by western blot with the indicated antibodies. (B) p21 interacts with endogenous CUL2. Myc-p21 was expressed in HEK293T cells, and anti-Myc or anti-CUL2 immunoprecipitations were analyzed by probing with the indicated antibodies. (C) Overexpression of LRR1 stimulates the in vivo ubiquitylation of endogenous p21 in HeLa cells. FLAG-LRR1 or FLAG-LRR1Δ (with deletion of the VHL-box motif that is required for binding Elongin C) were expressed in HeLa cells with HA-ubiquitin (noted above the lanes) in the presence of MG132. Endogenous p21 was immunoprecipitated under denaturing conditions, and analyzed by western blot with the indicated antibodies. (D) In vitro ubiquitylation of p21 by the CRL2LRR1 complex. FLAG-LRR1 and p21 were expressed in HEK293T cells (noted at the top), in the presence of MG132. The CRL2LRR1 complex was immunoprecipitated with anti-FLAG antibody (first IP), which co-immunoprecipitated p21. The CRL2LRR1 complex, with bound p21, was subjected to an in vitro ubiquitylation reaction. p21 was subsequently re-immunoprecipitated under denaturing conditions, followed by immunoblot with antibodies to detect HA-ubiquitin and p21. Asterisks mark non-specific bands.
As a final test of whether p21 is a CRL2LRR1 substrate, we determined if CRL2LRR1 ubiquitylates p21. Expression of FLAG-LRR1 in HeLa cells significantly enhanced the in vivo ubiquitylation of endogenous p21, while a FLAG-LRR1Δ mutant protein that lacks the VHL-box, so that it cannot bind to the CRL2 complex, did not stimulate p21 ubiquitylation (Fig. 4C). The CRL2LRR1 complex was also able to ubiquitylate p21 in vitro (Fig. 4D). Overall, our data indicate that the human CRL2LRR1 complex directly ubiquitylates p21 to target it for proteasome-mediated degradation.
Human CRL2LRR1 regulates p21 levels in the cytoplasm
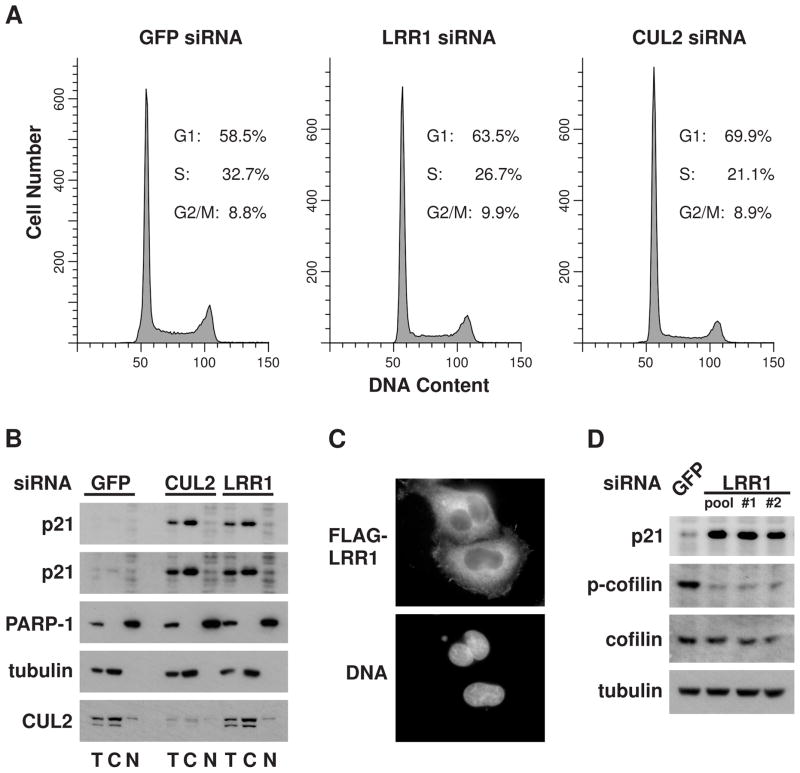
Unlike C. elegans germ cells that undergo G1 phase arrest upon inactivation of LRR-1 or CUL-2, we did not observe an obvious effect on the proliferation of human cells from knockdown of LRR1 or CUL2, despite the significantly elevated p21 levels. The cell cycle distribution of LRR1 knockdown HeLa cells was not appreciably different from control cells, while CUL2 knockdown cells had only a small increase in the percentage of G1 phase cells (Fig. 5A). Thus, the CRL2LRR1 complex does not appear to significantly affect cell cycle progression.
Figure 5.
CRL2LRR1 targets the degradation of cytoplasmic p21, which is associated with changes in the phosphorylation of cofilin. (A) Flow cytometry profiles of DNA content of HeLa cells transfected with siLRR1, siCUL2, or siGFP. Cell cycle phase distributions were determined using the ModFit program. (B) Subcellular fractionation of HeLa cells transfected with LRR1 or CUL2 siRNA. Total (T), cytoplasmic (C), or nuclear (N) fractions of cell lysates were analyzed by western blot with the indicated antibodies. A longer exposure of the anti-p21 blot (2nd panel from the top) is presented to show the p21 levels in control cells. Immunoblots of anti-α-tubulin (a cytoplasmic marker) and anti-PARP-1 (a nuclear marker) demonstrate the purity of the fractionation. (C) Cytoplasmic localization of LRR1. HeLa cells were transiently transfected with FLAG-LRR1 and examined by immunofluorescence with anti-FLAG antibody. DNA was stained with Hoechst. (D) Ser3-phosphorylation of cofilin is reduced in LRR1 knockdown cells. HeLa cells were transfected with LRR1 siRNAs (pooled or individual siRNAs) or with GFP siRNA, and whole-cell lysates were analyzed by immunoblot for p21, cofilin, and phospho-cofilin (Ser3). See also Suppl. Fig. S1.
CDK-cyclin activity and cell cycle progression is regulated by p21 that is nuclear localized (Abukhdeir and Park, 2008). In contrast, p21 localized to the cytoplasm has functions that are independent of cell cycle regulation (Besson et al., 2008). To distinguish which subcellular pool of p21 was affected by loss of the CRL2LRR1 complex, we analyzed nuclear and cytoplasmic fractions of HeLa cells. Nuclear p21 levels were not significantly increased upon LRR1 or CUL2 knockdown; however, the level of cytoplasmic p21 was markedly elevated (Fig. 5B). This elevation does not require the PIP-box motif of p21, which is required for PCNA-dependent CRL4Cdt2-mediated degradation of nuclear p21 (Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008) (Suppl. Fig. S1). Consistent with its role in regulating cytoplasmic p21, FLAG-LRR1 localizes to the cytoplasm in HeLa cells and is largely excluded from the nucleus (Fig. 5C). CUL2 is also predominantly cytoplasmic (Fig. 5B).
Human CRL2LRR1 degrades p21 to control actin cytoskeleton remodeling
Cytoplasmic p21 has been shown to bind and inhibit ROCK1 and thereby block the RhoA/ROCK/LIMK pathway from inhibiting cofilin via phosphorylation (Lee and Helfman, 2004). To determine if LRR1 inactivation produces a similar effect on the RhoA pathway, we asked whether LRR1 knockdown affected the level of cofilin phosphorylation on Ser3, the target site of LIM kinase. Indeed, phospho(Ser3)-cofilin levels (but not total cofilin protein levels) decreased significantly in LRR1 knockdown HeLa cells (Fig. 5D). Similar results were obtained upon LRR1 knockdown in the glioblastoma tumor cell line T98-G (data not shown).
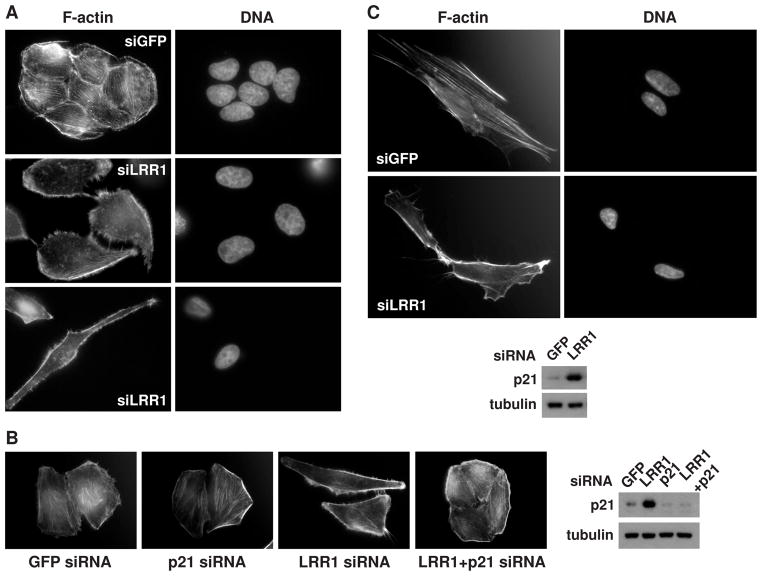
The RhoA/ROCK/LIMK pathway protects stress fibers from the depolymerizing activity of cofilin. When the pathway is inactivated, cofilin can induce the disassembly of F-actin in stress fibers. The released G-actin is used at the cell periphery for new filament formation via Rac/Arp2/3-dependent processes to form membrane protrusions (Kedrin et al., 2007; Pollard and Borisy, 2003; Ridley et al., 2003). Thus, we sought to determine if stress fibers are affected by LRR1 knockdown. We observed that LRR1 knockdown cells indeed had a marked decrease in the abundance of stress fibers, with a concomitant increase in F-actin staining at the cell periphery when compared to control cells (Fig. 6). The expression of an LRR1 silent mutant construct that is resistant to LRR1 siRNA suppressed the loss of stress fibers in HeLa cells treated with LRR1 siRNA, thereby confirming the specificity of the siRNA effect (Suppl. Table S1 and Fig. 3D).
Figure 6.
LRR1 knockdown is associated with remodeling of the actin cytoskeleton in HeLa cells and in IMR-90 normal human fetal lung fibroblasts. (A) HeLa cells transfected with LRR1 siRNA were stained with rhodamine-phalloidin to visualize the actin cytoskeleton, and with Hoechst to visualize DNA. The lower panel presents an elongated cell in the LRR1 knockdown cell population. (B) The loss of stress fibers in LRR1 knockdown HeLa cells is suppressed by co-knockdown of p21. A western blot (bottom right) demonstrates the levels of p21 in the knockdown cell populations. (C) IMR-90 cells transfected with LRR1 or GFP siRNA were stained as in (A). Western blot analysis of the cell lysate (presented below) demonstrates elevated p21 in siLRR1 treated IMR-90 cells. See also Suppl. Fig. S2 and Tables S1 and S2.
We noticed that HeLa cells treated with LRR1 siRNA did not contact each other as closely as control cells, being situated an appreciable distance from each other, while control cells were tightly associated in clusters (Fig. 6A). CUL2 knockdown HeLa cells had a similar cytoskeleton appearance and lack of cell-cell contacts (data not shown). Similar actin cytoskeleton rearrangements were also observed in T98-G glioblastoma fibroblasts treated with LRR1 or CUL2 siRNA (data not shown). The separation of cells suggests increased motility after cell division, analogous to the scattering effect (Grotegut et al., 2006).
In addition to the cytoskeleton rearrangements, we observed morphological changes upon LRR1 knockdown in HeLa cells: 19.4% (n=290 cells) possessed an abnormally elongated form with a length-to-width ratio at least three times greater than that of control cells; no such elongated cells were observed in the control culture (n=450 cells) (Fig. 6A). Similar observations were made in multiple experiments. Cell elongation is a known phenotype associated with inhibiting the RhoA pathway, arising as a consequence of actin cytoskeleton rearrangement (Arthur and Burridge, 2001).
Importantly, double knockdown of LRR1 and p21 suppressed the LRR1 knockdown phenotypes. Stress fiber abundance was restored in the cells treated with LRR1 siRNA plus p21 siRNA, to an extent comparable to control cells (Fig. 6B; Suppl. Table S2). p21 siRNA alone did not affect the abundance of stress fibers relative to control cells (Fig. 6B; Suppl. Table S2). Cell populations with double knockdown of LRR1 and p21 lacked elongated cells, and cells were predominantly in colonies with close cell contact. Thus, our data supports the model that CRL2LRR1 acts to maintain the integrity of actin stress fibers and cell morphology of stationary cells by regulating p21 levels.
Our initial experiments with LRR1 siRNA used cancer cell lines. To determine if CRL2LRR1 has a similar role in regulating the actin cytoskeleton in normal human cells, we analyzed IMR-90 normal human fetal lung fibroblasts. IMR-90 cells showed a similar elevation of p21 levels upon treatment with LRR1 siRNA or CUL2 siRNA (Fig. 6C; data not shown). LRR1 knockdown IMR-90 cells continued to actively divide, and did not have major deviations from the normal cell cycle distribution (Suppl. Fig. S2). LRR1 knockdown also decreased stress fiber abundance and increased the level of peripheral F-actin in IMR-90 cells. Significantly, the LRR1 knockdown IMR-90 cells had prominent filopodia-like actin-rich membrane protrusions that were rare in control cells (Fig. 6C). Cells of such atypical morphology comprised a significant proportion of the LRR1 knockdown cell population: 64.9% (n=111) versus 7.1% (n=113) in the control population. Therefore CRL2LRR1 regulates the actin cytoskeleton in both normal and cancer cells.
Human CRL2LRR1 regulates cell motility
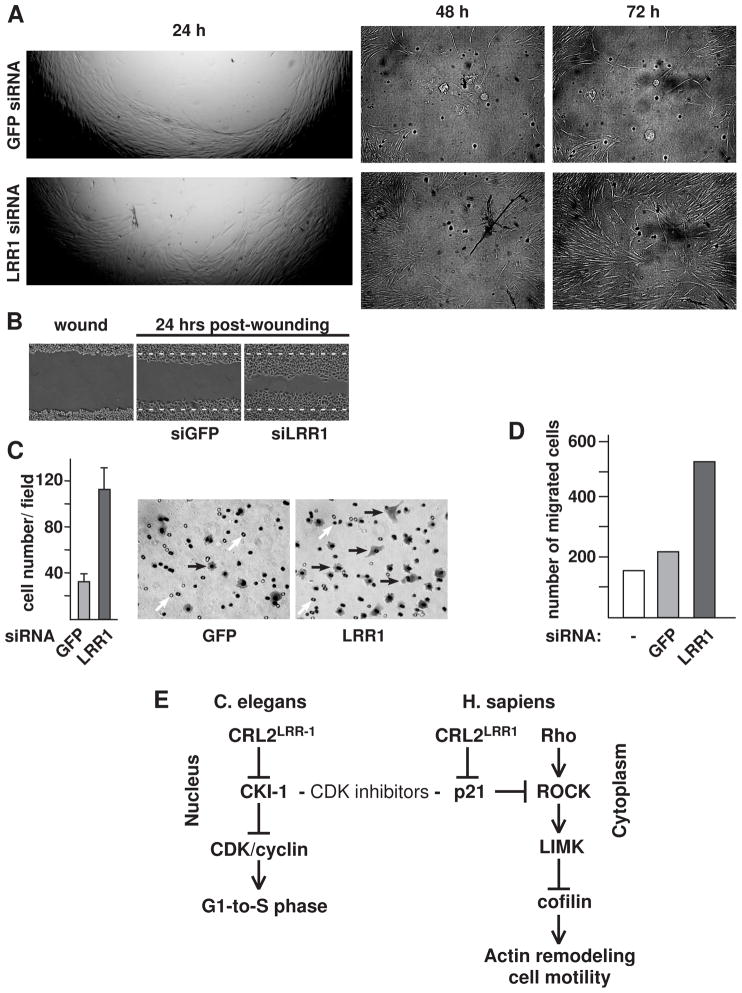
While the inhibition of the RhoA pathway is associated with increased cell motility (Ridley et al., 2003), there are no published reports on whether the accumulation of cytoplasmic p21 activates cell motility. We sought to determine if LRR1 knockdown (with its concomitant higher level of cytoplasmic p21) increases cell motility. Using a transwell assay, in which cells migrated through a membrane in response to a serum gradient, we observed a greater than 3-fold increase in the number of LRR1 knockdown HeLa cells that migrated across the membrane compared to control cells (Fig. 7C). LRR1 siRNA knockdown of IMR-90 cells also produced a 2-to-3-fold increase in motility in the transwell assay (Fig. 7D).
Figure 7.
LRR1 knockdown increases cell motility. (A) Wound-healing assay (Oris cell migration assay). IMR-90 cells were transfected with LRR1 or GFP siRNA and were placed into wells containing disks in the center for the creation of a migration detection zone. After cells reached confluency, they were treated with mitomycin C to prevent cell division, and the disks were removed to allow cells to migrate to the center of the well. The edge of the cleared area at 24 hrs post-disk removal is shown at 25× magnification, and the center of the cleared area is shown at 48 and 72 hours post-disk removal (50× magnification, DIC). (B) Wound-healing cell migration assay with LRR1 knockdown HeLa cells. Confluent HeLa control or LRR1 knockdown cells were treated with mitomycin C and cells were scraped to form a 0.8 mm-wide lane lacking cells. The images are 24 hrs post-wound healing. White dotted lines indicate the borders of the wound. (C) LRR1 knockdown cells have increased motility in a transwell assay. The graph presents the number of GFP siRNA or LRR1 siRNA treated HeLa cells that migrated across the transwell membrane in seven random field-of-views. Error bars are SD. Panels on the right are representative images of cells that migrated across the transwell membrane (Phase contrast, 200× magnification). Cells are indicated by black arrows; the filter pores are indicated by white arrows. (D) Graph of transwell cell-migration assay of IMR-90 cells, performed as described in (C). (E) Flow chart reflecting the functions of CRL2LRR1 uncovered in this study. See text for details. See also Suppl. Figs S3 and S4.
In a wound-healing assay, LRR1 knockdown IMR-90 cells (treated with mitomycin C to block proliferation) demonstrated noticeably faster migration into a circular cleared area as early as 24 hours post-wounding, and at 72 hours, the LRR1 knockdown cells had filled-in most of the previously empty area, while the area in the control cell culture was still largely empty (Fig. 7A). Analogous results were obtained with HeLa cells in the wound-healing assay with either a circular cleared area, or scratches (Fig. 7B; data not shown). LRR1 inactivation therefore stimulates the motility of both normal and cancer cells. Importantly, the increased motility of LRR1 knockdown cells was dependent on p21, as double knockdown of LRR1 and p21 suppressed the cell motility associated with LRR1 knockdown in both the transwell and wound-healing assays (Suppl. Fig. S3).
Analysis of cell migration in C. elegans lrr-1 mutants
Given the role that human LRR1 has in regulating cell motility, we asked whether cell migration was altered in C. elegans somatic cells. We analyzed the migration of three cell types: distal tip cell (DTC), sex myoblast, and touch receptor neurons. The DTC migration pattern was normal in 93% (28/30) of lrr-1 mutants, while in 2/30 lrr-1 mutants, the two DTCs migrated on the same lateral side of the animal (rather than on different lateral sides as in wild type). A similar defect was observed in 1/30 lrr-1(RNAi) animals. This defect of the gonad on the same lateral side is not observed in other DTC migration mutants (Hedgecock et al., 1987; Nishiwaki, 1999), and may reflect a defect in the shape of the initial gonad primordium rather than a migration defect. In 95% (36/38) of lrr-1 mutants, the vulva muscles were positioned normally, indicating that the precursor cell, the sex myoblast, had migrated properly. In the two cases of mispositioning, the vulva muscle cells were localized both anterior and posterior to the vulva, implying that the sex myoblasts had initially migrated properly to the mid-gonad region. Finally, we analyzed the positions of five of the six touch receptor neurons (TRNs), which undergo migration and axon projections during the embryonic and larval stages. In 10/10 lrr-1 mutants, the TRNs and their axons were normally positioned (Suppl. Fig. S4). Further, it is unlikely that there are extensive defects in neuron and axon migrations in lrr-1 mutants, as such defects produce an uncoordinated (Unc) phenotype (Hedgecock et al., 1987) while lrr-1 mutants have normal locomotion (data not shown). We therefore conclude that LRR-1 is not required for proper cell migration in C. elegans.
DISCUSSION
C. elegans CRL2LRR-1 targets the degradation of the CDK-inhibitor CKI-1 in germ cells to allow G1 phase progression
We identified LRR-1 as a protein that physically associates with CUL-2 in C. elegans. LRR-1 binds to the adaptor protein for CRL2 complexes, as expected for an SRS (Bosu and Kipreos, 2008). Both lrr-1 and cul-2 mutants share the phenotype of G1-phase cell cycle arrest in germ cells. Further, our data suggests that the accumulation of the CDK-inhibitor CKI-1 in lrr-1 and cul-2 mutants causes the G1 arrest, as the phenotype is suppressed by lowering the cki-1 gene copy number.
CKI-1 appears to be a direct substrate of the CRL2LRR-1 complex. The co-expression of LRR-1 and CKI-1 in human cells leads to the proteasome-mediated degradation of CKI-1, presumably via the incorporation of C. elegans LRR-1 into a CRL2 complex with core human CRL2 components (Starostina et al., 2007). Collectively, our data indicate that LRR-1 functions as the SRS for a CRL2LRR-1 ubiquitin ligase complex that targets CKI-1 for degradation to ensure G1-to-S-phase progression in C. elegans germ cells.
Human CRL2LRR1 targets the CDK-inhibitor p21Cip1 for degradation to regulate the actin cytoskeleton
In this work, we show that the human CRL2LRR1 complex has the conserved function of targeting a Cip/Kip CDK-inhibitor for degradation. Human LRR1 regulates the abundance of the CKI p21Cip1, but does not affect the levels of the related CKIs p27Kip1 and p57Kip2. CUL2 and LRR1 physically interact with p21, and the overexpression of LRR1 stimulates the in vivo ubiquitylation of p21. Further, the CRL2LRR1 complex ubiquitylates p21 in vitro. Thus, our data indicates that p21 is a direct substrate for human CRL2LRR1. However, the CRL2LRR1-mediated degradation of the CDK-inhibitor p21 does not appear to regulate the cell cycle.
Cytoplasmic p21 has been shown to have important cellular roles that are independent of cell-cycle regulation: it can act as an anti-apoptotic factor or as a regulator of the actin cytoskeleton. These alternative functions appear to underlie its role as an oncoprotein (Besson et al., 2008; Blagosklonny, 2002). Elevated levels of cytoplasmic p21 are associated with highly aggressive cancers (Abukhdeir and Park, 2008). Significantly, high cytoplasmic p21 levels are observed in cancers that are induced by the Ras and HER2/neu oncogenes (Lee and Helfman, 2004; Xia et al., 2004). The accumulation of cytoplasmic p21 has been linked to the loss of stress fibers in Ras transformed cells (Lee and Helfman, 2004). Elevated cytoplasmic p21 levels have been attributed in part to transcriptional stimulation (e.g. via Ras or p53) (Heliez et al., 2003; Lee and Helfman, 2004), and have been linked to Akt/PKB- or PKC-mediated phosphorylation of p21 (Child and Mann, 2006; Rodriguez-Vilarrupla et al., 2005; Zhou et al., 2001). How cytoplasmic p21 protein stability is controlled has not previously been studied, although it has been implicated to have an important role. In Ras-transformed mouse fibroblasts, the accumulation of cytoplasmic p21 requires Ras/Raf/MEK activity (to induce p21 transcription) and Ras/PI3K/AKT activity (to promote p21 cytoplasmic retention), but the inhibition of either pathway does not produce a decline in p21 levels if the proteasome is inhibited, suggesting a critical role for proteolysis (Lee and Helfman, 2004).
We present evidence for a ubiquitin ligase that specifically regulates the level of cytoplasmic p21. CRL2LRR1 is likely to target p21 only in the cytoplasm because both LRR1 and CUL2 proteins are predominantly cytoplasmically localized in HeLa cells. Cytoplasmic p21 regulates the actin cytoskeleton by binding and inhibiting the Rho-associated kinase ROCK. ROCK functions in the Rho/ROCK/LIMK pathway to activate LIM kinase, which phosphorylates cofilin on its serine 3 residue in order to inhibit cofilin. When ROCK is inhibited, the actin-depolymerization function of cofilin is activated, and the subsequent disassembly of actin filaments facilitates actin cytoskeleton remodeling (Besson et al., 2008; Lee and Helfman, 2004; Tanaka et al., 2002). In this study, we demonstrate that LRR1 inactivation causes a dramatic reduction in the inhibitory phosphorylation of cofilin (Fig. 7E).
The major cellular processes driven by cytoskeleton remodeling are morphogenesis and cell motility. The inhibition of the Rho/ROCK pathway results in loss of stress fibers, and is often associated with cell elongation (including axon extension in neurons), and with increased cell motility (Fournier et al., 2003; Kedrin et al., 2007; Ridley et al., 2003). Elevated cytoplasmic p21 has been shown to produce a loss of stress fibers in NIH3T3 fibroblasts, and to promote neurite extension in neuroblastoma cells and hippocampal neurons (Lee and Helfman, 2004; Tanaka et al., 2002). However, direct experiments showing the effect of cytoplasmic p21 on cell motility have not been reported. Here we demonstrate that in both cancer and normal human cells, the accumulation of cytoplasmic p21 (upon LRR1 inactivation) facilitates cytoskeleton rearrangement, including: loss of stress fibers; enrichment of actin filaments at the cell periphery; the formation of membrane protrusions; changes in cell morphology; and increased cell motility.
The contrast between the conservation of the CRL2LRR-1 substrate and divergence in cellular roles for CRL2LRR-1 between C. elegans germ cells and human cells is striking. This difference in function is linked to targeting different subcellular pools of the CDK inhibitor. C. elegans LRR-1 is nuclear localized in germ cells and regulates the abundance of nuclear CKI-1 to control the cell cycle, while human LRR1 is cytoplasmic localized and regulates the abundance of cytoplasmic p21 to control the actin cytoskeleton and cell motility. The cell migration events that we assessed appear to largely occur normally in C. elegans lrr-1 mutants, suggesting that LRR-1 does not regulate cell motility in nematodes. An interesting question is how the degradation of CDK inhibitors by the same E3 complex evolved to regulate such divergent functions. A corollary to this is the question of when the regulation of the cytoskeleton by Cip/Kip CKIs evolved. Currently, Cip/Kip CKIs have only been linked to the regulation of the actin cytoskeleton in mammals. It is possible that the ability of CKIs to regulate the actin cytoskeleton occurred later in evolution, and when this new function arose, an already-existing means of regulating the CKI (via CRL2LRR-1-mediated degradation) was employed to regulate this new function.
Human LRR1 is expressed in most tissues (Jang et al., 2001), and therefore LRR1-mediated regulation of the cytoskeleton and cell motility may occur in a broad range of cell types. Elevated levels of cytoplasmic p21 are correlated with a number of developmental events: the differentiation of monocytes and neurons; neurite extension and axonal regeneration; and the conversion of pancreatic myofibroblasts to fibroblasts during pancreatic fibrosis (Asada et al., 1999; Manapov et al., 2005; Tanaka et al., 2002; Tanaka et al., 2004). In non-motile cells, our work suggests that LRR1 prevents the inhibition of the Rho/ROCK/LIMK pathway by maintaining low levels of cytoplasmic p21. In cancer cells, elevated cytoplasmic p21 levels are associated with poor cancer prognosis (McBride et al., 2002; Winters et al., 2001; Xia et al., 2004). The inactivation of the CRL2LRR1 pathway may contribute to the accumulated cytoplasmic p21 levels in tumors, thereby increasing cell motility and metastasis, particularly for those cancer cells that utilize a mesenchymal type of migration (Sanz-Moreno et al., 2008; Wolf et al., 2003). Our work introduces the possibility that CRL2LRR1 functions as a safeguard to prevent metastasis. In this light, it is interesting that the human chromosomal region containing the LRR1 gene, 14q21.3, is preferentially lost in metastatic cancer relative to non-metastatic cancer for lung adenocarcinoma; colorectal carcinoma; and breast cancer (Goeze et al., 2002; Knosel et al., 2004; Richard et al., 2000); and its loss is associated with poor prognosis for squamous cell carcinomas of the head and neck (Pehlivan et al., 2008). Defining how the CRL2LRR1-mediated degradation of p21 is regulated is an important future goal, with implications for both cancer and development.
EXPERIMENTAL PROCEDURES
C. elegans analysis
RNAi experiments were performed by the feeding RNAi method, as described (Timmons et al., 2001), using full-length lrr-1 or cki-1 cDNAs (from the start codon to the stop codon) cloned between double T7 promoters in the vector pPD129.36. The dsRNA was expressed in E. coli strain HT115. For lrr-1 RNAi, young adults were placed on plates containing dsRNA-expressing HT115, and then transferred to fresh RNAi plates at 12 hrs (2nd plate) and 24 hrs (3rd plate). The progeny from the second plate were examined by DIC microscopy. For cki-1 RNAi, L2-stage larvae were placed on dsRNA-expressing HT115, and the numbers of germ cells were analyzed in young adult progeny by counting Hoechst-stained germ cell nuclei with epifluorescence. Young adults were frozen, fixed in acetone for 2 min, and stained with 1 μg/ml Hoechst 33258. The DNA content of Hoechst 33258-stained germ cells in dissected gonads was determined as described (Kim et al., 2008) and internally standardized to the 2C signal from the somatic gonadal Distal Tip Cell (DTC).
Affinity purification of CUL-2::FLAG-associated proteins was performed as previously described (Starostina et al., 2007). Anti-CKI-1 immunofluorescence of dissected gonads was as described (Feng et al., 1999). Please see Supplementary Information for more details.
siRNA and cell cycle analysis
HeLa cells were transfected with 100 nM of siRNAs (for the pooled LRR1 siRNA, each siRNA was at 25 nM) using Oligofectamine (Invitrogen) or X-tremeGENE transfection reagent (Roche). T98-G cells were transfected with 100nM of siLRR1#1 and siCUL2 using X-tremeGENE. IMR-90 cells (used at 3–4 passages after receiving from ATCC) were transfected by siLRR1#1 or siCUL2 at 50 nM using jetPRIME transfection reagent (Polypus Transfection Inc.). X-tremeGENE transfection reagent was used for co-transfection of siRNA and DNA. The transfection reagents were used in accordance with the manufacturers’ instructions. Transfections were performed two or three times sequentially. Cells were collected 60–72 hrs after the first transfection and analyzed by western blot, microscopy, or cell migration assay.
DNA content was determined in siRNA-transfected HeLa cells using flow cytometry. Cells were fixed with 70% ethanol, treated with 20 μg/ml RNAse (Sigma) and stained with 0.5 mg/ml propidium iodide (Sigma) followed by analysis with a FACSCalibur flow cytometer (Beckton-Dickinson). ModFit software (Verity) was used to determine cell cycle phase distributions.
Immunofluorescence and immunoprecipitation
Immunoprecipitation of proteins ectopically expressed in HEK293T or HeLa cells was performed as described (Kim et al., 2008; Starostina et al., 2007). For immunofluorescence, HeLa, T98G, and IMR-90 cells were fixed in 4.0% paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.4% Triton X-100 in PBS, and stained with anti-FLAG M2 antibodies (1:600) followed by incubation with Alexa-Fluor 488 secondary antibodies, and/or with Rhodamine-phalloidin (1:500), and with Hoechst dye.
Pulse-chase and cell fractionation experiments
For pulse-chase experiments, 18–20 hrs after transfection, cell cultures were supplemented with 10 μM of the proteasome inhibitor MG132 (Calbiochem); 6 hrs later, MG132 was removed and 25 μg/ml cycloheximide (Sigma) was added (time 0 in the assay). Cells were lysed by boiling with 1.5% SDS, and analyzed by western blot. Cell fractionation was performed as in Sarcinella et al., 2007 (Sarcinella et al., 2007). See Supplementary Information for details.
Ubiquitylation assays
For in vivo ubiquitylation, FLAG-LRR1 or FLAG-LRR1Δ (VHL box deletion) was co-expressed with HA-ubiquitin in HeLa cells. 14 hrs after transfection, cells in a 6-well plate were subcultured into a 60 mm plate. 24 hrs later, the cultures were supplemented with 10 μM MG132, and 5 hrs later, cells were lysed by boiling with 1% SDS, and diluted 10 times with the binding buffer: 50 mM Tris-HCl (pH 7.6); 150 mM NaCl; 0.1% NP-40; 2 mM DTT; 1 mM EDTA; protease inhibitor cocktail (Roche); phosphatase inhibitor cocktail (Roche); 15 μM MG132; and 5 mM iodoacetamide (Sigma). Endogenous p21 was precipitated with anti-p21 antibodies, and analyzed by western blot for p21 and for HA-ubiquitin covalently bound to p21.
For in vitro ubiquitylation, FLAG-LRR1 and p21 were co-expressed in HEK293T cells for 26 hr; for the last 6 hr, in the presence of 10 μM MG132. Cells were lysed with NP-40 buffer: 0.5% NP-40; 50 mM Tris-HCl (pH 7.6); 150 mM NaCl; 10% glycerol; protease inhibitor cocktail; 2 mM DTT; and 15 μM MG132; supplemented with 100 U/ml OmniCleave endonuclease (Epicentre Biotechnologies) and 10 mM MgCl2. FLAG-LRR1 was precipitated with anti-FLAG antibody in the presence of 1 mM EDTA, 2.5 mM iodoacetamide, and 2.5 mM o-phenanthroline. The precipitated immunocomplex was incubated with 30 μl of ubiquitylation reaction mix containing 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 0.6 mM DTT, 2 mM ATP, 0.6 μl of energy generation solution (Biomol Int., LP), 1 μg HA-ubiquitin (Boston Biochem), 100 ng E1 (Biomol Int., LP), 400 ng E2-UbcH5a (Biomol Int., LP), 2 μM ubiquitin aldehyde (Biomol Int., LP), and 15 μM MG132. Reactions were incubated at 30°C for 90 min and terminated by boiling with 1% SDS. p21 was immunoprecipitated in a buffer containing 0.1% SDS and 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.1% NP-40, 2 mM DTT, 1 mM EDTA, protease inhibitor cocktail, phosphatase inhibitor cocktail, 15 μM MG132, and 5 mM iodoacetamide (Sigma), and analyzed by western blot for HA-ubiquitin and for p21.
Cell migration assays
For the transwell assay, 5×104 cells suspended in DMEM with 0.1% FBS were placed onto a Transwell filter with 8.0 μm pores (BD Biosciences), and allowed to migrate towards 10% FBS in DMEM for 8 hrs at 37°C in 5% CO2. The filter was then fixed with 4% formaldehyde in PBS for 20 min at room temperature and stained with Trypan blue or Coomassie Blue. After removing nonmigrated cells on the inner side of the filter with a cotton swab, cells that had migrated across the membrane to the outer side of the filter were counted at 100× magnification, and representative fields-of-view images were captured at 200× magnification. Cell counts for seven to ten random fields of view for each condition were recorded, and the experiment was repeated twice.
For the wound-healing assay, we used the Oris cell migration assay kit (Platipus Technologies). Cells were placed, at 80% confluency, into wells containing a round disk in the center, for creation of the detection zone. After the cells reached confluency, they were treated with 5 μg/ml of mitomycin C for 2 hrs (to block cell proliferation), and the disks were removed, thus allowing cells to migrate to the center of the well. At 0, 24, 48, and 72 hr time points, images were taken at 25× and 50× magnification. The experiment was performed twice with similar results. Analogous results were obtained when the experiment was performed in the absence of mitomycin C.
For the scratching wound-healing assay, confluent HeLa cells, after mitomycin C treatment, were scraped away with a plastic tip in 0.8 mm wide lanes. The cells were then incubated in DMEM with 10% FBS at normal growth conditions, and the extent of cell migration into the wound was followed by microscopic observation of five wounds per condition every 12 hrs, until 48 hrs. Images were taken at 50× magnification.
Supplementary Material
Acknowledgments
We thank Jae-min Lim and Lance Wells for mass spectrometry analysis; Christopher Dowd for technical assistance; Byoung S. Kwon, Kei-Ichi Nakayama, Yue Xiong, and Bruce E. Clurman for reagents; Shohei Mitani and the National Bioresource Project for the Animal Nematode C. elegans (Japan) for the lrr-1(tm3543) deletion allele; Julie G. Nelson and the UGA Flow Cytometry facility for DNA content analysis; and Jacek Gaertig, Marcus Fechheimer, and members of Kipreos Laboratory for critical reading of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by a grant from the NIH National Institute of General Medical Sciences (NIGMS) 1R01GM074212 with supplement S1 to E.T.K.
Footnotes
Supplemental information includes four figures, two tables, Experimental Procedures, and References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell. 2001;12:2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. Embo J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/Cofilin: a functional node in cell biology. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A, Assoian RK, Roberts JM. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors? Nat Rev Cancer. 2004a;4:948–955. doi: 10.1038/nrc1501. [DOI] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Besson A, Gurian-West M, Schmidt A, Hall A, Roberts JM. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004b;18:862–876. doi: 10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Are p27 and p21 cytoplasmic oncoproteins? Cell Cycle. 2002;1:391–393. doi: 10.4161/cc.1.6.262. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child ES, Mann DJ. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell Cycle. 2006;5:1313–1319. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM. Rho kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci. 2003;23:1416–1423. doi: 10.1523/JNEUROSCI.23-04-01416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeze A, Schluns K, Wolf G, Thasler Z, Petersen S, Petersen I. Chromosomal imbalances of primary and metastatic lung adenocarcinomas. J Pathol. 2002;196:8–16. doi: 10.1002/path.1009. [DOI] [PubMed] [Google Scholar]
- Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. Embo J. 2006;25:3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Turck CW, Morgan DO. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Heliez C, Baricault L, Barboule N, Valette A. Paclitaxel increases p21 synthesis and accumulation of its AKT-phosphorylated form in the cytoplasm of cancer cells. Oncogene. 2003;22:3260–3268. doi: 10.1038/sj.onc.1206409. [DOI] [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jang LK, Lee ZH, Kim HH, Hill JM, Kim JD, Kwon BS. A novel leucine-rich repeat protein (LRR-1): potential involvement in 4-1BB-mediated signal transduction. Mol Cells. 2001;12:304–312. [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE. Imaging tumor cell movement in vivo. Curr Protoc Cell Biol. 2007;Chapter 19(Unit 19):17. doi: 10.1002/0471143030.cb1907s35. [DOI] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knosel T, Schluns K, Stein U, Schwabe H, Schlag PM, Dietel M, Petersen I. Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer. Neoplasia. 2004;6:23–28. doi: 10.1016/s1476-5586(04)80050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Helfman DM. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J Biol Chem. 2004;279:1885–1891. doi: 10.1074/jbc.M306968200. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Ehrlich M, Lu T, Kloog Y, Weinberg RA, Lodish HF, Henis YI. Disruption of TGF-beta growth inhibition by oncogenic ras is linked to p27Kip1 mislocalization. Oncogene. 2000;19:5926–5935. doi: 10.1038/sj.onc.1203991. [DOI] [PubMed] [Google Scholar]
- Manapov F, Muller P, Rychly J. Translocation of p21(Cip1/WAF1) from the nucleus to the cytoplasm correlates with pancreatic myofibroblast to fibroblast cell conversion. Gut. 2005;54:814–822. doi: 10.1136/gut.2003.036491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SS, Becker-Hapak M, Pintucci G, Pagano M, Dowdy SF. Novel p27(kip1) C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol Cell Biol. 2003;23:216–228. doi: 10.1128/MCB.23.1.216-228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SR, Leonard N, Reynolds NJ. Loss of p21(WAF1) compartmentalisation in sebaceous carcinoma compared with sebaceous hyperplasia and sebaceous adenoma. J Clin Pathol. 2002;55:763–766. doi: 10.1136/jcp.55.10.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics. 1999;152:985–997. doi: 10.1093/genetics/152.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Pehlivan D, Gunduz E, Gunduz M, Nagatsuka H, Beder LB, Cengiz B, Rivera RS, Fukushima K, Palanduz S, Ozturk S, et al. Loss of heterozygosity at chromosome 14q is associated with poor prognosis in head and neck squamous cell carcinomas. J Cancer Res Clin Oncol. 2008;134:1267–1276. doi: 10.1007/s00432-008-0423-1. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12:1959–1964. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Richard F, Pacyna-Gengelbach M, Schluns K, Fleige B, Winzer KJ, Szymas J, Dietel M, Petersen I, Schwendel A. Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer. 2000;89:305–310. [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Vilarrupla A, Jaumot M, Abella N, Canela N, Brun S, Diaz C, Estanyol JM, Bachs O, Agell N. Binding of calmodulin to the carboxy-terminal region of p21 induces nuclear accumulation via inhibition of protein kinase C-mediated phosphorylation of Ser153. Mol Cell Biol. 2005;25:7364–7374. doi: 10.1128/MCB.25.16.7364-7374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmandke A, Strittmatter SM. ROCK and Rho: biochemistry and neuronal functions of Rho-associated protein kinases. Neuroscientist. 2007;13:454–469. doi: 10.1177/1073858407303611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shimokawa H, Rashid M. Development of Rho-kinase inhibitors for cardiovascular medicine. Trends Pharmacol Sci. 2007;28:296–302. doi: 10.1016/j.tips.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol. 2002;158:321–329. doi: 10.1083/jcb.200202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Yachi K, Fujiwara T, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) enhances axonal regeneration and functional recovery after spinal cord injury in rats. Neuroscience. 2004;127:155–164. doi: 10.1016/j.neuroscience.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Starostina NG, Kipreos ET. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 2007;8:279–286. doi: 10.1038/sj.embor.7400895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters ZE, Hunt NC, Bradburn MJ, Royds JA, Turley H, Harris AL, Norbury CJ. Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer. association with prognosis. Eur J Cancer. 2001;37:2405–2412. doi: 10.1016/s0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, Zhou BP, Hung MC. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res. 2004;10:3815–3824. doi: 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- Yokoo T, Toyoshima H, Miura M, Wang Y, Iida KT, Suzuki H, Sone H, Shimano H, Gotoda T, Nishimori S, et al. p57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J Biol Chem. 2003;278:52919–52923. doi: 10.1074/jbc.M309334200. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.