RNA interference (RNAi) inhibits transforming growth factor β-induced protein (TGFBIp) production by human corneal epithelial cells, indicating that RNAi represents a potential means to inhibit primary and recurrent dystrophic deposit formation in the corneas of patients with TGFBI dystrophies.
Abstract
Purpose.
To report the increased production of extracellular transforming growth factor β-induced protein (TGFBIp) by human corneal epithelial cells (HCECs) after induction by TGFB1 and the inhibition of TGFBIp production in induced and noninduced HCECs by RNA interference (RNAi).
Methods.
HCECs were cultured in serum-free medium and treated with 0 or 10 ng/mL TGFB1 over a period of 72 hours. Commercially available siRNAs targeting TGFBI mRNA were mixed with a transfection reagent and used to reverse transfect TGFB1-induced and noninduced HCECs. Extracellular and intracellular concentrations of TGFBIp were measured by ELISA and Western blot analysis, respectively, and TGFBI RNA was assayed using semiquantitative RT-PCR.
Results.
HCECs constitutively express TGFBIp, and treatment with TGFB1 results in up to a fourfold increase in the amount of extracellular TGFBIp. Four commercially available siRNAs targeting TGFBI mRNA produced a >70% decrease in extracellular TGFBIp within 48 hours after transfection of noninduced HCECs but a <25% decrease in extracellular TGFBIp by 48 hours after transfection of TGFB1-induced HCECs. The suppression of extracellular TGFBIp production correlated with a decrease in intracellular TGFBIp production and TGFBI mRNA expression after transfection.
Conclusions.
Extracellular TGFBIp expression by HCECs is increased several fold after exposure to TGFB1. Both HCEC-constitutive and HCEC-induced TGFBIp production can be inhibited with RNA interference, though the effect was greater and lasted longer for constitutive than induced TGFBIp production. Given that the corneal deposits in the TGFBI dystrophies consist of TGFBIp derived from HCECs, RNAi represents a potential means to inhibit primary dystrophic deposit formation and recurrence after surgical intervention.
Of the 35,000 to 40,000 corneal transplants performed in the United States annually, approximately 15% to 23% are performed for management of a corneal dystrophy.1–4 The genetic basis of two-thirds of the approximately 30 corneal dystrophies has been elucidated, with five of the most common dystrophies associated with dominant mutations in the transforming growth factor, β-induced gene (TGFBI; MIM 601692).5 Mutations in TGFBI result in the deposition of dysfunctional TGFBI protein (TGFBIp) in the corneal stroma in the form of discrete or confluent dystrophic deposits.6,7 If the dystrophic corneal deposits are superficially located, painful recurrent epithelial erosions may develop. Management of corneal stromal opacification or recurrent corneal erosions is typically achieved with laser phototherapeutic keratectomy (PTK), lamellar keratoplasty, or penetrating keratoplasty (PK). Although PTK is an effective technique to remove superficial dystrophic corneal deposits, it is not effective in a large percentage of patients with TGFBI dystrophies caused by the presence of visually significant deposits in the mid and posterior stroma. Additionally, PTK is associated with several potential complications such as induced corneal scarring, irregular astigmatism, and hyperopia. Penetrating and lamellar keratoplasty are also associated with a multitude of potential intraoperative and postoperative complications, including recurrence of the dystrophic deposits in the transplanted cornea, and thus are reserved for patients in whom more conservative therapies have failed.
In patients with TGFBI corneal dystrophies, the dystrophic deposits typically recur after both PTK and PK.8–10 In four published series documenting the recurrence of the TGFBI dystrophies after PK, the percentage of patients who experienced recurrence of the dystrophic deposits in the transplanted cornea was approximately 43% for granular corneal dystrophy (GCD), 48% to 60% for lattice corneal dystrophy (LCD), and 88% to 100% for corneal dystrophy of Bowman layer type I (CBD I) and CBD II.10–13 The median time to recurrence is highly variable but was estimated to be approximately 2 years for CBD I and II and 8 years for LCD.10 The incidence and rate of recurrence of the TGFBI corneal dystrophies after PTK vary widely, likely secondary to the differences in the number of patients, lengths of follow-up, and definitions of recurrence in the various reports. If the data from each of the reports are combined, recurrent deposits developed in approximately 52% of patients with CDB I and II, 25% of patients with LCD, 38% of patients with GCD, and 100% of patients with combined granular lattice corneal dystrophy (CGLCD) after corneal transplantation.8,9,13–16 Mean time to recurrence was shortest in patients who were homozygous for the mutation associated with CGLCD (9.5 months) and in patients with CDB I and II (26 months).
The need for surgical management of the dystrophic corneal deposits in most patients with TGFBI dystrophies has led to interest in nonsurgical means to prevent the development or the recurrence of the dystrophic corneal deposits, which may be associated with significant visual morbidity in affected patients. Immunohistochemical analysis of the corneal buttons removed from affected patients at the time of PK has shown that the deposits consist of mutated TGFBIp.17–20 Interestingly, dystrophic deposits are limited to the corneas of affected persons, as confirmed by clinical and autopsy studies.21 TGFBIp is constitutively produced by human corneal epithelial cells (HCECs),22 although the amount produced may be significantly increased in HCECs and in corneal stromal keratocytes in response to injury or surgery, as has been reported after LASIK procedures.23–28
We investigated the usefulness of RNA interference (RNAi) to inhibit or impede the formation of visually significant dystrophic corneal deposits in patients with TGFBI dystrophies and as a means to prevent the recurrence of visually significant dystrophic corneal deposits after PTK and corneal transplantation. Evidence to support the usefulness of this approach comes from previous in vitro and in vivo studies using RNAi to inhibit pathologic processes involving the cornea. This therapeutic strategy has been shown to significantly suppress the expression of vascular endothelial growth factor (VEGF) induced by hypoxia in HCECs and to inhibit injury-induced murine corneal neovascularization in vivo.29 RNAi has also been used in in vitro (cultured human corneal fibroblasts) and in vivo murine models of ocular inflammation to suppress the activity of TGF-β.30,31 In addition, RNAi has been shown to effectively decrease the expression of TGFBIp in an experimentally transformed cell line (HEK 293) transfected with plasmids expressing TGFBIp and TGFBIp-specific shRNA-generating plasmids.32 We report the efficacy of RNAi in decreasing TGFBIp expression in both TGFB1-induced and noninduced HCECs, which are the primary source of the dystrophic TGFBIp in persons with a TGFBI dystrophy.
Materials and Methods
Cell Line and Cultivation
An HCEC line (CRL-11515) was obtained from American Tissue Culture Collection (Manassas, VA) and cultured in serum-free medium (Keratinocyte-SFM; Invitrogen, Carlsbad, CA) supplemented with 5 ng/mL human recombinant endothelial growth factor (Invitrogen), 0.05 mg/mL bovine pituitary extract (Invitrogen), 0.005 mg/mL insulin, 500 ng/mL hydrocortisone (both from Sigma-Aldrich, St. Louis, MO). and 1× penicillin-streptomycin mix (Invitrogen). All flasks and plates used for experiments were coated with the following cocktail for 2 hours at 37°C: 0.01 mg/mL fibronectin (Fisher Scientific, Pittsburgh, PA), 0.01 mg bovine serum albumin (Sigma-Aldrich), and 0.03 mg/mL type I collagen (Sigma-Aldrich) prepared in sterile phosphate-buffered saline. Cells were cultured at 37°C in the presence of 5% CO2.
TGFB1-Mediated Induction of TGFBIp
To stimulate the production of TGFBI, TGFB1 (Millipore, Billerica, MA) was dissolved in 10 mM citrate buffer at pH 3.5 and added to keratinocyte-SFM at 10 ng/mL just before use. Enzyme-linked immunosorbent assays (ELISAs) were used to quantitate extracellular TGFBIp. To reduce the denaturation and aggregation of TGFBIp, we collected spent culture supernates using low-speed centrifugation (200g) at 4°C for 5 minutes. The supernates were used directly in sandwich-ELISAs with mouse monoclonal anti-βIGH3 as the capture antibody (R&D Systems, Minneapolis, MN) and biotinylated goat anti-βIGH3 as the detection antibody (R&D Systems). This was followed by the addition of streptavidin-horseradish peroxidase (HRP; R&D Systems) and o-phenylenediamine dihydrochloride (Sigma-Aldrich) as a soluble substrate. The reactions were then stopped with sulfuric acid and read at 490 nm. Measurement of TGFBIp was calibrated using recombinant human βIG-H3 (R&D Systems) as a standard to a sensitivity of 2 ng/mL.
Testing and Selection of Predesigned siRNAs
To identify the most effective predesigned siRNAs, we performed an initial experiment using seven commercially available siRNAs targeting various portions of the coding region (siRNAs 138708, 138709, s14068, s14069, s14070, and 8327; Ambion, Austin, TX) or the 3′ UTR (siRNA 8518; Ambion) of TGFBI mRNA. Transfection was performed by the reverse transfection approach in which the siRNA and transfection reagent (siPORT NeoFX; Ambion) were mixed with keratinocyte-SFM and added to the plates, after which the HCECs were added. Each siRNA, obtained as 5 nmol dried-down RNA, was dissolved in 1 mL nuclease-free water. To achieve a final concentration of 30 nM in a 500-μL system, 1.5 μL of each predesigned siRNA was then added to 50 μL keratinocyte-SFM and mixed with an equal volume of medium containing 1.5 μL transfection reagent (siPORT NeoFX; Ambion). The mixes were allowed to form complexes for 10 minutes at room temperature. HCECs used in all experiments were between passages 45 and 50 and had been actively growing for at least three passages. Next, 4 × 104 HCECs in 400 μL medium were added to 100 μL transfection reagent (siPORT NeoFX; Ambion) complexes in precoated 24-well plates (Becton Dickinson, Franklin Lakes, NJ) in quadruplicate. The cultures were incubated at 37°C in a 5% CO2 environment. The medium was replaced after 24 hours with medium that did not contain either an siRNA or the transfection reagent. The efficacy of each predesigned siRNA in decreasing the amount of extracellular TGFBI protein was determined at 24, 48, and 72 hours after transfection. The four siRNAs with the highest average knockdown percentage were then selected for use in subsequent experiments.
Determination of Cytotoxicity of siRNAs
To identify and quantify possible cytotoxic effects of the siRNA treatment, Trypan blue dye (Sigma-Aldrich) exclusion was used to compare the number of viable HCECs between a duplicate set of siRNA-treated and untreated samples. The HCECs were trypsinized, washed once with phosphate-buffered saline (PBS), and resuspended in PBS containing 0.2% Trypan blue and counted using a hemocytometer. A second method of identifying siRNA-mediated cytotoxicity assayed intracellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) enzyme levels for differences between siRNA-treated and untreated HCECs. GAPDH enzyme levels were measured in HCECs collected 48 hours after transfection using a GAPDH assay kit (KDalert; Ambion) according to the manufacturer's instructions.
Determination of Efficacy of siRNAs Targeting TGFBI
Transfection of HCECs and Measurement of Extracellular TGFBI Protein in Noninduced HCECs.
Each of the four predesigned siRNAs that provided the greatest suppression of extracellular TGFBI protein expression in the initial experiment was added to 50 μL keratinocyte-SFM to obtain five different final concentrations (15 nM, 30 nM, 50 nM, 75 nM, and 100 nM). These dilutions were then mixed with 50 μL transfection reagent (siPORT NeoFX; Ambion)/keratinocyte-SFM in quadruplicate in precoated 24-well plates and allowed to form complexes for 10 minutes at room temperature. Then 4 × 104 HCECs in 400 μL medium were added to each well. The cultures were incubated at 37°C in a 5% CO2 environment. The medium was replaced after 24 hours with medium that did not contain either an siRNA or the transfection reagent. The efficacy of each predesigned siRNA in decreasing the amount of extracellular TGFBI protein was determined at 24, 48, and 72 hours after transfection.
ELISAs were performed as described previously to quantitate extracellular TGFBIp. Experiments were performed three times to obtain a mean knockdown percentage for each siRNA at each of the five concentrations 24, 48, and 72 hours after transfection. To identify the optimal concentration of each siRNA to use in subsequent experiments, the mean knockdown percentages (averaging the knockdown percentages at 24, 48, and 72 hours after transfection) for each siRNA at each of the five concentrations were compared. A concentration that gave >5% additional knockdown when compared with the next lower concentration of a particular siRNA was selected as the concentration of each siRNA that would be used for subsequent experiments (termed the optimal concentration).
Transfection of HCECs and Measurement of Extracellular TGFBI Protein in TGFB1-Induced HCECs.
Transfection of induced HCECs was performed in the same manner as described for noninduced HCECs, with the inclusion of 10 ng/mL TGFB1 in the medium. Only the optimal concentration of the each siRNA, as determined in the previous experiment, was used.
Measurement of Intracellular TGFBI Protein in Noninduced HCECs.
Transfection of HCECs was performed using the same reverse transfection technique described previously. Each experiment was performed using the four predesigned siRNAs that provided the greatest suppression of extracellular TGFBI protein expression in the initial experiment at the five different concentrations (15 nM, 30 nM, 50 nM, 75 nM, and 100 nM). The efficacy of each of the four siRNAs in decreasing the amount of intracellular TGFBIp was determined 24 and 48 hours after transfection. Protein samples were extracted from the HCECs using RIPA lysis and extraction buffer containing Halt proteinase inhibitor cocktail (Pierce, Rockford, IL). After denaturation in Laemmli sample buffer (Bio-Rad, Richmond, CA), the samples (6 μg as determined by the microBCA kit; Pierce) were loaded onto 8% to 16% polyacrylamide gradient gels (Ready-Gels; Bio-Rad) and electrophoresed at 200 V for 45 minutes. Also loaded in one lane as an external standard was 1 ng pure TGFBIp (Millipore). The gels were then transferred to polyvinylidene difluoride membranes at 100 V for 1 hour at 4°C. The membranes were probed using reagent transport (SNAP i.d. Protein Detection System; Millipore), 0.4 μg/mL biotinylated goat anti–human βIG-H3 antibody (R&D Systems), and 1:500 dilution of streptavidin-HRP (R&D Systems). Chemiluminescence was used to detect bound antibody using extended-duration substrate (Super Signal West Dura; Pierce) and film (CL-Xposure; Pierce). GAPDH quantification in the HCEC lysate served as an internal control. To confirm that an equivalent amount of total protein was loaded in each lane, each blot was stripped and probed with a 1:6000 dilution of anti–GAPDH antibody (4 mg/mL; Ambion) using a similar chemiluminescent detection method.
Measurement of TGFBI mRNA in Noninduced HCECs.
Two-step qRT-PCR was performed to monitor TGFBI mRNA levels in HCECs using GAPDH mRNA levels as the reference. Total RNA was isolated from HCECs using a purification kit (GenElute Mammalian Total RNA Miniprep Kit; Sigma-Aldrich) according to the manufacturer's instructions. Extracted mRNA was first converted to cDNA (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Foster City, CA). The cDNA was then used in the RT-PCR experiments (SYBR Fast qPCR Kit; Kapa Biosystems, Woburn, MA). Exonic primers (TGFBI forward, 5′-AGATCGAGGACACCTTTGAG-3′; TGFBI reverse, 5′-TTGTTCAGCAGGTCTCTCAG-3′; GAPDH forward, 5′-CTGGCCAAGGTCATCCATG-3′; GAPDH reverse, 5′-GCCATGCCAGTGAGCTTCC-3′) used to amplify TGFBI and GAPDH cDNA were designed to selectively amplify cDNA rather than genomic DNA. The percentage knockdown of TGFBI mRNA was determined using the δ-Ct method, in which the percentage knockdown = 100 × 2−ΔΔCT, where ΔΔCT = (Ct for TGFBI experimental sample − Ct for GAPDH experimental sample) − (Ct for TGFBI control sample − Ct for GAPDH control).33
Positive and Negative Controls.
Positive and negative controls were used to determine the degree and specificity of the effect of each siRNA on the production of extracellular TGFBIp. The measurement of intracellular GAPDH after transfection of HCECs with a siRNA complementary to GAPDH mRNA (AM4631; Ambion) served as a positive control (data not shown). The measurement of extracellular TGFBIp was performed as well using several negative controls: after transfection of HCECs with a siRNA that does not target any human genes (AM4636; Ambion) (scrambled control); after transfection of HCECs with the transfection reagent but without the addition of an siRNA (no siRNA control); and using HCECs that were not exposed to either the transfection reagent or an siRNA (untreated control).
Results
TGFB1-Mediated Induction of TGFBIp
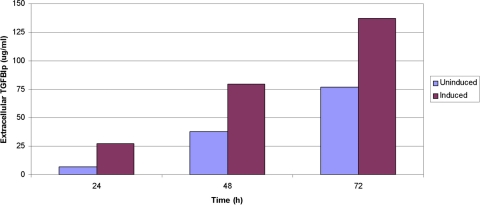
Cultured HCECs constitutively expressed TGFBIp, which was transported out of the cells into the surrounding medium, and were measured by ELISA 24 hours after plating. The amount of TGFBIp produced by HCECs increased in an exponential fashion, with extracellular levels more than doubling every 24 hours for the first 3 days (Fig. 1). Exposure of the cells to TGFB1 in the media resulted in an increased expression of TGFBIp at 24, 48, and 72 hours after the initial exposure, with levels increased approximately fourfold at 24 hours and twofold at 48 hours compared with TGFBIp expression by the uninduced cells (Fig. 1).
Figure 1.
TGFB1 induces TGFBIp expression in the HCEC line. Constitutive expression of TGFBIp by uninduced HCECs is shown at 24, 48, and 72 hours after plating. The amount of TGFBIp produced by HCECs is increased up to fourfold (at 24 hours) after the addition of TGFB1 to the media.
Testing and Selection of Predesigned siRNAs
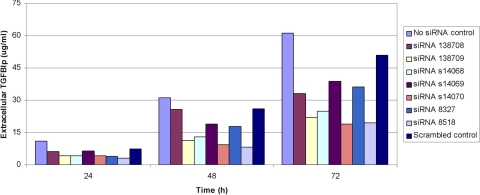
Transfection of noninduced cells with each of the predesigned siRNAs targeting TGFBI mRNA at 30 nM concentration resulted in decreased expression of extracellular TGFBIp at all measured time points for all seven of the siRNAs tested (Fig. 2). The greatest knockdown effect was obtained with siRNA 8518, which demonstrated the suppression of extracellular TGFBIp levels by 73%, 74%, and 68% at 24, 48, and 72 hours after transfection, respectively. Three other siRNAs—138709, s14068, and s14070—produced knockdown of >58% at each time point and gave an average knockdown across the three time points of >59%. Only one of the other three siRNAs produced knockdown of >58% at any time point (64% knockdown with siRNA 8327 at 24 hours). A reduction in the extracellular TGFBIp level was also seen in cells exposed to the nontargeted (scrambled) siRNA when compared with the no siRNA control, though the reduction was much less than that seen with the targeted siRNAs.
Figure 2.
Extracellular TGFBIp production by HCECs is reduced after transfection with siRNAs targeting TGFBI mRNA. Each of the seven commercially available siRNAs (at 30 nM concentration) reduced the amount of extracellular TGFBIp compared with HCECs that were exposed to the transfection reagent but not to an siRNA (no siRNA control). Additionally, the inhibition of TGFBIp production by several of the siRNAs was much greater than that observed with the scrambled siRNA control.
Determination of Cytotoxicity of siRNAs
Viable cell counts and intracellular GAPDH enzyme levels measured in both treated and untreated cells showed no evidence of cytotoxicity by any of the siRNAs (data not shown).
Determination of Efficacy of siRNAs Targeting TGFBI
Measurement of Extracellular TGFBI Protein in Noninduced HCECs.
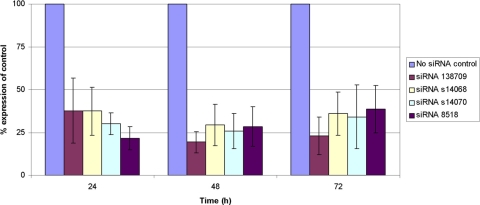
HCECs transfected with siRNAs 138709, s14068, s14070, and 8518 targeting TGFBI mRNA produced less extracellular TGFBIp at each measured time point compared with the no siRNA control. Determination of the mean knockdown percentage for each siRNA at each of the five concentrations tested revealed that siRNA 138709 was the most effective siRNA tested, achieving the greatest average knockdown at each concentration, with maximal average knockdown of 76% at 75 nM and the greatest knockdown at any time point (85% at 75 nM 48 hours after transfection) (Fig. 3). For three of the four siRNAs tested, maximal knockdown of extracellular TGFBI protein was observed 48 hours after transfection of HCECs, whereas for siRNA 8518, maximal knockdown was observed 24 hours after transfection (Fig. 3). The optimal concentration of each siRNA, defined as the concentration that gave >5% additional knockdown when compared with the next lowest concentration of the siRNA, was determined to be 30 nM for siRNAs 138709 and s14068, 50 nM for siRNA s14070, and 75 nM for siRNA 8518.
Figure 3.
Selection of the most effective siRNAs targeting TGFBI mRNA and optimization of the concentration of each maximize the inhibition of TGFBIp production by HCECs. Data shown reflect the mean reduction in extracellular TGFBIp production (knockdown) by HCECs after exposure to the optimal concentration of each siRNA compared with the extracellular TGFBIp production by HCECs exposed to the transfection reagent but not to an siRNA (no siRNA control). Experiments were performed three times to obtain a mean knockdown percentage for each siRNA at each of the five concentrations tested.
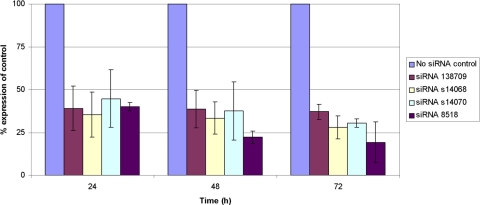
Measurement of Extracellular TGFBI Protein in TGFBI-Induced HCECs.
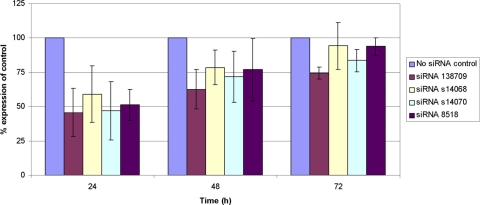
Transfection of TGFB1-induced cells with siRNAs 138709, s14068, s14070, and 8518 reduced the expression of extracellular TGFBIp at each measured time point after transfection (Fig. 4). As was observed in the noninduced cells, the greatest decrease relative to the amount of extracellular TGFBIp produced by the control cells at each of the measured time points was observed in the cells transfected with siRNA 138709, which demonstrated the suppression of extracellular TGFBIp levels by 54%, 37%, and 26% at 24, 48, and 72 hours after transfection, respectively.
Figure 4.
Extracellular TGFBIp production by TGFB1-induced HCECs is reduced after transfection with siRNAs targeting TGFBI mRNA. Data shown reflect the mean reduction in extracellular TGFBIp production (knockdown) by HCECs after exposure to the optimal concentration of each siRNA compared with extracellular TGFBIp production by HCECs exposed to the transfection reagent but not to an siRNA (no siRNA control).
Measurement of Intracellular TGFBI Protein in Noninduced HCECs.
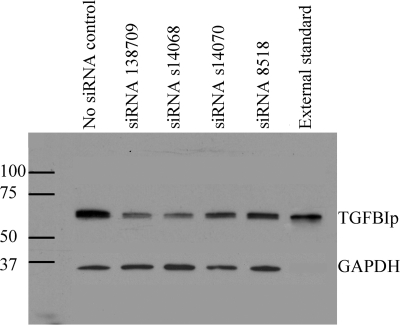
Transfection of noninduced HCECs with siRNAs 138709, s14068, s14070, and 8518 resulted in decreased expression of intracellular TGFBIp at 24 hours after transfection for each of the four siRNAs (Fig. 5). The greatest intracellular knockdown was observed with siRNAs 138709 and s14068, which demonstrated 46% and 43% expression of TGFBIp compared with untreated HCECs, respectively. siRNAs s14070 and 8518 showed only slightly decreased intracellular TGFBIp 24 hours after siRNA transfection (93% and 91% expression compared with control HCECs, respectively).
Figure 5.
Intracellular TGFBIp production by HCECs is reduced after transfection with siRNAs targeting TGFBI mRNA. Western blot analysis of intracellular TGFBIp and GAPDH protein (internal control) isolated from HCECs 24 hours after transfection with the optimal concentration of four different siRNAs targeting TGFBI mRNA. Also shown is intracellular TGFBIp production by HCECs exposed to the transfection reagent but not to an siRNA (no siRNA control) and pure TGFBIp (external standard).
Measurement of TGFBI mRNA in Noninduced HCECs.
Transfection of noninduced HCECs with siRNA 138709 at 30 nM, siRNA s14068 at 30 nM, siRNA s14070 at 50 nM, and siRNA 8518 at 75 nM resulted in a reduction of expression of TGFBI mRNA by ≥60% at 24, 48, and 72 hours after transfection for each of the siRNAs except siRNA s14070 at 24 hours (Fig. 6). Maximum knockdown of expression at 24 hours (66.5%) was observed with siRNA s14068 and at 48 hours (78%) and 72 hours (81%) with siRNA 8518 (Fig. 6).
Figure 6.
TGFBI mRNA expression by HCECs is reduced after transfection with siRNAs targeting TGFBI mRNA. Data shown reflect the mean reduction in TGFBI mRNA expression in HCECs after exposure to the optimal concentration of each siRNA compared with the TGFBI mRNA expression in HCECs exposed to the transfection reagent but not to an siRNA (no siRNA control).
Discussion
We report the successful inhibition of extracellular TGFBIp production by HCECs using RNA interference, a process wherein intracellular double-stranded RNA (dsRNA) elicits the selective elimination of mRNA with sequence homology to the dsRNA. We have demonstrated >70% knockdown of extracellular TGFBIp production using four commercially available siRNAs at either 24, 48, or 72 hours after transfection of HCECs and have correlated this decrease with a decrease in the intracellular TGFBIp and TGFBI RNA. At three of the five concentrations tested (15 nM, 30 nM, and 50 nM), the knockdown values obtained with siRNA 8518 were lower than the other three siRNAs (12%–19% lower than the values obtained with siRNA 138709 at these concentrations). The fact that this siRNA targets the 3′UTR while the other three siRNAs against which it was tested target exonic regions of TGFBI is suggestive that the most effective means of inhibiting expression of the TGFBI transcript is by targeting the coding region. We also noted that the scrambled siRNA used in these experiments produced a modest reduction in TGFBIp production by HCECs, 43% less than the average knockdown seen with each of the four siRNAs targeting TGFBI. Multiple mechanisms have been proposed to explain the nonspecific inhibitory effects that a scrambled siRNA control may have on expression of the target gene. Activation of the host cell's microRNA pathway by the introduced foreign siRNA and partial sequence homology between the scrambled siRNA and a portion of the target gene's mRNA allowing binding of the siRNA are just two such proposed mechanisms.34,35
The study design we used to transfect HCECs with nonselective siRNAs limited our ability to speculate on the potential clinical usefulness of topical siRNA therapy targeting the TGFBI dystrophies. Although the HCECs in this study were exposed to the siRNAs targeting TGFBI for 24 hours before replacing the medium, in the clinical setting, a topically applied siRNA would likely remain in contact with the ocular surface for a shorter period. The length of exposure of a cell to a transfection reagent does affect the amount of knockdown of the targeted gene transcript, with gene silencing in cell cultures exposed to a transfection reagent for 4 hours found to be 30% less than that in cell cultures exposed to an siRNA in a medium that was not changed (Applied Biosystems TechNotes volume 12:1; www.ambion.com/techlib/tn/121/9.html; accessed July 2, 2010). Just how long HCECs will be exposed to a transfection reagent contained in a topical preparation containing a vehicle that allows for prolonged retention on the ocular surface is unknown. Additionally, because we transfected only a single HCEC line and because we did not measure the degree of knockdown beyond 72 hours, the generalizability of the results we report to HCECs from many different persons and the duration of the knockdown effect are not known. Finally, whether nonselective suppression of the TGFBI transcript, both wild-type and mutant, may lead to a compensatory increase in the production of both wild-type and mutant TGFBIp by HCECs and keratocytes, negating the effect of RNA interference, will have to be determined by monitoring intracellular and extracellular TGFBIp production for a much longer period than the 72 hours used in this study.
The pathogenic corneal deposits that characterize the TGFBI dystrophies represent a gain of function, or dominant negative effect, with the dysfunctional protein product, leading to loss of vision and painful corneal erosions. In patients reported to be homozygous for mutations in TGFBI, in whom both copies of the TGFBI gene are mutated, dystrophic deposits appear earlier in life and are more dense than in patients who are heterozygous for these mutations.36,37 However, the dystrophic deposits are the only cause of corneal opacification in these patients because the stroma between, posterior, and peripheral to these deposits remains clear. Therefore, it can be inferred that the expression of wild-type TGFBI may not be necessary for the maintenance of corneal clarity and that a loss of function of the TGFBI transcript would not be expected to lead to a loss of corneal clarity. Thus, although the roles that TGFBIp plays in the corneal stroma have yet to be defined, we are not aware of evidence that indicates inhibition of the expression of both wild-type and mutant TGFBIp would necessitate the restoration of expression of the wild-type TGFBIp or that an allele-specific siRNA that targets only the mutant form of the TGFBI mRNA would be necessary to avoid inhibition of the wild-type TGFBI transcript.
Our results indicate that RNAi appears to be more effective in inhibiting constitutively expressed TGFBIp than TGFB1-induced TGFBIp expression by HCECs. The potential clinical implications of these results are that a topically applied RNAi targeting the TGFBI transcript is likely to be more effective in inhibiting or impeding dystrophic TGFBIp deposition that occurs primarily or secondarily after corneal transplantation and LASIK procedures, which are not associated with significant increases of TGFB1 expression.38–41 In contrast, topically applied RNAi targeting the TGFBI transcript is expected to be less effective after procedures that are associated with an increased expression of TGFB1 by stromal keratocytes, such as photorefractive keratectomy/LASEK and phototherapeutic keratectomy, given the TGFB1-induced expression of TGFBIp by HCECs, stromal keratocytes, or both.40,42,43
Although Huang et al.32 have previously demonstrated the suppression of intracellular TGFBI-GFP fusion protein expression in human kidney cells using RNAi, we have now demonstrated the efficacy of RNAi in decreasing extracellular TGFBIp expression by HCECs, which are the source of most of the TGFBIp in the corneas of affected persons. However, we will not be able to evaluate the possible efficacy of RNAi in the management of the TGFBI dystrophies in humans until an animal model of the TGFBI dystrophies is developed. Attempts to create a knock-in murine model of a TGFBI corneal dystrophy have so far been unsuccessful in producing transgenic expression in the cornea.44 Additionally, the safety of nonselective inhibition of the TGFBIp transcript will have to be demonstrated in an animal model before human trials, especially in light of the in vitro and in vivo evidence that TGFBI acts as a tumor suppressor gene, the loss of which has been shown to lead to spontaneous tumor development.45 Regardless, the demonstrated efficacy of RNAi in inhibiting extracellular TGFBIp production by HCECs is an important step in the process of developing gene-based therapeutic strategies to prevent the development or recurrence of the dystrophic corneal deposits that characterize the TGFBI dystrophies.
Footnotes
Supported by the Stein-Oppenheimer Research Foundation and National Institutes of Health Grant K08 EY016079 (AJA).
Disclosure: V.S. Yellore, None; S.A. Rayner, None; A.J. Aldave, None
References
- 1. Brady SE, Rapuano CJ, Arentsen JJ, Cohen EJ, Laibson PR. Clinical indications for and procedures associated with penetrating keratoplasty. 1983–1988. Am J Ophthalmol. 1989;108(2):118–122 [DOI] [PubMed] [Google Scholar]
- 2. Cosar CB, Sridhar MS, Cohen EJ, et al. Indications for penetrating keratoplasty and associated procedures: 1996–2000. Cornea. 2002;21(2):148–151 [DOI] [PubMed] [Google Scholar]
- 3. Dobbins KR, Price FW, Jr, Whitson WE. Trends in the indications for penetrating keratoplasty in the midwestern United States. Cornea. 2000. ;19(6):813–816 [DOI] [PubMed] [Google Scholar]
- 4. Lois N, Kowal VO, Cohen EJ, et al. Indications for penetrating keratoplasty and associated procedures: 1989–1995. Cornea. 1997;16(6):623–629 [PubMed] [Google Scholar]
- 5. Aldave AJ, Sonmez B. Elucidating the molecular genetic basis of the corneal dystrophies: are we there yet? Arch Ophthalmol. 2007;125(2):177–186 [DOI] [PubMed] [Google Scholar]
- 6. Konishi M, Yamada M, Nakamura Y, Mashima Y. Immunohistology of kerato-epithelin in corneal stromal dystrophies associated with R124 mutations of the BIGH3 gene. Curr Eye Res. 2000;21(5):891–896 [DOI] [PubMed] [Google Scholar]
- 7. Korvatska E, Munier FL, Chaubert P, et al. On the role of kerato-epithelin in the pathogenesis of 5q31-linked corneal dystrophies. Invest Ophthalmol Vis Sci. 1999. ;40(10):2213–2219 [PubMed] [Google Scholar]
- 8. Dinh R, Rapuano CJ, Cohen EJ, Laibson PR. Recurrence of corneal dystrophy after excimer laser phototherapeutic keratectomy. Ophthalmology. 1999;106(8):1490–1497 [DOI] [PubMed] [Google Scholar]
- 9. Inoue T, Watanabe H, Yamamoto S, et al. Recurrence of corneal dystrophy resulting from an R124H Big-h3 mutation after phototherapeutic keratectomy. Cornea. 2002;21(6):570–573 [DOI] [PubMed] [Google Scholar]
- 10. Marcon AS, Cohen EJ, Rapuano CJ, Laibson PR. Recurrence of corneal stromal dystrophies after penetrating keratoplasty. Cornea. 2003;22(1):19–21 [DOI] [PubMed] [Google Scholar]
- 11. Lyons CJ, McCartney AC, Kirkness CM, Ficker LA, Steele AD, Rice NS. Granular corneal dystrophy: visual results and pattern of recurrence after lamellar or penetrating keratoplasty. Ophthalmology. 1994;101(11):1812–1817 [DOI] [PubMed] [Google Scholar]
- 12. Meisler DM, Fine M. Recurrence of the clinical signs of lattice corneal dystrophy (type I) in corneal transplants. Am J Ophthalmol. 1984;97(2):210–214 [DOI] [PubMed] [Google Scholar]
- 13. Sorour HM, Yee SB, Peterson NJ, et al. Recurrence of chromosome 10 Thiel-Behnke corneal dystrophy (CDB2) after excimer laser phototherapeutic keratectomy or penetrating keratoplasty. Cornea. 2005;24(1):45–50 [DOI] [PubMed] [Google Scholar]
- 14. Dogru M, Katakami C, Nishida T, Yamanaka A. Alteration of the ocular surface with recurrence of granular/avellino corneal dystrophy after phototherapeutic keratectomy: report of five cases and literature review. Ophthalmology. 2001;108(4):810–817 [DOI] [PubMed] [Google Scholar]
- 15. Ellies P, Bejjani RA, Bourges JL, Boelle PY, Renard G, Dighiero P. Phototherapeutic keratectomy for BIGH3-linked corneal dystrophy recurring after penetrating keratoplasty. Ophthalmology. 2003;110(6):1119–1125 [DOI] [PubMed] [Google Scholar]
- 16. Orndahl MJ, Fagerholm PP. Treatment of corneal dystrophies with phototherapeutic keratectomy. J Refract Surg. 1998;14(2):129–135 [DOI] [PubMed] [Google Scholar]
- 17. Aldave AJ, Yellore VS, Sonmez B, et al. A novel variant of combined granular-lattice corneal dystrophy associated with the Met619Lys mutation in the TGFBI gene. Arch Ophthalmol. 2008;126(3):371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawasaki S, Nishida K, Quantock AJ, Dota A, Bennett K, Kinoshita S. Amyloid and Pro501 Thr-mutated (beta)ig-h3 gene product colocalize in lattice corneal dystrophy type IIIA. Am J Ophthalmol. 1999;127(4):456–458 [DOI] [PubMed] [Google Scholar]
- 19. Klintworth GK, Valnickova Z, Enghild JJ. Accumulation of beta ig-h3 gene product in corneas with granular dystrophy. Am J Pathol. 1998;152(3):743–748 [PMC free article] [PubMed] [Google Scholar]
- 20. Streeten BW, Qi Y, Klintworth GK, Eagle RC, Jr, Strauss JA, Bennett K. Immunolocalization of beta ig-h3 protein in 5q31-linked corneal dystrophies and normal corneas. Arch Ophthalmol. 1999;117(1):67–75 [DOI] [PubMed] [Google Scholar]
- 21. El Kochairi I, Letovanec I, Uffer S, Munier FL, Chaubert P, Schorderet DF. Systemic investigation of keratoepithelin deposits in TGFBI/BIGH3-related corneal dystrophy. Mol Vis. 2006;12:461–466 [PubMed] [Google Scholar]
- 22. Akhtar S, Meek KM, Ridgway AE, Bonshek RE, Bron AJ. Deposits and proteoglycan changes in primary and recurrent granular dystrophy of the cornea. Arch Ophthalmol. 1999;117(3):310–321 [DOI] [PubMed] [Google Scholar]
- 23. Aldave AJ, Sonmez B, Forstot SL, Rayner SA, Yellore VS, Glasgow BJ. A clinical and histopathologic examination of accelerated TGFBIp deposition after LASIK in combined granular-lattice corneal dystrophy. Am J Ophthalmol. 2007;143(3):416–419 [DOI] [PubMed] [Google Scholar]
- 24. Jun RM, Tchah H, Kim TI, et al. Avellino corneal dystrophy after LASIK. Ophthalmology. 2004;111(3):463–468 [DOI] [PubMed] [Google Scholar]
- 25. Kim TI, Roh MI, Grossniklaus HE, et al. Deposits of transforming growth factor-beta-induced protein in granular corneal dystrophy type II after LASIK. Cornea. 2008;27(1):28–32 [DOI] [PubMed] [Google Scholar]
- 26. Kim TI, Kim T, Kim SW, Kim EK. Comparison of corneal deposits after LASIK and PRK in eyes with granular corneal dystrophy type II. J Refract Surg. 2008;24(4):392–395 [DOI] [PubMed] [Google Scholar]
- 27. Wan XH, Lee HC, Stulting RD, et al. Exacerbation of Avellino corneal dystrophy after laser in situ keratomileusis. Cornea. 2002;21(2):223–226 [DOI] [PubMed] [Google Scholar]
- 28. Banning CS, Kim WC, Randleman JB, Kim EK, Stulting RD. Exacerbation of Avellino corneal dystrophy after LASIK in North America. Cornea. 2006;25(4):482–484 [DOI] [PubMed] [Google Scholar]
- 29. Singh N, Higgins E, Amin S, et al. Unique homologous siRNA blocks hypoxia-induced VEGF upregulation in human corneal cells and inhibits and regresses murine corneal neovascularization. Cornea. 2007;26(1):65–72 [DOI] [PubMed] [Google Scholar]
- 30. Cordeiro MF, Mead A, Ali RR, et al. Novel antisense oligonucleotides targeting TGF-beta inhibit in vivo scarring and improve surgical outcome. Gene Ther. 2003;10(1):59–71 [DOI] [PubMed] [Google Scholar]
- 31. Nakamura H, Siddiqui SS, Shen X, et al. RNA interference targeting transforming growth factor-beta type II receptor suppresses ocular inflammation and fibrosis. Mol Vis. 2004;10:703–711 [PubMed] [Google Scholar]
- 32. Yuan C, Zins EJ, Clark AF, Huang AJ. Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro. Mol Vis. 2007;13:2083–2095 [PubMed] [Google Scholar]
- 33. Pfaffl MW, Georgieva TM, Georgiev IP, Ontsouka E, Hageleit M, Blum JW. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest Anim Endocrinol. 2002;22(2):91–102 [DOI] [PubMed] [Google Scholar]
- 34. Couzin J. Molecular biology. RNAi shows cracks in its armor. Science. 2004;306(5699):1124–1125 [DOI] [PubMed] [Google Scholar]
- 35. Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637 [DOI] [PubMed] [Google Scholar]
- 36. Kannabiran C, Sridhar MS, Chakravarthi SK, Vemuganti GK, Lakshmipathi M. Genotype-phenotype correlation in 2 Indian families with severe granular corneal dystrophy. Arch Ophthalmol. 2005;123(8):1127–1133 [DOI] [PubMed] [Google Scholar]
- 37. Moon JW, Kim SW, Kim TI, Cristol SM, Chung ES, Kim EK. Homozygous granular corneal dystrophy type II (Avellino corneal dystrophy): natural history and progression after treatment. Cornea. 2007;26(9):1095–1100 [DOI] [PubMed] [Google Scholar]
- 38. Zou J, Chu RY, Zhou XT, Dai JH. [The effects on corneal wound healing of LASIK with ultra-thin corneal flaps and LASEK in rabbits]. Zhonghua Yan Ke Za Zhi. 2009;45(7):594–600 [PubMed] [Google Scholar]
- 39. Ivarsen A, Laurberg T, Moller-Pedersen T. Role of keratocyte loss on corneal wound repair after LASIK. Invest Ophthalmol Vis Sci. 2004;45(10):3499–3506 [DOI] [PubMed] [Google Scholar]
- 40. Kaji Y, Soya K, Amano S, Oshika T, Yamashita H. Relation between corneal haze and transforming growth factor-beta1 after photorefractive keratectomy and laser in situ keratomileusis. J Cataract Refract Surg. 2001;27(11):1840–1846 [DOI] [PubMed] [Google Scholar]
- 41. King WJ, Comer RM, Hudde T, Larkin DF, George AJ. Cytokine and chemokine expression kinetics after corneal transplantation. Transplantation. 2000;70(8):1225–1233 [DOI] [PubMed] [Google Scholar]
- 42. Long Q, Chu R, Zhou X, et al. Correlation between TGF-beta1 in tears and corneal haze following LASEK and epi-LASIK. J Refract Surg. 2006;22(7):708–712 [DOI] [PubMed] [Google Scholar]
- 43. Lee JB, Choe CM, Kim HS, Seo KY, Seong GJ, Kim EK. Comparison of TGF-beta1 in tears following laser subepithelial keratomileusis and photorefractive keratectomy. J Refract Surg. 2002;18(2):130–134 [DOI] [PubMed] [Google Scholar]
- 44. Bustamante M, Tasinato A, Maurer F, et al. Overexpression of a mutant form of TGFBI/BIGH3 induces retinal degeneration in transgenic mice. Mol Vis. 2008;14:1129–1137 [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Y, Wen G, Shao G, et al. TGFBI deficiency predisposes mice to spontaneous tumor development. Cancer Res. 2009;69(1):37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]