This study provides evidence that, after 16 hours, diffusion of fatty acids from the aqueous humor is limited to the outer 25% to 30% of the rat lens radius. It also demonstrates that incorporation of these fatty acids into molecular phospholipids is limited, supporting the hypothesis that there is no active remodeling of fiber cell phospholipids.
Abstract
Purpose.
Phospholipids are a major component of lens fiber cells and influence the activity of membrane proteins. Previous investigations of fatty acid uptake by the lens are limited. The purpose of the present study was thus to determine whether exogenous fatty acids could be taken up by the rat lens and incorporated into molecular phospholipids.
Methods.
Lenses were incubated with fluorescently labeled palmitic acid and then analyzed by confocal microscopy. Concurrently, lenses incubated with either fluorescently labeled palmitic acid or the more physiologically relevant 13C18-oleic acid were sectioned into nuclear and cortical regions and analyzed by highly sensitive and structurally selective electrospray ionization tandem mass spectrometry techniques.
Results.
The detection of fluorescently labeled palmitic acid, even after 16 hours of incubation, was limited to approximately the outer 25% to 30% of the rat lens. Mass spectrometry also revealed the presence of free 13C18-oleic acid in the cortex but not the nucleus. No evidence could be found for incorporation of fluorescently labeled palmitic acid into phospholipids; however, a low level of 13C18-oleic acid incorporation into phosphatidylethanolamine (PE), specifically PE (PE 16:0/13C18 18:1) was detected in the lens cortex after 16 hours.
Conclusions.
These data demonstrate that uptake of exogenous (e.g., dietary fatty acids) by the lens and their incorporation into phospholipids is minimal, most likely occurring only during de novo synthesis in the outermost region of the lens. This finding adds support to the hypothesis that once synthesized there is no active remodeling or turnover of fiber cell phospholipids.
Phospholipids are a major component of lens fiber cells and also interact with integral membrane proteins such as Ca2+-ATPase,1,2 caveolin 1 and 2,3,4 and aquaporin-0 (AQP0),5,6 as well as α-crystallin.7,8 This interaction is crucial for the structure and function of membrane proteins. For example, phospholipids are thought to play a vital role in the conversion of nonjunctional- to junctional-AQP0 as fiber cells age.8 Both the phospholipid9,10 and protein11 composition of the lens is specific to the particular animal species. It is therefore reasonable to surmise that membrane phospholipid composition is optimized to maintain cellular function.
We have recently demonstrated that altering dietary fatty acid composition is unable to influence the phospholipid composition of the rat lens.12 This finding suggests that, once synthesized, lens phospholipid composition remains unchanged. An inability to replace oxidatively damaged phospholipids is likely to have significant consequences for membrane integrity (and thus function) over an animal's lifespan. Borchman et al.13 have previously hypothesized that a lack of cellular machinery may result in a lack of active lipid turnover in the lens center. This, however, does not explain the lack of dietary influence on the outer, metabolically active fiber cells. One explanation for this observation is that phospholipid synthesis is highly regulated. Another possible explanation is that fatty acids obtained from the diet are not taken up by the lens; however, previous studies suggest that lipids enter the aqueous humor from the blood stream bound to albumin14 or as part of high-density lipoproteins.15 From there, it is believed that they are taken up by the lens epithelium as free fatty acids16 or bound to albumin17 by a mechanism that may involve caveolae and clathrin-coated vesicles.18 It is also possible that fatty acid binding proteins play a role in the delivery of fatty acids to the lens, and these have been identified in mouse,19 cow,20 and rat lenses21; however, no studies have been conducted into its function in the lens.
The purpose of the present study was to determine whether exogenous fatty acids could be taken up by the rat lens and incorporated into molecular phospholipids. To investigate this, confocal microscopy was used to track the movement of BODPIY palmitate (BODIPY C16) into intact rat lenses. To rule out any influence on diffusion and enzyme specificity associated with the use of a relatively large fluorophore (palmitic acid has a mass of 256 Da vs. 474 Da for BODIPY palmitate), we performed complementary experiments where 13C-labeled oleic acid (13C18-oleic acid) was added to intact lenses and its movement and incorporation into phospholipids were determined by analysis of sectioned lenses using electrospray ionization tandem mass spectrometry. Palmitate and oleate were chosen, as they are abundant fatty acids present in rat lens phospholipids.10
Methods
Reagents
HPLC grade chloroform, methanol, and analytical grade ethanol were purchased from Crown Scientific (Moorebank, NSW, Australia). Analytical grade butylated hydroxytoluene (BHT), fatty acid free bovine serum albumin (BSA), and 13C18-oleic acid (99%)were purchased from Sigma-Aldrich (Castle-Hill, NSW, Australia). 4,4-Difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid (BODIPY C16) and Medium 199 without phenol red were purchased from Invitrogen-Molecular Probes (Mount Waverley, NSW, Australia). OCT (Tissue-Tek) compound was purchased from ProSci Tech (Thuringowa, QLD, Australia). Unless stated, all other chemicals were purchased from Sigma-Aldrich.
Lens Incubation
Fatty acid-free BSA was added to Medium 199 at a concentration of 1 mg/mL and filter-sterilized using a 0.22-μm filter (Sartoriu-Stedim Biotech, Dandenong South, VIC, Australia). BODIPY C16 or 13C18-oleic acid were dissolved separately in ethanol and vortexed. Supplemented medium was pre-equilibrated at 37°C for 30 minutes before addition of BODIPY C16 to a final concentration of 110 μM and incubated for a further 30 minutes before addition of the lens. For confocal microscopy, lenses were obtained from 18-week-old male Sprague-Dawley rats after euthanatization by a peritoneal injection of sodium pentobarbitone (400 mg/kg body weight) and immediately incubated for 1, 4, 6, and 16 hours at 37°C. For mass spectrometric analysis, lenses were incubated in supplemented medium 199 as described above with either 13C18-oleic acid or BODIPY C16 added to a final concentration of 110 μM for 16 hours. After incubation, the lenses were washed with PBS (0.15 M NaCl and 0.01 M Na3PO4 [pH 7.4]) and frozen immediately at −80°C for mass spectrometric analysis or fixed for histologic analysis.
All experiments were approved by the Animal Ethics Committee of the University of Wollongong and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Confocal Microscopy
Lenses used for confocal microscopy after incubation were prepared as reported previously.22,23 In brief, the lenses were fixed in 0.75% aqueous paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 24 hours. They were washed three times in PBS, followed with cryoprotection by incubation in 10% sucrose in PBS for 1 hour at room temperature, 20% sucrose in PBS for 1 hour at room temperature, and 30% sucrose in PBS overnight at 4°C.
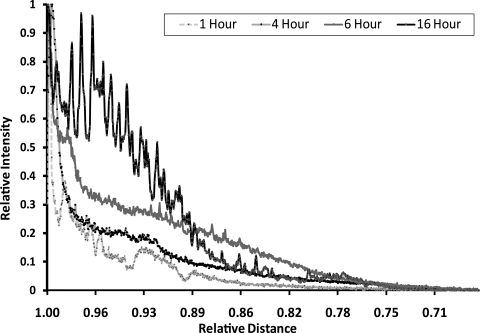
For sectioning, the lenses were positioned in an equatorial orientation so that the posterior surface of the lens was mounted onto prechilled chucks. The lens was then encased in OCT compound. Sections were cut at −20°C on a cryotome with disposable blades (Thermo Shandon; ThermoFisher Scientific, Adelaide, SA, Australia). The lenses were sliced into 18-μm sections and transferred to poly-l-lysine-coated microscope slides in PBS and then washed three times with PBS. Cell membranes were stained by incubation with wheat germ agglutinin (WGA) conjugated with Alexa Fluor 555 (Invitrogen-Molecular Probes) for 1.5 hours, in the dark at room temperature. Antifade reagent (10 μL) was added to each section and viewed with a Pascal confocal system (LSM5; Carl Zeiss Meditec, Inc., North Ryde, NSW, Australia). BODIPY C16 was imaged at 488 nm with an argon laser at 34%, with a BP505-530 filter and a 52-μm pinhole. WGA was imaged at 543 nm using an HeNe laser at 100% with an LP 560 filter and a 54-μm pinhole. Images were processed by Image J Software 1.41 (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html). To create a plot of pixel intensity versus distance (see Fig. 2), we highlighted a section of the image from the epithelium to the central core (the same number of pixels were selected for each image for consistency of comparison), and a plot profile was generated within the Image J software, listing a distance and the corresponding pixel intensity at that distance. These data were then normalized for both intensity (with the pixel intensity at the edge of the epithelium being set to 1) and distance (with the center of the lens set to 0 and the capsule set to 1).
Figure 2.
Diffusion of BODIPY C16 into the rat lens as a function of time. The intensity of fluorescence versus distance from the lens epithelium was plotted after 1, 4, 6, and 16 hours. C16, palmitic acid.
Lipid Extraction
After incubation, lenses were frozen at −80°C, and sectioned into nucleus and cortex with a 3-mm trephine. Lenses were homogenized in chloroform-methanol (2:1 vol/vol) with 0.01% BHT at a ratio of 20:1 solvent to tissue. Total lipids were extracted as described elsewhere10 and stored at −80°C until analysis.
Mass Spectrometry
All mass spectra were obtained with a triple quadrapole mass spectrometer (QuattroMicro; Waters, Manchester, UK) equipped with a z-spray electrospray ion source and controlled by allied software (Micromass Masslynx, ver. 4.0; Waters). Capillary voltage was set to 3000 V, source temperature to 80°C, and desolvation temperature to 120°C. Cone voltage was set to 50 V in negative ion mode and 35 V in positive ion mode. Nitrogen was used as the drying gas at a flow rate of 320 L/h. Samples were infused into the electrospray ion source at a flow rate of 10 μL/min by the instrument's on-board syringe pump. Formation of ions was enhanced by the addition of aqueous ammonium acetate to a final concentration of approximately 50 mM.24 Negative ion scans were performed to identify the presence of free [M-FH2]− BODIPY C16 and 13C18-oleic acid in the mass range of m/z 250 to 900.
Precursor ion scans for m/z 453.5 (BODIPY C16), 299.3 (13C18-oleic acid), and 281.4 (unlabeled oleic acid) were performed in negative ion mode at collision energies of 20, 35, and 35 eV, respectively. Precursor ion scanning in positive ion mode for m/z 184.1 (phosphocholine) using a collision energy of 35 eV was performed to determine incorporation of labeled fatty acids into phosphatidylcholines and sphingomyelins. Product ion scans were performed at a collision energy of 35 eV to further validate labeled fatty acid incorporation. In all precursor ion and product ion scans, argon was used as the collision gas at a pressure of 3 mTorr.
Results
Incubation of Intact Lenses with BODIPY Palmitate
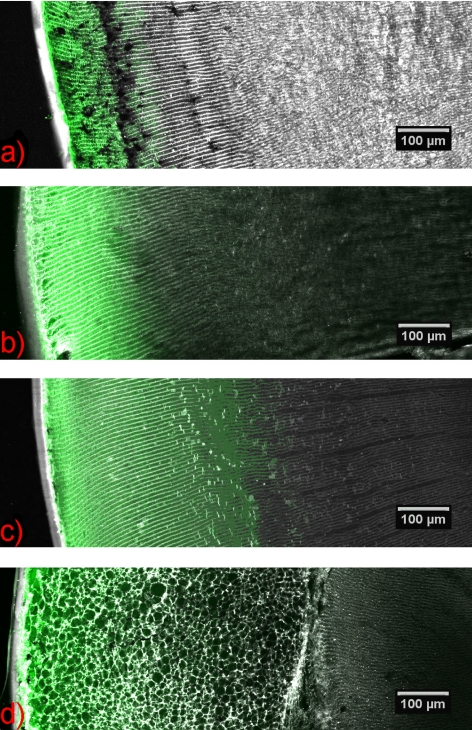
Incubation of rat lenses with BODIPY C16 showed the presence of the fluorescently labeled fatty acid in the lens cortex after 1 hour (Fig. 1a). The diffusion distance was extended slightly with 4 and 6 hours of incubation by comparison to the 1-hour sample (Figs. 1b, 1c).
Figure 1.
Diffusion of labeled fatty acid into rat lens. Equatorial cryosections showing the diffusion of BODIPY C16 into a rat lens after (a) 1, (b) 4, (c) 6, and (d) 16 hours. Green fluorescence corresponds to BODIPY C16 and the gray fluorescence corresponds to WGA Alexa Fluor 555. C16, palmitic acid; WGA, wheat germ agglutinin.
BODIPY C16 did not penetrate deeper into the lens nucleus with increased times of incubation, up to 16 hours. A plot distribution was obtained by averaging the pixel intensity across this distance with Image J software. As shown in Figure 2, even after a 16-hour incubation, BODIPY C16 (474 Da) was observed only in the outer 25% to 30% of the lens. The confocal image at 16 hours was notably irregular compared with the earlier time points, which may reflect a disruption of the fiber cell membranes by BODIPY C16 (Fig. 1d).
Further investigation of the distribution of BODIPY C16 into the lens was performed by separating the lens into nucleus and cortex and then analyzing lipid extracts from these sections by electrospray ionization tandem mass spectrometry. A negative ion mass spectrum of the rat cortex after 16 hours showed an abundant ion at m/z 453.5, which corresponds to the [M-FH2]− free form of BODIPY C16 (data not shown). Further interrogation of the ion at m/z 453.5 using collision-induced dissociation in a tandem mass spectrometry experiment revealed fragmentation that was identical with that obtained from an authentic sample of BODIPY C16, confirming the presence of the fluorescent probe in the cortex. No such signal was detected in the nucleus.
No evidence was obtained for incorporation of BODIPY C16 into phospholipids in the range of m/z 600 to 1000 as assessed by performing a negative ion precursor scan (m/z 453.5) selective for any molecule containing BODIPY C16 and a positive ion precursor scan for phosphatidylcholines and sphingomyelins, which contain a phosphocholine head group (m/z 184.1).
Analysis of the rat lens nucleus by mass spectrometry confirmed the data obtained by confocal microscopy in that no evidence of free or phospholipid-incorporated BODIPY C16 was found, even after 16 hours of incubation.
13C18-oleic Acid Incubation
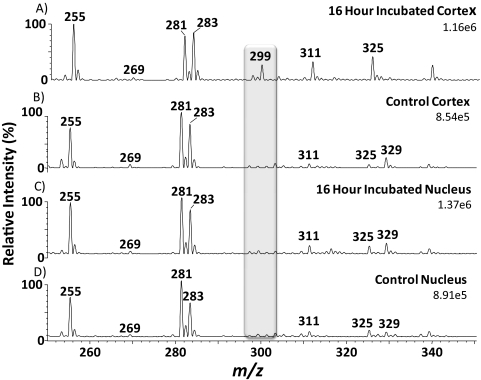
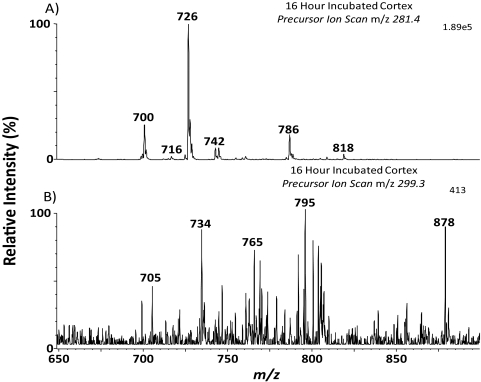
Negative ion spectra of lipid extracts obtained from the cortex and nucleus of a lens incubated in 13C18-oleic acid and also a control lens cortex and nucleus are shown in Figure 3. The spectra show 13C18-oleic acid (Fig. 3A; m/z 299.5,) present only in the incubated cortex at an ion abundance approximately one third that of the native oleic acid (Fig. 3A; m/z 281.4). These data confirm the results shown in Figure 1, where free BODIPY C16 was detected in the cortex, but not in the nucleus.
Figure 3.
Negative ion mass spectra of control lenses and lenses incubated with 13C18-oleic acid. (A) Cortex incubated with 13C18-oleic acid for 16 hours, (B) control cortex, (C) nucleus incubated with 13C18-oleic acid for 16 hours, (D) control nucleus. Shaded region: illustrates the position of 13C18-oleic acid at m/z 299.5.
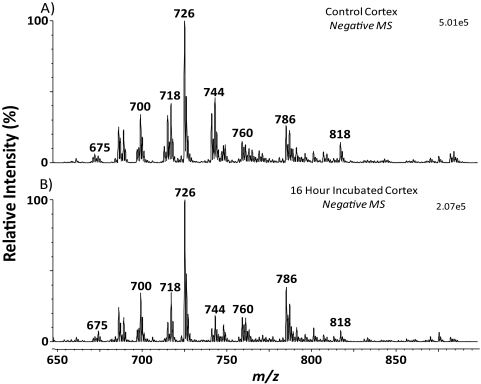
Figure 4A shows mass spectra of the naturally occurring phospholipids found in the cortex of a rat lens. No major differences in anionic phospholipid profile were observed with the incubated cortex (Fig. 4B) in comparison to the control cortex. Furthermore, scans were performed to determine whether 13C18-oleic acid had been incorporated into phospholipids. An initial precursor ion scan (m/z 281.4) shown in Figure 5A, was performed to identify phospholipids containing unlabeled 18:1. It would be expected that any incorporation of 13C18-oleic acid would occur in these highly abundant phospholipids. The corresponding precursor ion scan for 13C18-oleic acid (m/z 299.3) is shown in Figure 5B. The signal obtained from the 13C18-oleic acid precursor ion scan is approximately 500 times lower than that obtained for precursors of native oleic acid. 13C18-oleic acid was detected in one phospholipid present at m/z 734.6. This ion was tentatively identified as phosphatidylethanolamine (PE) (16:0/13C1818:1), as it was 18 Da higher than PE (16:0/18:1; m/z 716.6), which is observed as a low-abundance ion in Figure 5A and has been identified in the rat lens.12 The identification of PE (16:0/13C18; 18:1) was confirmed using tandem mass spectrometry by the presence of an ion at m/z 255.4 corresponding to the 16:0 fatty acid, and an ion at m/z 299.5 corresponding to the labeled 18:1 fatty acid (data not shown). Phosphatidylcholines and sphingomyelins were analyzed using the precursor ion scan m/z 184.1, and no evidence of 13C18-oleic acid incorporation was observed.
Figure 4.
Negative ion mass spectra of lipid extracts from a control rat lens cortex (A) and a 13C18-oleic acid–incubated rat lens cortex (B), showing the profile of acidic phospholipids present.
Figure 5.
Profile of oleic acid-containing phospholipids in a lipid extract from a rat lens cortex after incubation with 13C18-oleic acid for 16 hours. (A) A negative precursor ion scan for m/z 281.4 showing the profile of acidic phospholipids containing native oleic acid. (B) A negative precursor ion scan for m/z 299.3 showing the profile of acidic phospholipids containing 13C18-oleic acid.
Discussion
The present study suggests that movement of fatty acids into the lens over a 16-hour period is restricted to the outer 25% to 30% of the rat lens radius. Moreover, a minimal amount of exogenous fatty acid incorporation into molecular phospholipids was detected.
To the authors' knowledge only two previous studies have examined the uptake of fatty acids into the lens and their incorporation into fiber cell phospholipids. After incubating rabbit lenses in radio-labeled fatty acids for 3.5 hours, Broekhuyse and Bögemann16 found most of the radioactivity in the epithelium and the equator, with small amounts in the peripheral, anterior, and posterior cortex. No radioactivity was observed in the lens center. A more recent study indicates that fluorescently labeled lauric acid (BODIPY C12) can enter the nucleus of cultured rat lenses when the incubation period is extended to 12 hours.17
The current finding that BODIPY or 13C-labeled fatty acids did not enter the rat lens nucleus after 16 hours is in contrast to that reported by Sabah et al.17 An explanation for the discrepancy may be the significantly increased binding efficiency to BSA of both palmitic and oleic acid compared with lauric acid.25 After incubating lenses in BODIPY C16 for 16 hours, we observed a disruption of fiber cell structure (Fig. 1d). Nevertheless, it did not result in further diffusion of fluorescently labeled palmitic acid into the lens. Similar vesiculation of fiber cells has been observed in rat lenses after an 18-hour incubation with the chloride channel inhibitor tamoxifen, probably owing to an impairment of cell volume regulation.26 Disruption of fiber cell membrane structure by free BODIPY C16 could also impair cell volume regulation and may explain fiber cell swelling in the present study. From the low-magnification images it cannot be determined whether a similar phenomenon occurred in the study of Sabah et al.17 and whether this had an impact of fatty acid diffusion. The plot shown in Figure 2 suggests that the fatty acids entered the rat lens via diffusion, and therefore a longer incubation period may have resulted in the labeled fatty acids penetrating farther into the lens. Given the cellular damage observed after 16 hours, we were reluctant to incubate the lenses for longer periods.
This study provides, for the first time, structural evidence, obtained with confocal microscopy and electrospray ionization tandem mass spectrometry, of both the extent and specificity of fatty acid diffusion and incorporation into lens phospholipids. We could find no evidence for incorporation of BODIPY C16 into lens phospholipids using mass spectrometry. This suggests that this modified fatty acid is either not inserted into phospholipids in the lens or that the degree of incorporation is too low to detect. Sabah et al.17 observed co-migration of fluorescence with that of phospholipids by thin-layer chromatography (TLC), which they interpreted as incorporation of BODIPY C12 into sphingomyelin, phosphatidylcholine, and phosphatidylethanolamine. Only nonlabeled phospholipid standards were run on these TLC plates and the effect on migration of adding the large fluorophore was not shown.
In the present study, a very low level of 13C18-oleic acid incorporation into PE (16:0/18:1) was detected in the rat lens cortex. This low level was near the limit of detection of the mass spectrometer (∼550 picomoles/g tissue) and indicates that over the period of the experiment (16 hours) the extent of incorporation into phospholipids was low. This low level is not due to the availability of free fatty acids, because the ion abundance of free oleic acid was approximately 3 times that of free 13C18-oleic acid (see Fig. 3), compared with approximately 500 times for phospholipids containing native versus 13C18-oleic acid (see Fig. 5). Broekhuyse and Bögemann16 also reported incorporation of radio-labeled fatty acids into phospholipids in the outer cortex regions of the rabbit lens after 3.5 hours; however, no molecular characterization was conducted. Previous work suggests that the de novo synthesis of fiber cell membrane components (fatty acids, cholesterol, and AQP0) is restricted to the outer 3% to 6% of the lens radius.27 This localization is also likely to be the case for phospholipids and would explain the low levels of fatty acid incorporation that we observed when analyzing the whole cortex, if incorporation occurred exclusively during de novo synthesis in the very outer cortical region. This result implies that, at least over a 16-hour period, fatty acids from external sources (for example, diet), do not replace fatty acids that have already been incorporated into the membrane. These findings do not rule out replacement of membrane fatty acids occurring over longer periods, such as months or years. If this were true it would make the lens very different from other tissues as intravenously administered fatty acids are incorporated into phospholipids within 2 to 10 minutes in brain,28 heart, kidney, and liver.29 In vitro studies of cultured neuroblastoma cells have also demonstrated incorporation of fatty acids within 2 to 10 minutes.30
In summary, the present study adds support to the hypothesis that, once synthesized, there is no active remodeling or turnover of fiber cell phospholipids.
Acknowledgments
The authors thank Kerry Walker and Julie Lim (University of Auckland) for assistance in the study.
Footnotes
Supported by Australian Research Council Discovery Project Grant DP0986628 and National Institutes of Health Grant EY013570. JRN is supported by an Australian Postgraduate Award. SJB and TWM were supported by the University of Wollongong and the Australian Research Council. PJD received financial support from the Health Research Council (HRC) of New Zealand International Investment Opportunities Fund and the HRC Australia Collaboration Award. RJWT is a National Health and Medical Research Council Fellow and received financial support from the National Health and Medical Research Council and National Institute of Health.
Disclosure: J.R. Nealon, None; S.J. Blanksby, None; P.J. Donaldson, None; R.J.W. Truscott, None; T.W. Mitchell, None
References
- 1. Tang D, Dean WL, Borchman D, Paterson CA. The influence of membrane lipid structure on plasma membrane Ca2+-ATPase activity. Cell Calcium. 2006;39:209–216 [DOI] [PubMed] [Google Scholar]
- 2. Zeng J, Zhang Z, Paterson CA, Ferguson-Yankey S, Yappert MC, Borchman D. Ca2+-ATPase activity and lens lipid composition in reconstituted systems. Exp Eye Res. 1999;69:323–330 [DOI] [PubMed] [Google Scholar]
- 3. Lo W-K, Zhou C-j, Reddan J. Identification of caveolae and their signature proteins caveolin 1 and 2 in the lens. Exp Eye Res. 2004;79:487–498 [DOI] [PubMed] [Google Scholar]
- 4. Lin D, Zhou J, Zelenka PS, Takemoto DJ. Protein kinase Cγ regulation of gap junction activity through caveolin-1-containing lipid rafts. Invest Ophthalmol Vis Sci. 2003;44:5259–5268 [DOI] [PubMed] [Google Scholar]
- 5. Baumann CG, Malewicz B, Anderson WH, Lampe PD, Johnson RG, Baumann WJ. Lipid differentiation in MP26 junction enriched membranes of bovine lens fiber cells. Biochim Biophys Acta. 1996;1303:145–153 [DOI] [PubMed] [Google Scholar]
- 6. Gonen T, Cheng Y, Sliz P, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borchman D, Tang D. Binding capacity of α-crystallin to bovine lens lipids. Exp Eye Res. 1996;63:407–410 [DOI] [PubMed] [Google Scholar]
- 8. Zhang ZZ, Zeng JJ, Yin HH, Tang DD, Borchman DD, Paterson CAC. Membrane lipid a-crystallin interaction and membrane Ca2+-ATPase activities. Curr Eye Res. 1999;18:56–61 [DOI] [PubMed] [Google Scholar]
- 9. Borchman D, Yappert MC, Afzal M. Lens lipids and maximum lifespan. Exp Eye Res. 2004;79:761–768 [DOI] [PubMed] [Google Scholar]
- 10. Deeley JM, Mitchell TW, Xiaojia W, et al. Human lens lipids differ markedly from those of commonly used experimental animals. Biochim Biophys Acta Mol Cell Biol Lipids. 2008;1781:288–298 [DOI] [PubMed] [Google Scholar]
- 11. Truscott RJW. Age-related nuclear cataract: oxidation is the key. Exp Eye Res. 2005;80:709–725 [DOI] [PubMed] [Google Scholar]
- 12. Nealon JR, Blanksby SJ, Abbott SK, Hulbert AJ, Mitchell TW, Truscott RJW. Phospholipid composition of the rat lens is independent of diet. Exp Eye Res. 2008;87:502–514 [DOI] [PubMed] [Google Scholar]
- 13. Borchman D, Tang D, Yappert MC. Lipid composition, membrane structure relationships in lens and muscle sarcoplasmic reticulum membranes. Biospectroscopy. 1999;5:151–167 [DOI] [PubMed] [Google Scholar]
- 14. Broekhuyse RM, Soeting WJ. Lipids in tissues of the eye, XV: essential fatty acids in lens lipids. Exp Eye Res. 1976;22:653–657 [DOI] [PubMed] [Google Scholar]
- 15. Cenedella RJ. Lipoproteins and lipids in cow and human aqueous humor. Biochim Biophys Acta Lipids Lipid Metab. 1984;793:448–454 [DOI] [PubMed] [Google Scholar]
- 16. Broekhuyse RM, Bögemann B. Lipids in tissues of the eye, XVI: uptake of lipids by the rabbit lens in vitro. Exp Eye Res. 1978;26:567–572 [DOI] [PubMed] [Google Scholar]
- 17. Sabah J, McConkey E, Welti R, Albin K, Takemoto LJ. Role of albumin as a fatty acid carrier for biosynthesis of lens lipids. Exp Eye Res. 2005;80:31–36 [DOI] [PubMed] [Google Scholar]
- 18. Sabah JR, Schultz BD, Brown ZW, Nguyen AT, Reddan J, Takemoto LJ. Transcytotic passage of albumin through lens epithelial cells. Invest Ophthalmol Vis Sci. 2007;48:1237–1244 [DOI] [PubMed] [Google Scholar]
- 19. Ueda Y, Duncan MK, David LL. Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci. 2002;43:205–215 [PubMed] [Google Scholar]
- 20. Jaworski C, Wistow G. LP2, a differentiation-associated lipid-binding protein expressed in bovine lens. Biochem J. 1996;320:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wen Y, Li G-W, Chen P, Wong E, Bekhor I. Lens epithelial cell mRNA, II: expression of a mRNA encoding a lipid-binding protein in rat lens epithelial cells. Gene. 1995;158:269–274 [DOI] [PubMed] [Google Scholar]
- 22. Grey AC, Jacobs MD, Gonen T, Kistler J, Donaldson P. Insertion of MP20 into lens fibre cell plasma membranes correlates with the formation of an extracellular diffusion barrier. Exp Eye Res. 2003;77:567–574 [DOI] [PubMed] [Google Scholar]
- 23. Jacobs MD, Soeller C, Cannell MB, Donaldson PJ. Quantifying changes in gap junction structure as a function of lens fiber cell differentiation. Cell Commun Adhes. 2001;8:349–353 [DOI] [PubMed] [Google Scholar]
- 24. Thai TP, Rodemer C, Worsch J, Hunziker A, Gorgas K, Just WW. Synthesis of plasmalogens in eye lens epithelial cells. FEBS Lett. 1999;456:263–268 [DOI] [PubMed] [Google Scholar]
- 25. Spector AA, John K, Fletcher JE. Binding of long-chain fatty acids to bovine serum albumin. J Lipid Res. 1969;10:56–67 [PubMed] [Google Scholar]
- 26. Merriman-Smith BR, Young MA, Jacobs MD, Kistler J, Donaldson PJ. Molecular identification of P-glycoprotein: a role in lens circulation? Invest Ophthalmol Vis Sci. 2002;43:3008–3015 [PubMed] [Google Scholar]
- 27. Cenedella RJ. Apparent coordination of plasma membrane component synthesis in the lens. Invest Ophthalmol Vis Sci. 1993;34:2186–2194 [PubMed] [Google Scholar]
- 28. Shetty HU, Quentin RS, Kazushige W, Stanley IR. Identification of two molecular species of rat brain phosphatidylcholine that rapidly incorporate and turn over arachidonic acid in vivo. J Neurochem. 1996;67:1702–1710 [DOI] [PubMed] [Google Scholar]
- 29. Thies F, Pillon C, Moliere P, Lagarde M, Lecerf J. Preferential incorporation of sn-2 lysoPC DHA over unesterified DHA in the young rat brain. Am J Physiol Regul Integr Comp Physiol. 1994;267:R1273–1279 [DOI] [PubMed] [Google Scholar]
- 30. Chakravarthy BR, Spence MW, Cook HW. Turnover of phospholipid fatty acid chains in cultured neuroblastoma cells: involvement of deacylation-reacylation and de novo synthesis in plasma membranes. Biochim Biophys Acta Biomem. 1986;879:264–277 [DOI] [PubMed] [Google Scholar]