The pentapeptide repeat protein AlbG, provides self-resistance to the nonribosomally encoded hybrid polyketide-peptide termed albicidin. Analysis of the AlbG three-dimensional structure and the sequences of other pentapeptide repeat proteins that confer resistance to topiosomerase poisons suggests they have a similar dimer interface which may be critical to their interaction with topoisomerases.
Keywords: pentapeptide-repeat proteins, topoisomerase poison resistance factors, dimer interfaces, AlbG
Abstract
The protein AlbG is a self-resistance factor against albicidin, a nonribosomally encoded hybrid polyketide-peptide with antibiotic and phytotoxic properties produced by Xanthomonas albilineans. Primary-sequence analysis indicates that AlbG is a member of the pentapeptide-repeat family of proteins (PRP). The structure of AlbG from X. albilineans was determined at 2.0 Å resolution by SAD phasing using data collected from a single trimethyllead acetate derivative on a home source. AlbG folds into a right-handed quadrilateral β-helix composed of approximately eight semi-regular coils. The regularity of the β-helix is blemished by a large loop/deviation in the β-helix between coils 4 and 5. The C-terminus of the β-helix is capped by a dimerization module, yielding a dimer with a 110 Å semi-collinear β-helical axis. This method of dimer formation appears to be common to all PRP proteins that confer resistance to topoisomerase poisons and contrasts with most PRP proteins, which are typically monomeric.
1. Introduction
The pathogenic bacterium Xanthomonas albilineans is the causative agent of sugar cane leaf scald, a yellowing of leaf tissue arising from a lack of chlorophyll (chlorosis; Rott & Davis, 2000 ▶). The small molecule albicidin is a key pathogenesis factor in host-cell invasion by X. albilineans (Birch & Patil, 1987a ▶,b ▶). Albicidin inhibits DNA replication by stabilizing the covalent protein–DNA (phosphotyrosine) cleavage complex of DNA gyrase, which is analogous to the action of fluoroquinolones (Hashimi et al., 2007 ▶). Albicidin is of pharmacological interest as it is noncytotoxic to mammalian cells at 8 µM concentration (Birch, 1983 ▶) but inhibits DNA gyrase at nanomolar concentrations and is bactericidal to a range of Gram-negative and Gram-positive organisms (Birch & Patil, 1985b ▶; Hashimi et al., 2007 ▶). The albicidin-biosynthetic cluster includes polyketide and nonribosomal peptide synthases (Rott et al., 1996 ▶; Huang et al., 2001 ▶; Royer et al., 2004 ▶; Vivien et al., 2005 ▶), suggesting a hybrid polyketide-peptide backbone; however, because of the low amounts of albicidin produced by the host organism its chemical structure has not been elucidated. Initial studies indicate that the most prevalent isoform has a molecular weight of ∼842 Da with approximately 38 C atoms and several aromatic groups (Birch & Patil, 1985a ▶; Huang et al., 2001 ▶).
Resistance of Escherichia coli to albicidin is typically through alteration of the Tsx nucleotide-uptake channel, which is an especially effective transporter for albicidin (the IC50 for E. coli to albicidin is ∼1 nM; Birch et al., 1990 ▶). Other resistance mechanisms in nonproducing organisms include sequestration through tight binding (AlbA or AlbB; Basnayake & Birch, 1995 ▶; Zhang et al., 1998 ▶) and cleavage by an esterase (AlbD; Zhang & Birch, 1997 ▶). Three modes of albicidin resistance have been determined for the host organism. Firstly, reconstituted X. albilineans DNA gyrase exhibits 20–25-fold higher resistance to albicidin than E. coli DNA gyrase (Hashimi et al., 2008 ▶). Secondly, an efflux pump (AlbF) encoded within the X. albilineans albicidin-biosynthetic cluster is responsible for actively removing albicidin from the cytoplasm (Bostock et al., 2006 ▶). Finally, the X. albilineans albicidin-biosynthetic cluster encodes a topoisomerase-interacting protein termed AlbG. When expressed in E. coli, AlbG conferred a 4–30-fold increase in resistance to albicidin (Hashimi et al., 2007 ▶).
Primary-sequence analysis of AlbG suggests that it belongs to a family of proteins termed pentapeptide-repeat proteins (PRPs). PRPs are mainly composed of tandemly encoded five-amino-acid repeats with consensus sequence (STAV)1(DN)2(LF)3(STR)4(G)5 and fold into a structure termed a right-handed quadrilateral β-helix (Bateman et al., 1998 ▶; Hegde et al., 2005 ▶; Buchko et al., 2006 ▶; Vetting et al., 2006 ▶). There is a distinct clustering of PRP sequences in cyanobacteria, although they are also found in a diverse set of prokaryotes and eukaryotes (Bateman et al., 1998 ▶; Vetting et al., 2006 ▶). Only a small number of PRPs have been assigned a biological function. The protein HglK from the cyanobacterium Nostoc sp. strain PCC 7120 has been linked to the localization of glycolipid components required for the production of heterocysts (Black et al., 1995 ▶). The RfrA protein from Synechocystis sp. strain 6803 has been linked to regulation of manganese transport (Chandler et al., 2003 ▶). A subset of PRPs confer resistance to topoisomerase poisons, including fluoroquinolone resistance by the proteins Qnr and MfpA (Montero et al., 2001 ▶; Tran & Jacoby, 2002 ▶), microcin B17 resistance by the protein McbG (Garrido et al., 1988 ▶) and albicidin resistance by the protein AlbG (Hashimi et al., 2007 ▶). These topoisomerase poisons stabilize the tyrosyl-DNA covalent adduct, producing a rapid bacteriostatic effect through inhibition of DNA topoisomerases and a slower bactericidal effect through the production of double-stranded chromosomal breaks (reviewed in Deweese & Osheroff, 2009 ▶). The chromosomally encoded protein MfpA from Mycobacterium tuberculosis (MtMfpA) was the first experimentally determined structure of a PRP topoisomerase poison resistance factor (TPRF; Hegde et al., 2005 ▶). It has been proposed that PRP TPRFs bind to the topoisomerase DNA-binding saddle, preventing formation of the topoisomerase–DNA covalent complex and subsequent DNA damage (Hegde et al., 2005 ▶). While MtMfpA and EfsQnr confer a reduced fluoroquinolone-susceptibility phenotype when overexpressed in the native organism or recombinantly in E. coli (Montero et al., 2001 ▶; Hegde et al., 2005 ▶; Rodríguez-Martínez et al., 2008 ▶), their true cellular function has yet to be discovered.
Here, we present the structure of the albicidin self-immunity factor AlbG from Xanthomonas albilineans at 2 Å resolution. This is the first structure of a chromosomally encoded PRP protein with a demonstrated biological function and is therefore an important confirmation of the structural features that are important for PRP TPRFs (i.e. MtMfpA and EfsQnr). Of special note is our proposal that all PRP TPRFs sequenced to date will be found to exist as dimers with coaxial β-helices, which has probable and important implications for binding across the α2β2 topoisomerase II oligomers.
2. Materials and methods
2.1. Cloning and purification
All chemicals and reagents were purchased from Sigma–Aldrich Chemical Co. Enzymes used in molecular biology were supplied by New England Biolabs. Plasmid pET-28a and E. coli strains Nova Blue and BL21 (DE3) were obtained from Novagen.
The open reading frame of AlbG was amplified by standard PCR techniques using X. albilineans (ATCC 29184) chromosomal DNA as template. The oligonucleotides AlbGF (5′-ATCCCGCTCATATGCCGGCCAAGACCCTTG-3′) and AlbGR (5′-ATCCCGCTCTC GAGTCAATCGGACAGCTCGATATC-3′) containing NdeI and XhoI restriction sites, respectively (shown in bold), were used. The PCR fragment was cloned into pET-28a(+) and recombinant AlbG bearing a thrombin-cleavable N-terminal His6 tag was expressed in E. coli strain BL21 (DE3). For shake-flask growth, 1 l Luria broth medium supplemented with kanamycin (35 µg ml−1) was inoculated with 10 ml of an overnight culture and incubated at 310 K. The culture was grown to mid-log phase (A 600 of ∼0.8), cooled to 293 K, induced with 0.5 mM IPTG and further incubated overnight at 293 K.
All purification procedures were carried out at 277 K. The cells were collected by centrifugation at 3000g, resuspended in buffer A [50 mM Tris–HCl pH 7.8 containing 300 mM NaCl, protease inhibitors, lysozyme (5 µg ml−1) and DNase I (0.1 µg ml−1)] and stirred for 20 min. The cells were then lysed by sonication and cell debris was removed by centrifugation at 10 000g for 30 min. The supernatant was loaded onto an Ni–NTA column pre-equilibrated with buffer A and washed with ten column volumes of the same buffer. The bound proteins were eluted with a linear 0–0.3 M imidazole gradient and the peak fractions were pooled and concentrated by ultrafiltration. The His6 tag was then cleaved using thrombin (2 U per milligram of protein) and the solution was dialyzed overnight against 50 mM Tris–HCl pH 7.8 containing 300 mM NaCl and 2 mM CaCl2. The dialysate was loaded onto a Superdex S75 column pre-equilibrated with 50 mM Tris–HCl pH 7.8 containing 300 mM NaCl and pure fractions, as determined by SDS–PAGE, were pooled and concentrated by ultrafiltration. Protein concentrations were estimated by the Bio-Rad protein-assay method using bovine serum albumin as a standard.
AlbGΔ91–97 was constructed using overlap extension PCR. Initial fragments encoding residues 1–90 and 97–200 were generated using two pairs of primers, namely AlbGF and −A90R (5′-CAGCGCTCGAACGACAGCGCCCCCGCTTGTGCGCTGGTCCAGTTGAC-3′), and AlbGR and A90F (5′-GTCAACTGGACCAGCGCACAAGCGGGGGCGCTGTCGTTCGAGCGCTG-3′), respectively. In the second step, primer pair AlbGF and AlbGR were amplified using the fragments generated as templates. The amplified PCR fragment was cloned as described for the wild-type protein and the deletion was confirmed by DNA sequencing. Expression and purification of the Δ91–96 mutant was carried as described above for the wild-type protein.
2.2. Molecular-size analysis
Analytical gel filtration was performed using a Superose 12 10/30 FPLC column (Pharmacia) equilibrated with 50 mM Tris pH 8.5 containing 100 mM ammonium sulfate. The flow rate was 0.5 ml min−1 and A 280 was monitored continuously. The molecular-sizing standards (Bio-Rad) thyroglobulin (670 000 Da), bovine gamma globulin (158 000 Da), ovalbumin (44 000 Da), myoglobin (17 000 Da) and vitamin B (1350 Da) were used to calibrate the column. Dynamic light scattering was measured with a DynaPro MS/X dynamic light-scattering instrument (Protein Solutions) with AlbG at 35 mg ml−1 in 20 mM Tris pH 8.5 containing 200 mM ammonium sulfate. The buried surface area at the dimer interface was calculated in PISA (Krissinel & Henrick, 2007 ▶).
2.3. Crystallization and data collection
Crystallization experiments were performed under 150 µl silicon oil (Fisher) in 96-well round-bottom plates stored exposed to room humidity at 291 K (vapour diffusion under oil). The protein was tested against several commercially available crystallization screens from Hampton Research (Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2, Grid Screen PEG 6000, Grid Screen PEG/LiCl, Grid Screen Ammonium Sulfate and Grid Screen Sodium Malonate).
2.3.1. Native protein
Combining 2 µl native protein solution (20 mg ml−1 in 20 mM Tris pH 8.0, 1 mM DTT) with 2 µl precipitant solution (1 M MgSO4, 100 mM Li2SO4, 100 mM ADA pH 6.3) produced crystals with a thick plate (0.3 × 0.3 × 0.1 mm) or cube (0.15 × 0.15 × 0.15 mm) morphology that grew over a period of 5–7 d. Both crystal habits exhibited the same space group and similar unit-cell parameters. Crystals were streaked through a cryoprotectant solution consisting of 2 M MgSO4, 100 mM Li2SO4, 100 mM ADA pH 6.3, 15%(w/v) trehalose prior to vitrification by immersion in liquid nitrogen.
2.3.2. AlbGΔ91–97
Combining 2 µl of AlbGΔ91–97 protein solution (15 mg ml−1 in 20 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT, 1 mM EDTA) with 2 µl precipitant solution (100 mM ammonium tartrate pH 7.0, 12% PEG 3350) produced crystals with a bipyramidal morphology (0.2 × 0.2 × 0.2 mm) over 2–3 d. The crystals were transferred into 30 µl of the original precipitant solution, which was then allowed to dehydrate to approximately one-third of its original volume (15–30 min). Slow dehydration was required to increase the PEG 3350 concentration to the point where it could act as a cryoprotectant (>25%) and to prevent crystal cracking. Attempts to serially transfer crystals into higher levels of PEG 3350 to achieve the same effect resulted in higher levels of mosaicity and poor diffraction. Crystals were vitrified by immersion in liquid nitrogen prior to data collection.
Data were collected on an R-AXIS IV image-plate detector using X-rays generated from an RU-H3R rotating-anode generator (Cu Kα wavelength) operating at 50 kV and 100 mA. Data were processed and scaled utilizing MOSFLM (Leslie, 2006 ▶) and SCALA (Evans, 2006 ▶), respectively. The crystals of native AlbG belonged to space group C2 with a dimer in the asymmetric unit, yielding a solvent content of 51.4%, while those of the AlbGΔ91–97 mutant belonged to space group P212121 with a dimer in the asymmetric unit, yielding a solvent content of 47.4%.
2.4. Structure determination
A single crystal of AlbG was soaked in a trimethyllead acetate (TMLA) doped solution (2 M MgSO4, 100 mM Li2SO4, 100 mM ADA pH 6.3, 100 mM TMLA) for 5 min and was then streaked through cryoprotectant solution and vitrified by immersion in liquid nitrogen. The derivative was not isomorphous with the native data set, so the phases were determined by single-wavelength anomalous dispersion (SAD). The program PHENIX (Adams et al., 2004 ▶) was utilized to locate 21 Pb3+-binding sites and to produce initial solvent-flattened SAD phases. The program ARP/wARP (Perrakis et al., 1997 ▶) was used for automated model building to the experimentally determined phases. The initial ARP/wARP model, which consisted of approximately 80% of the final model, underwent rigid-body refinement to determine phases against a higher resolution, but somewhat non-isomorphous, native data set. Cycles of model building in the molecular-graphics program Coot (Emsley & Cowtan, 2004 ▶) followed by refinement in PHENIX were then performed to arrive at the final model. Waters were added to map peaks (F o − F c > 3.5 σ and 2F o − F c > 1.0σ) when the peaks were located adjacent to atoms with proper hydrogen-bonding potential. After converging on a stable R factor, the last rounds of model building included translation–libration–screw (TLS) refinement (Winn et al., 2001 ▶). TLS bodies were determined by submitting refined coordinates to the TLSMD server (Painter & Merritt, 2006 ▶) with a cutoff of five TLS bodies per monomer. The structure of the AlbGΔ91–97 deletion mutant was determined by molecular replacement utilizing the native dimeric AlbG structure as a search model and the molecular-replacement module within PHENIX. The structure of the AlbGΔ91–97 deletion mutant was refined using a similar protocol as used for the native protein; however, residues 90–100 were not modeled until the remaining structure had converged upon a stable R factor/R free. Data-collection and refinement statistics are listed in Table 1 ▶. Structure figures were generated using PyMOL (DeLano, 2002 ▶)
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Data set | Native | TMLA† | AlbGΔ91–97 |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | P212121 |
| Unit-cell parameters (Å, °) | a = 88.8, b = 90.2, c = 56.0, β = 100.9 | a = 88.7, b = 90.8, c = 56.6, β = 102.1 | a = 58.5, b = 65.6, c = 105.9 |
| Resolution (Å) | 29.0–2.0 (2.11–2.0) | 35.0–2.2 (2.32–2.2) | 30–2.4 (2.52–2.39) |
| Completeness (%) | 99.4 (98.7) | 99.0 (94.1) | 99.4 (96.0) |
| Multiplicity | 2.9 (2.7) | 6.6 (5.7) | 4.1 (3.7) |
| Mean I/σ(I) | 24.8 (8.7) | 33.5 (12.2) | 15.7 (3.3) |
| Rmerge | 0.032 (0.11) | 0.036 (0.13) | 0.038 (0.31) |
| Wilson B factor (Å2) | 25.5 | 25.4 | 62.5 |
| Model and refinement data | |||
| Resolution (Å) | 29–2.0 (2.0–2.07) | 30–2.4 (2.55–2.34) | |
| Unique reflections | 27662 (2625) | 16585 (1550) | |
| Rcryst (%) | 16.3 (19.2) | 20.4 (35.7) | |
| Rfree (5% of data) (%) | 21.5 (25.0) | 29.3 (44.6) | |
| Contents of model | |||
| Residues (1–200) | A3–A199, B6–B199 | A9–A199, B1–B199 | |
| Waters | 198 | 25 | |
| Other | SO42− (4) | — | |
| Total atoms | 3344 | 3018 | |
| Average B factor (Å2) | |||
| Protein | 29.5 | 53.1 | |
| Waters | 36.0 | 46.9 | |
| R.m.s.d. | |||
| Bond lengths (Å) | 0.010 | 0.012 | |
| Angles (°) | 1.20 | 1.36 | |
| MolProbity statistics | |||
| Ramachandran favored (%) | 97.7 | 94.7 | |
| Ramachandran outliers (%) | 0.0 | 0.27 | |
| Rotamer outliers (%) | 1.7 | 2.4 | |
| Clashscore‡ | 5.26 (97th percentile) | 19.38 (66th percentile) | |
| Overall score‡ | 1.53 (96th percentile) | 2.44 (73rd percentile) | |
Bijvoets merged.
Scores are ranked according to structures of similar resolution as formulated in MolProbity (Chen et al., 2010 ▶).
3. Results and discussion
3.1. AlbG protein analysis
AlbG was cloned and purified to homogeneity by Ni–NTA column chromatography. AlbG is a 200-amino-acid protein with a calculated molecular weight of 22 750 Da. Analytical gel filtration and dynamic light scattering indicated molecular weights of 56 and 50 kDa, respectively, which are consistent with AlbG being a dimer in solution (data not shown). The structure of native AlbG was solved by single-wavelength anomalous dispersion and refined at 2 Å resolution (Table 1 ▶). There is a dimer in the asymmetric unit consistent with the biologically observed oligomer (see below). The final model contains all 200 amino acids minus a small number of N- and C-terminal residues that were not observed in the electron density (Table 1 ▶). Atomic coordinates and experimental structure factors for AlbG and AlbGΔ91–97 have been deposited in the PDB (PDB codes 2xt2 and 2xt4, respectively).
3.2. Overall structure
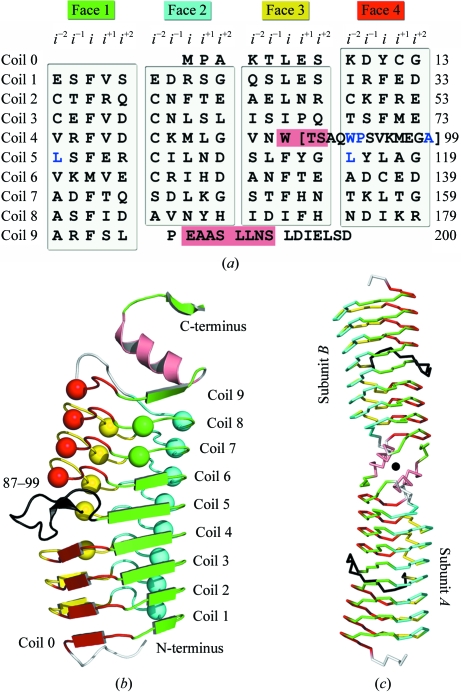
As suggested by primary-sequence analysis, AlbG folds as a right-handed quadrilateral β-helix typical of PRPs. Each coil/revolution of the β-helix (coils 0–9) is composed of four pentapeptide repeats, with each repeat forming a face (face 1–4) of a generally regular quadrilateral and each coil separated by ∼4.8 Å (Figs. 1 ▶ a and 1 ▶ b). For PRPs the typical repeat consists of two residues (termed i and i −2) whose side chains form the hydrophobic interior of the β-helix and three external residues (termed i −1, i +1 and i +2) whose side chains form the exterior of the β-helix (Hegde et al., 2005 ▶; Buchko et al., 2006 ▶; Vetting et al., 2006 ▶). The repeats are in one of two conformations, with one maximizing β-sheet interactions between coils followed by a type IV turn and one that utilizes a type II turn and produces isolated β-bridges (shown as spheres in Fig. 1 ▶ b). The composition and conformation of the AlbG pentapeptide repeats are unremarkable when contrasted with MtMfpA and EfsQnr (Hegde et al., 2005 ▶; Vetting et al., 2009 ▶) and will not be discussed further other than to note that all three resistance proteins have greater diversity in both sequence composition and pentapeptide conformation than non-PRP TPRFs and have a clustering of type IV turn-utilizing repeats in the N-terminal coils and type II turn-utilizing repeats in the C-terminal coils. The β-helix of AlbG is capped at the N-terminal end by a small N-terminal extension (residues 1–8) that transverses the quadrilateral from face 4 to face 1 and nontypical residues at the i −2 positions of coil 0 and 1, while at the C-terminus the β-helix is capped by a dimerization module (Fig. 1 ▶ c; see below).
Figure 1.
Crystal structure of AlbG from X. albilineans. (a) Sequence of AlbG mapped onto a right-handed quadrilateral β-helix diagram illustrating the position of sequence elements by coil (coil 0–9) and face (face 1–4). Residues that loop out from the β-helix are shown in brackets and α-helical residues are shown on a salmon background. (b) Subunit structure of AlbG. Pentapeptides that utilize a type IV turn are shown as β-strands, whilst those that utilize a type II turn are shown as spheres. Pentapeptides are colored by face and residues of the loop excursion are colored black. (c) Cα trace of the AlbG dimer. Residues maintain the coloring scheme of Fig. 1 ▶(b); the rotation axis is shown as a sphere.
3.3. β-Helical disruption
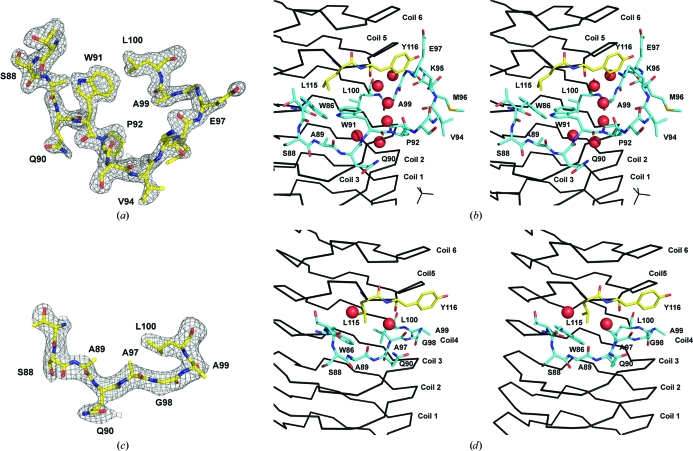
Loop disruptions of the PRP coils, while infrequent, have been visualized in the structures of Rfr23 (one 12-residue loop) and HetL (two loops of five and eight residues) (Buchko et al., 2008 ▶; Ni et al., 2009 ▶). However, these loops always connect the i +2 residue of one face to the i −2 residue of the directly preceding face, such that the β-helix is not disrupted. In AlbG there is a complete loss of one pentapeptide repeat and partial disruption of another. Between Trp86 (the i residue of face 3/coil 4; F3/C4) and Leu100 (the i −2 residue of face1/coil5; F1/C5) there are 13 residues that deviate from the repeating structure and loop out towards the solvent (Figs. 1 ▶, 2 ▶ a and 2 ▶ b). There is a break in the typical hydrogen-bonding scheme between coils 3/4/5 on face 4 and a change in the β-helical axis, producing a visually distinct lower domain (C0–C4) and upper domain (C5–C9). A similar β-helix kink at the exact same location is found in MtMfpA but is not accompanied by an insertion loop. The loop and β-helical kink are stabilized by a number of noncanonical PRP residues (highlighted in blue in Fig. 1 ▶ a). Trp86, Leu100 and Leu115 fill the hydrophobic void left by the deviation from the β-helix and create a pocket for the side chain of Trp91 (from the loop). Six waters are captured between the inner surface of the loop and the β-helix and fill the intercoil hydrogen bonding lost by the loop-out.
Figure 2.
Structure of the AlbG β-helix loop excursion. (a) Shaken F o − F c OMIT map (2.5σ) for the region bounding the loop adjoining coils 4 and 5 on face 4. (b) Stereo diagram of the loops position relative to the β-helix. Residues from the loop (86–100) are shown as sticks with cyan C atoms, whilst structural residues (Leu115 and Tyr116) from coil 5 are shown as sticks with yellow C atoms. Waters are shown as red spheres. (c) Shaken F o − F c OMIT map (2.5σ) for corresponding residues after reconstruction of the sequence to remove the β-helix loop excursion (AlbGΔ91–97). (d) Stereo diagram of the AlbGΔ91–97 structure about the deletion. For the OMIT maps, residues 87–100 were removed from the structure and the coordinates were randomly shaken by 0.3 Å, which was followed by a round of steepest-gradient refinement in PHENIX.
3.4. AlbGΔ91–97 deletion mutant
An attempt was made to replace the loop with a normal PRP repeat. There are six excess residues in the loop, so residues Trp91–Glu97 were replaced with a single alanine, such that coil 4 consisted of face 3, V84N85W86T87S88, and face 4, A89Q90A97G98A99. AlbGΔ91–97 crystallized in a different space group to the wild type and its structure was determined by molecular replacement. The creation of a normal PRP repeat on face 4 by excision of the loop was successful, with the new PRP repeat participating in normal coil–coil hydrogen bonding with coil 3 (Figs. 2 ▶ b and 2 ▶ c) in a type IV turn conformation. However, the deletion has not significantly repaired the gap between coils 4 and 5, where a lack of hydrogen bonding remains. For example, the intercoil distance between the i +2 residues of coils 3 and 5 on face 4 is 16 Å, whereas the normal two-coil PRP span would be ∼9.6 Å. For AlbGΔ91–97 this distance is 13.8 Å, indicating a minor closing of the intercoil gap. The AlbGΔ91–97 structure indicates that the noncanonical PRP residues that remain in the deletion structure (Trp86, Leu100 and Leu115) are a large factor in the helical kink. Unfortunately, the gyrase poison that AlbG confers immunity to, albicidin, is not commercially available so the effect of the loop removal on activity was not determined.
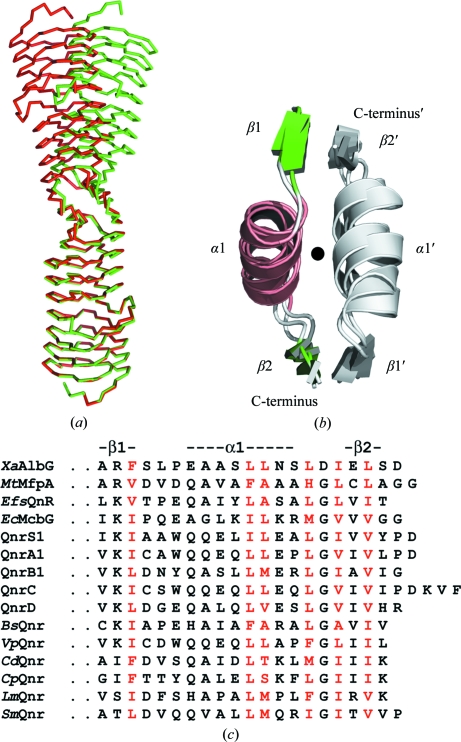
3.5. A common dimer interface for PRP TPRFs
All PRP TPRF structures solved to date (MtMfpA, EfsQnr and AlbG) form a highly asymmetric dimer (∼15–30 Å diameter, 110 Å in length) mediated by a conserved C-terminal dimerization module consisting of a strand (β1)/helix (α1)/strand (β2) (Figs. 1 ▶ c, 3 ▶ a and 3 ▶ b). The module forms an extended structure that interacts with its twofold-related partner. The main interactions are between hydrophobic side chains along the interface and main-chain hydrogen bonds between β2 in one subunit and β1 in the opposing subunit (parallel fashion). The PRP dimer interface is quite small, with only 900 Å2 buried upon dimer formation per AlbG subunit. Flipping and superposition of the dimer upon itself (subunit A to subunit B; rotation, ω = 144.9°, ϕ = −26°, κ = 169.4°) revealed that the dimer in the asymmetric unit of the C2 crystal form (native AlbG) is imperfect. There is a large deviation of up to an 18 Å Cα difference at the N-terminal end in the location of the dimer-related subunit (Fig. 3 ▶ a). This asymmetry is generated by flexibility at the dimer interface, with α1 translating (∼3 Å) along hydrophobic interactions made with its dimer-related helix (α1′) and a change in the angle between α1 and β2 (∼5°). This asymmetry is presumably supported by crystal contacts to the individual subunits, resulting in a captured asymmetric dimer. The A and B subunits make crystal contacts that bury 1383 Å2 (four contact points) and 1090 Å2 (two contact points), respectively. These contact points are distributed at several points throughout the dimer surface, so it is speculative to speak of what effect each may have on the asymmetry of the dimer. In contrast, for AlbGΔ91–97, which crystallized in a different space group, the molecular dimer is nearly symmetric, with maximal Cα deviations of ∼3 Å at the N-terminus. Though not previously noted, a similar dimer asymmetry can be found in MtMfpA (PDB entry 2bm5; maximal Cα deviation of ∼11 Å; Hegde et al., 2005 ▶). Based on the similar position of hydrophobic residues within the C-terminal sequences of other PRP topoisomerase poison resistance factors, it is likely that they will all contain the same β1/α1/β2 dimerization module (Fig. 3 ▶ c). This is in contrast to other members of the PRP family, which do not contain similar sequence markers and to date have all been determined to be monomeric (Vetting et al., 2007 ▶; Buchko et al., 2008 ▶; Ni et al., 2009 ▶). This suggests that dimerization and perhaps flexibility of the dimer interface is a key factor in their binding to topoisomerases. Indeed, since topoisomerase II enzymes are α2β2 heterotetramers and undergo large conformational changes during the topoisomerase reaction, the dimerization of PRP TPRFs can yield higher binding affinity, while the flexibility of the PRP TPRF dimer can facilitate the capture of selected topoisomerase II conformations. The finding of a common feature amongst PRP TPRFs is a significant step forward in their analysis since it is difficult to discern which coils/repeats are structurally analogous for their interactions with toposiomerases owing to their low sequence homology . The PRP TPRF dimerization domain provides an anchor point for future structure-based sequence alignments.
Figure 3.
Description of the PRP TPRF dimerization module. (a) Illustration of the asymmetric dimer in the C2 crystal form. The AlbG dimer was superimposed on itself by aligning subunit A of one dimer (shown in red) with subunit B of the same structure (shown in green). (b) Structure alignment of the dimerization modules from AlbG, MtMfpA (PDB entry 2bm5) and EfsQnr (PDB entry 2w7z; Vetting et al., 2009 ▶). One module is colored by secondary structure, while its dimer-related partner is colored gray. (c) Comparison of amino-acid sequences of dimerization modules of PRP TPRFs: X. albilineans AlbG (XaAlbG; gi:46425373), Mycobacterium tuberculosis MfpA (MtMfpA; gi:15610497), Enterococcus faecalis Qnr (EfsQNR; gi:29342949), Escherichia coli McbG (EcMcbG; gi:218349863), QnrS1 (gi:156601490), QnrA1 (gi:169117961), QnrB1 (gi:110672126), QnrC (gi:218202688), QnrD (gi:221101560), Bacillus subtilis Qnr (BsQnr; gi:16078153), Vibrio parahaemolyticus Qnr (VpQnr; gi:153837620), Clostridium difficile Qnr (CdQnr; gi:182626580), C. perfringens (CpQnr; gi:182626580), Listeria monocytogenes Qnr (LmQnr; gi:255519898) and Stenotrophomonas maltophilia Qnr (SmQnr; gi:300245036). Protein names without organism identifiers are plasmid-encoded. Residues highlighted in red are conserved hydrophobic interfacial residues.
Supplementary Material
PDB reference: AlbG, 2xt2
PDB reference: AlbGΔ91–97, 2xt4
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (AI33696 to JSB).
References
- Adams, P. D., Gopal, K., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Pai, R. K., Read, R. J., Romo, T. D., Sacchettini, J. C., Sauter, N. K., Storoni, L. C. & Terwilliger, T. C. (2004). J. Synchrotron Rad. 11, 53–55. [DOI] [PubMed]
- Basnayake, W. V. & Birch, R. G. (1995). Microbiology, 141, 551–560. [DOI] [PubMed]
- Bateman, A., Murzin, A. G. & Teichmann, S. A. (1998). Protein Sci. 7, 1477–1480. [DOI] [PMC free article] [PubMed]
- Birch, R. G. (1983). PhD thesis. Honolulu: University of Hawaii.
- Birch, R. G. & Patil, S. S. (1985a). US Patent 4525354.
- Birch, R. G. & Patil, S. S. (1985b). J. Gen. Microbiol. 131, 1069–1075. [DOI] [PubMed]
- Birch, R. G. & Patil, S. S. (1987a). Physiol. Mol. Plant Pathol. 30, 199–206.
- Birch, R. G. & Patil, S. S. (1987b). Physiol. Mol. Plant Pathol. 30, 207–214.
- Birch, R. G., Pemberton, J. M. & Basnayake, W. V. (1990). J. Gen. Microbiol. 136, 51–58. [DOI] [PubMed]
- Black, K., Buikema, W. J. & Haselkorn, R. (1995). J. Bacteriol. 177, 6440–6448. [DOI] [PMC free article] [PubMed]
- Bostock, J. M., Huang, G., Hashimi, S. M., Zhang, L. & Birch, R. G. (2006). J. Appl. Microbiol. 101, 151–160. [DOI] [PubMed]
- Buchko, G. W., Ni, S., Robinson, H., Welsh, E. A., Pakrasi, H. B. & Kennedy, M. A. (2006). Protein Sci. 15, 2579–2595. [DOI] [PMC free article] [PubMed]
- Buchko, G. W., Robinson, H., Pakrasi, H. B. & Kennedy, M. A. (2008). J. Struct. Biol. 162, 184–192. [DOI] [PubMed]
- Chandler, L. E., Bartsevich, V. V. & Pakrasi, H. B. (2003). Biochemistry, 42, 5508–5514. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org. [DOI] [PubMed]
- Deweese, J. E. & Osheroff, N. (2009). Nucleic Acids Res. 37, 738–748. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Garrido, M. C., Herrero, M., Kolter, R. & Moreno, F. (1988). EMBO J. 7, 1853–1862. [DOI] [PMC free article] [PubMed]
- Hashimi, S. M., Huang, G., Maxwell, A. & Birch, R. G. (2008). Antimicrob. Agents Chemother. 52, 1382–1390. [DOI] [PMC free article] [PubMed]
- Hashimi, S. M., Wall, M. K., Smith, A. B., Maxwell, A. & Birch, R. G. (2007). Antimicrob. Agents Chemother. 51, 181–187. [DOI] [PMC free article] [PubMed]
- Hegde, S. S., Vetting, M. W., Roderick, S. L., Mitchenall, L. A., Maxwell, A., Takiff, H. E. & Blanchard, J. S. (2005). Science, 308, 1480–1483. [DOI] [PubMed]
- Huang, G., Zhang, L. & Birch, R. G. (2001). Microbiology, 147, 631–642. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Montero, C., Mateu, G., Rodriguez, R. & Takiff, H. (2001). Antimicrob. Agents Chemother. 45, 3387–3392. [DOI] [PMC free article] [PubMed]
- Ni, S., Sheldrick, G. M., Benning, M. M. & Kennedy, M. A. (2009). J. Struct. Biol. 165, 47–52. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006). J. Appl. Cryst. 39, 109–111.
- Perrakis, A., Sixma, T. K., Wilson, K. S. & Lamzin, V. S. (1997). Acta Cryst. D53, 448–455. [DOI] [PubMed]
- Rodríguez-Martínez, J. M., Velasco, C., Briales, A., García, I., Conejo, M. C. & Pascual, A. (2008). J. Antimicrob. Chemother. 61, 1240–1243 [DOI] [PubMed]
- Rott, P. & Davis, M. J. (2000). Leaf Scald. Montpellier, France: La Librairie du Cirad.
- Rott, P. C., Costet, L., Davis, M. J., Frutos, R. & Gabriel, D. W. (1996). J. Bacteriol. 178, 4590–4596. [DOI] [PMC free article] [PubMed]
- Royer, M., Costet, L., Vivien, E., Bes, M., Cousin, A., Damais, A., Pieretti, I., Savin, A., Megessier, S., Viard, M., Frutos, R., Gabriel, D. W. & Rott, P. C. (2004). Mol. Plant Microbe Interact. 17, 414–427. [DOI] [PubMed]
- Tran, J. H. & Jacoby, G. A. (2002). Proc. Natl Acad. Sci. USA, 99, 5638–5642. [DOI] [PMC free article] [PubMed]
- Vetting, M. W., Hegde, S. S. & Blanchard, J. S. (2009). Acta Cryst. D65, 462–469. [DOI] [PMC free article] [PubMed]
- Vetting, M. W., Hegde, S. S., Fajardo, J. E., Fiser, A., Roderick, S. L., Takiff, H. E. & Blanchard, J. S. (2006). Biochemistry, 45, 1–10. [DOI] [PMC free article] [PubMed]
- Vetting, M. W., Hegde, S. S., Hazleton, K. Z. & Blanchard, J. S. (2007). Protein Sci. 16, 755–760. [DOI] [PMC free article] [PubMed]
- Vivien, E., Megessier, S., Pieretti, I., Cociancich, S., Frutos, R., Gabriel, D. W., Rott, P. C. & Royer, M. (2005). FEMS Microbiol. Lett. 251, 81–89. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Zhang, L. & Birch, R. G. (1997). Proc. Natl Acad. Sci. USA, 94, 9984–9989. [DOI] [PMC free article] [PubMed]
- Zhang, L., Xu, J. & Birch, R. G. (1998). Microbiology, 144, 555–559. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AlbG, 2xt2
PDB reference: AlbGΔ91–97, 2xt4