TEM-72 is a quadruple mutant of TEM-1 and shows extended-spectrum β-lactamase properties. The present structure shows the presence of a citrate anion bound to the TEM-72 active site and supports the use of polycarboxylates as a scaffold for the design of broad-spectrum inhibitors of serine β-lactamases.
Keywords: class A β-lactamases, extended-spectrum β-lactamases, inhibitors, citrate, polycarboxylate
Abstract
TEM-72, a class A β-lactamase identified in isolates of Enterobacteriaceae, is a quadruple mutant of TEM-1 (Q39K, M182T, G238S and E240K) and shows extended-spectrum β-lactamase (ESBL) properties arising from the G238S and E240K substitutions. Although many structures of TEM variants have been published, they do not include an enzyme with the simultaneous presence of both of the ESBL-conferring G238S and E240K substitutions. Furthermore, the structure shows the presence of a citrate anion bound to the TEM-72 active site, where it interacts with all of the conserved residues of class A β-lactamases. The present structure supports the use of polycarboxylates as a scaffold for the design of broad-spectrum inhibitors of serine β-lactamases.
1. Introduction
The TEM family of β-lactamases belongs to Ambler class A and includes almost 200 variants of the original TEM-1/2 enzymes. These TEM variants, and especially those that show activity against oxyimino cephalosporins and a reduced susceptibility to mechanism-based inactivators (e.g. clavulanate and tazobactam), represent important bacterial resistance factors against β-lactam antibiotics (Bush et al., 1995 ▶; Bush & Jacoby, 1997 ▶, 2010 ▶; http://www.lahey.org/studies/). TEM-72, a quadruple mutant of TEM-1, has been identified in isolates of Enterobacteriaceae in Italian hospitals and shows extended-spectrum β-lactamase (ESBL) properties thanks to the presence of Gly238Ser and Glu240Lys substitutions (Perilli et al., 2000 ▶). To counter the activity of ESBLs several inhibitors have been developed such as clavulanate, tazobactam and sulbactam (Georgopapadakou, 2004 ▶), the function of which consists of acylating either the catalytic serine (Ser70) and/or the conserved Ser130 present in the active site of class A β-lactamases (Brown et al., 1996 ▶; Bush & Mobashery, 1998 ▶). However, inhibitor-resistant TEM β-lactamases (IRTs) have emerged clinically (Thomas et al., 2005 ▶). Thus, the search for new classes of molecules able to inhibit serine β-lactamases constitutes a very active research field (Morandi et al., 2008 ▶; Tondi et al., 2010 ▶). We have recently reported the crystal structure of the complex between the class D β-lactamase OXA-46 and tartrate, providing a structural explanation of the ability of polycarboxylates to act as inhibitors of this class of enzymes (Docquier et al., 2010 ▶). In the present work, we report the structural characterization of a class A TEM-72 enzyme with ESBL activity that presents an original set of substitutions (Q39K, M182T, G238S and E240K). Although many structures of TEM variants have been published, they do not include enzymes with the simultaneous presence of both of the ESBL-conferring G238S and E240K substitutions. Our data also support the capability of citrate anions to bind in the active site of class A β-lactamases, suggesting the possibility that polycarboxylates may act as broad-spectrum inhibitors of serine β-lactamases.
2. Experimental
2.1. Protein purification and crystallization
The TEM-72 β-lactamase from Morganella morganii was cloned, expressed and purified from Escherichia coli BL21 (DE3) [pET-TEM-72] strain as described previously (Perilli et al., 2000 ▶). The pure enzyme was concentrated to 10 mg ml−1 (in 10 mM HEPES pH 7.45) using ultrafiltration (Microcon concentrators, Millipore, Bedford, Massachusetts, USA) and used in screening crystallization conditions with Crystal Screen and Crystal Screen 2 (Hampton Research, Aliso Viejo, California, USA) in a sitting-drop setup. 2 µl protein solution was added to an equal amount of precipitant solution and equilibrated at 298 K. Crystals appeared in 4 d from Crystal Screen condition No. 9 [0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 30%(w/v) polyethylene glycol 4000] and Crystal Screen 2 condition No. 16 [0.5 M sodium chloride, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 2%(v/v) ethyleneimine polymer].
However, since these crystals were small and not suitable for X-ray diffraction analysis, further optimization of the initial crystallization conditions was carried out by changing some of the crystallization parameters. Optimization of crystallization conditions was performed at 298 K using a 24-well sitting-drop plate sealed with clear sealing tape (Cryschem, Hampton Research). Small plate-like crystals that were suitable for microseeding were grown in 4 µl drops containing a 1:1(v:v) mixture of 8 mg ml−1 protein solution and 0.2 M ammonium acetate, 0.1 M sodium citrate pH 5.5, 5.7 or 6.0 and polyethylene glycol 4000 at 20–30%(w/v). Plate-like crystals were crushed using a Seed Bead (Hampton Research) in the precipitant buffer. TEM-72 single crystals that were useful for data collection were obtained from a precipitant solution consisting of 0.1 M sodium citrate, 0.2 M ammonium acetate, 30% PEG 4000 pH 6.0 and 100-fold diluted seed solution at 298 K.
2.2. Data collection, structure determination and refinement
Before data collection, crystals were cryoprotected by adding 20% glycerol to the mother liquor and were flash-frozen in a cold (100 K) nitrogen stream. Data collection was performed on beamline ID23-1 at the European Synchrotron Radiation Facility, Grenoble, France. Data were indexed, integrated and scaled using MOSFLM and SCALA (Collaborative Computational Project, Number 4, 1994 ▶; Evans, 1997 ▶; Leslie, 2006 ▶). The structure was solved by molecular replacement with the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶; Vagin & Teplyakov, 1997 ▶) using the TEM-1 structure as a search model (PDB entry 1zg4; Stec et al., 2005 ▶). The TEM-72 model was built by a combination of automatic (ARP/wARP; Collaborative Computational Project, Number 4, 1994 ▶; Morris et al., 2003 ▶) and manual (Coot; Collaborative Computational Project, Number 4, 1994 ▶; Emsley & Cowtan, 2004 ▶) rebuilding programs. The refinement protocol involved a sequence of iterative manual rebuilding of the model and maximum-likelihood refinement (Collaborative Computational Project, Number 4, 1994 ▶; Murshudov et al., 1997 ▶) using TLS parametrization (Winn et al., 2001 ▶). Water molecules were added using standard procedures within the ARP/wARP suite. The stereochemical quality of the final model was checked using the program PROCHECK (Laskowski et al., 1993 ▶). Data-collection and refinement statistics are reported in Table 1 ▶. The coordinates and structure factors for TEM-72 have been deposited in the Protein Data Bank under accession code 3p98.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | ESRF ID23-1 |
| Wavelength (Å) | 1.008 |
| Data-collection temperature (K) | 100 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 60.63, b = 90.21, c = 96.05 |
| Subunits per asymmetric unit | 2 |
| Matthews coefficient (Å3 Da−1) | 2.22 |
| Solvent content (%) | 44.61 |
| Resolution limits (Å) | 37.64–2.10 (2.21–2.10) |
| Reflections measured | 80821 (11624) |
| Unique reflections | 30846 (4389) |
| Completeness (%) | 98.6 (98.0) |
| Rmerge (%) | 10.7 (34.3) |
| Multiplicity | 2.6 (2.6) |
| 〈I/σ(I)〉 | 9.0 (2.9) |
| Wilson B factor (Å2) | 17.59 |
| Rcryst (%) | 20.0 (25.5) |
| Rfree (%) | 28.3 (32.0) |
| Protein atoms | 4056 |
| Ligand atoms (two citrate molecules) | 26 |
| Other atoms (PEG fragments) | 14 |
| Water molecules | 252 |
| Average B factor (Å2) | 15.00 |
| R.m.s.d. bond lengths (Å) | 0.022 |
| R.m.s.d. bond angles (°) | 1.943 |
| R.m.s.d. planar groups (Å) | 0.009 |
| R.m.s.d. chiral centres (Å3) | 0.129 |
| E.s.d. on atomic positions (Å) | 0.162 |
| Ramachandran favoured (%) | 95.4 |
| Ramachandran allowed (%) | 4.6 |
| PDB code | 3p98 |
3. Results and discussion
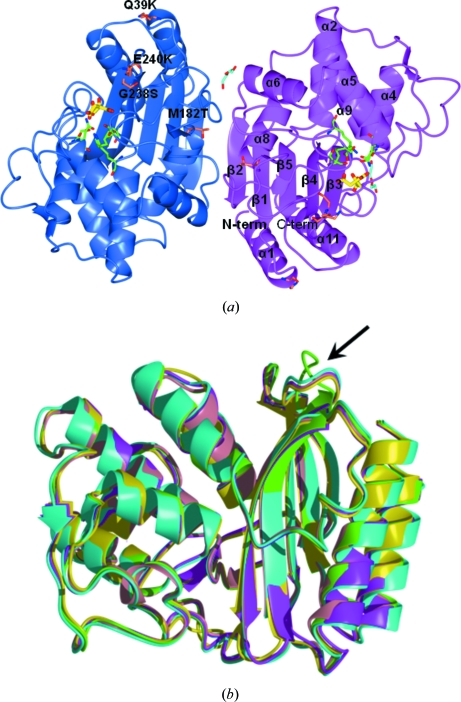
The numbering scheme used follows that proposed for class A β-lactamases (Ambler et al., 1991 ▶) and used for previously released TEM-type β-lactamase crystal structures. TEM-72 crystallizes in space group P212121 with two independent molecules in the asymmetric unit. Least-squares superposition shows that the two molecules are identical (r.m.s.d. of 0.26 Å on 263 Cα atoms, which is well within three times the expected coordinate error at this resolution) and adopt the same conformation in all parts of the molecule. A schematic representation of the three-dimensional structure of TEM-72 is shown in Fig. 1 ▶(a). The protein tertiary structure consists of an αβα sandwich characteristic of active-site serine β-lactamases, in which a five-stranded β-sheet is surrounded by eight helices on one side and three helices on the other that comprise the amino-terminal and carboxy-terminal helices. The disulfide bridge between Cys77 and Cys123 present in the other TEMs is conserved, as is the cis-peptide bond between Glu166 and Pro167. These two residues belong to a large loop (residues 155–183) that includes a short helix starting at Pro167 which shapes the active-site cleft. The two independent molecules are not related by noncrystallographic symmetry, but are quite close and interact by forming two intermolecular hydrogen bonds involving backbone atoms from residues belonging to the β-sheet motif. This appears to be sufficient to align the two molecules to form a loose intermolecular β-sheet as shown in Fig. 1(a) ▶. Analysis of the TEM-72 quaternary structure with the PISA tool (Krissinel & Henrick, 2007 ▶) suggests that the intermolecular assembly is a consequence of packing effects and does not represent a stable quaternary structure. Gel-filtration experiments confirmed that the dimer observed in the crystal is not maintained in solution. Superimposition with other enzymes of the TEM family of known structure (TEM-1, TEM-30, TEM-32, TEM-34, TEM-52, TEM-64 and TEM-76) shows that they are all very similar, with r.m.s.d.s on common Cα atoms in the 0.35–0.50 Å range. Interestingly, the only noticeable differences between TEM-72 and the other TEM-type enzymes are in the loop linking the β4 and β5 strands and in the conformation of the smallest β5 strand where, despite sequence conservation, deviations of up to about 6.0 Å are present (Fig. 1b) ▶. This might reflect the impact of the simultaneous presence of both of the ESBL-conferring substitutions at positions 238 and 240, which are observed for the first time in this structure. This observation (i.e. the alternate conformation of the β4–β5 loop) would be in agreement with the proposed increased flexibility of the enzyme arising from these ESBL-conferring mutations (Wang et al., 2002a ▶). However, crystallization and/or packing effects are also most probably involved considering the exposure of this loop at the protein surface.
Figure 1.
(a) View of the contents of the crystal asymmetric unit. The two independent TEM-72 molecules (subunit A, blue; subunit B, magenta) are related by an almost exact noncrystallographic twofold axis perpendicular to the plane of the figure. Secondary-structure elements are indicated on subunit B (α, α-helices; β, β-strands). For clarity, only the eight longest α-helices are labelled. The two citrate molecules found bound to the active site and the molecules interpreted as PEG fragments are shown as yellow and cyan sticks, respectively. The active-site residues relevant to catalysis are shown as green sticks. The positions of the typical TEM-72 amino-acid substitutions compared with TEM-1 are labelled on subunit A. (b) Superimposition of TEM-72 (green) with available structures of TEM-type enzymes: TEM-1 (PDB code 1btl, cyan; Jelsch et al., 1993 ▶), TEM-30 (PDB code 1lhy, pink; Wang et al., 2002b ▶), TEM-52 (PDB code 1htz, magenta; Orencia et al., 2001 ▶) and TEM-64 (PDB code 1jwz, yellow; Wang et al., 2002a ▶). The β4–β5 loop, the conformation of which is different from that of other TEM-type β-lactamase structures, is indicated by an arrow.
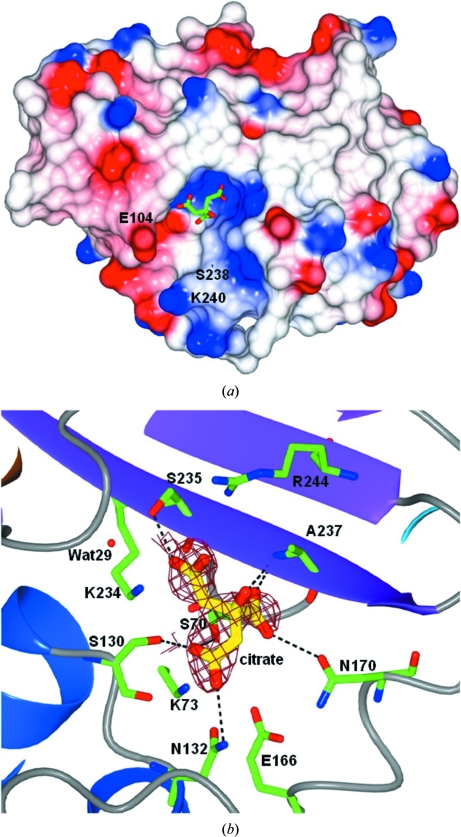
The active-site cavity is a shallow and wide opening on the side of the β-sheet delimited by the two helical domains and maintains the main features observed previously in TEM-1 and TEM-2 as shown in Fig. 2 ▶(a). The Gly238Ser and Glu240Lys mutations that are present in TEM-72 lie at one of the entrances to the active site (Fig. 2 ▶ a). While the Ser238 side chain points towards the core of the molecule and towards Met69, the side chain of Lys240 determines the charge of the rim of the active site opposite Glu104 (Fig. 2 ▶ a).
Figure 2.
(a) Molecular surface coloured by electrostatic potential (red, negative charge; blue, positive charge). The predominant positive charge present in the active-site cavity drives the binding of the citrate anion (green sticks). (b) View of the active-site cavity of one of the TEM-72 subunits showing the citrate anion (yellow sticks) with a superimposed OMIT map calculated using the refined phases obtained without the contribution of the citrate molecule (yellow sticks; map contoured at 2.5σ). The active-site residues are shown as green sticks. Figures were prepared using CCP4MG (Potterton et al., 2002 ▶).
As shown in Figs. 2 ▶(a) and 2 ▶(b), the TEM-72 crystal structure shows a citrate molecule from the crystallization buffer tightly bound in the active site of both independent molecules in the cell, in which it has been modelled in slightly different orientations. Citrate is engaged in several hydrogen bonds to the residues present in the active-site cavity, including the catalytic Ser70, Ser130, Asn132, Asn170, Ser235 and Ala237, and is within contact distance of Lys234 and Arg244 (see Fig. 2 ▶ b). One of the carboxylate groups of citrate occupies the ‘oxyanion hole’ where a water or a sulfate is observed in other TEM-type enzyme structures (Jelsch et al., 1993 ▶). However, the conformations of all of the residues involved in catalysis are not influenced by the binding of citrate as evidenced by structural comparison with other TEM-type enzymes such as TEM-1 (Jelsch et al., 1993 ▶; Maveyraud et al., 1998 ▶; Stec et al., 2005 ▶) and TEM-76 (Thomas et al., 2005 ▶). Interestingly, a citrate anion has been observed in the active sites of various β-lactamases, including the class A carbapenemase KPC-2 (Petrella et al., 2008 ▶; PDB entry 3c5a), the plasmid-encoded class C β-lactamase CMY-2 (PDB entry 1zc2; C. Bauvois, L. Jacquamet, S. Fieulaine, J.-M. Frere, M. Galleni & J.-L. Ferrer, unpublished work) and even a metallo-β-lactamase (PDB entry 1mqo; I. Garcia-Saez, L. Chantalat & O. Dideberg, unpublished work). In addition, binding of a tartrate molecule in the active site of the class D β-lactamase OXA-46 (Docquier et al., 2010 ▶) has also been observed. The binding of a citrate or tartrate molecule in the active site of β-lactamases relies on interactions involving invariant residues (or also metal ions in the case of class B enyzmes) which are relevant to catalysis of β-lactam hydrolysis. In conclusion, the binding mode of these closely related molecules in different classes of β-lactamases strongly supports the possibility of developing wide-spectrum inhibitors based on a polycarboxylate scaffold.
Supplementary Material
PDB reference: TEM-72, 3p98
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility (ESRF; Grenoble, France) for providing access to the ID23-1 beamline and the ESRF staff for excellent assistance. This work was funded in part by grants from the European Union and by the Italian Ministero dell’Istruzione, Università e Ricerca (MIUR; contract Nos. HPRN-CT-2002-00264 and 2005061894_004). SM also acknowledges the contribution of MIUR (PRIN2007 2007M5MWM9_001 research grant). VC was the recipient of a postdoctoral fellowship from the Italian CIRMPP consortium.
References
- Ambler, R. P., Coulson, A. F., Frère, J. M., Ghuysen, J. M., Joris, B., Forsman, M., Levesque, R. C., Tiraby, G. & Waley, S. G. (1991). Biochem. J. 276, 269–270. [DOI] [PMC free article] [PubMed]
- Brown, R. P., Aplin, R. T. & Schofield, C. J. (1996). Biochemistry, 35, 12421–12432. [DOI] [PubMed]
- Bush, K. & Jacoby, G. (1997). J. Antimicrob. Chemother. 39, 1–3. [DOI] [PubMed]
- Bush, K. & Jacoby, G. A. (2010). Antimicrob. Agents Chemother. 54, 969–976. [DOI] [PMC free article] [PubMed]
- Bush, K., Jacoby, G. A. & Medeiros, A. A. (1995). Antimicrob. Agents Chemother. 39, 1211–1233. [DOI] [PMC free article] [PubMed]
- Bush, K. & Mobashery, S. (1998). Adv. Exp. Med. Biol. 456, 71–98. [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Docquier, J.-D., Benvenuti, M., Calderone, V., Giuliani, F., Kapetis, D., De Luca, F., Rossolini, G.-M. & Mangani, S. (2010). Antimicrob. Agents Chemother. 54, 2167–2174. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 33, 22–24.
- Georgopapadakou, N. H. (2004). Expert Opin. Investig. Drugs, 13, 1307–1318. [DOI] [PubMed]
- Jelsch, C., Mourey, L., Masson, J.-M. & Samama, J.-P. (1993). Proteins, 16, 364–383. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Maveyraud, L., Pratt, R. F. & Samama, J.-P. (1998). Biochemistry, 37, 2622–2628. [DOI] [PubMed]
- Morandi, S., Morandi, F., Caselli, E., Shoichet, B. K. & Prati, F. (2008). Bioorg. Med. Chem. 16, 1195–1205. [DOI] [PMC free article] [PubMed]
- Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003). Methods Enzymol. 374, 229–244. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Orencia, M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. (2001). Nature Struct. Biol. 8, 238–242. [DOI] [PubMed]
- Perilli, M., Segatore, B., de Massis, M. R., Riccio, M. L., Bianchi, C., Zollo, A., Rossolini, G.-M. & Amicosante, G. (2000). Antimicrob. Agents Chemother. 44, 2537–2539. [DOI] [PMC free article] [PubMed]
- Petrella, S., Ziental-Gelus, N., Mayer, C., Renard, M., Jarlier, V. & Sougakoff, W. (2008). Antimicrob. Agents Chemother. 52, 3725–3736. [DOI] [PMC free article] [PubMed]
- Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K. & Noble, M. (2002). Acta Cryst. D58, 1955–1957. [DOI] [PubMed]
- Stec, B., Holtz, K. M., Wojciechowski, C. L. & Kantrowitz, E. R. (2005). Acta Cryst. D61, 1072–1079. [DOI] [PubMed]
- Thomas, V. L., Golemi-Kotra, D., Kim, C., Vakulenko, S. B., Mobashery, S. & Shoichet, B. K. (2005). Biochemistry, 44, 9330–9338. [DOI] [PMC free article] [PubMed]
- Tondi, D., Calò, S., Shoichet, B. K. & Costi, M. P. (2010). Bioorg. Med. Chem. Lett. 20, 3416–3419. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022–1025.
- Wang, X., Minasov, G. & Shoichet, B. K. (2002a). J. Mol. Biol. 320, 85–95. [DOI] [PubMed]
- Wang, X., Minasov, G. & Shoichet, B. K. (2002b). J. Biol. Chem. 277, 32149–32156. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: TEM-72, 3p98