This work describes the purification and preliminary crystallographic analysis of the CBS-pair regulatory domain of the human ancient domain protein 4 (ACDP4), also known as CNNM4.
Keywords: Mg2+ transporters, CBS domains, Jalili syndrome, biomineralization, sensory transduction, vision, cyclin M4, ion transport, magnesium, ancient conserved domain-containing protein 4
Abstract
This work describes the purification and preliminary crystallographic analysis of the CBS-pair regulatory domain of the human ancient domain protein 4 (ACDP4), also known as CNNM4. ACDP proteins represent the least-studied members of the eight different types of magnesium transporters that have been identified in mammals to date. In humans the ACDP family includes four members: CNNM1–4. CNNM1 acts as a cytosolic copper chaperone and has been associated with urofacial syndrome, whereas CNNM2 and CNNM4 have been identified as magnesium transporters. Interestingly, mutations in the CNNM4 gene have clinical consequences that are limited to retinal function and biomineralization and are considered to be the cause of Jalili syndrome, which consists of autosomal recessive cone-rod dystrophy and amelogenesis imperfecta. The truncated protein was overexpressed, purified and crystallized in the orthorhombic space group C222. The crystals diffracted X-rays to 3.6 Å resolution using synchrotron radiation. Matthews volume calculations suggested the presence of two molecules in the asymmetric unit, which were likely to correspond to a CBS module of the CBS pair of CNNM4.
1. Introduction
Magnesium, which is the second most abundant cation within the human body, is an essential element that is required for the catalytic activity of numerous metalloenzymes and can also serve a purely structural role by stabilizing the conformation of certain metal-dependent protein domains and transcriptional regulators (Quamme, 2010 ▶). Deficiency of this essential metal has been implicated in many diseases ranging from hypertension, cardiac arrhythmias and asthma to migraines and bone disorders (Agus, 1999 ▶; Agus & Agus, 2001 ▶; Quamme, 2010 ▶). The intracellular Mg2+ level is maintained well below the concentration predicted from the transmembrane electrochemical potential, being transported into and out of a variety of intracellular organelle compartments, most likely by the action of multidomain proteins known as magnesium transporters, which have just begun to be identified at the molecular level (Günther, 1993 ▶; Quamme, 1997 ▶, 2010 ▶; Cefaratti et al., 2000 ▶; Romani & Scarpa, 2000 ▶; Tashiro et al., 2000 ▶; Watanabe et al., 2005 ▶; Schweigel et al., 2008 ▶). Taking into account the notable physical properties of the Mg2+ cation [the unhydrated Mg2+ has the smallest diameter (0.65 Å) of all biological ions, whereas the fully hydrated cation (5.0 Å) is the largest], there are some challenges in explaining how Mg2+ moves across cellular membranes. Recently, the crystal structures of the CorA (Eshaghi et al., 2006 ▶; Lunin et al., 2006 ▶) and MgtE Mg2+ transporters have been reported (Hattori et al., 2007 ▶). In mammals, eight different types of magnesium transporters have been identified to date (Quamme, 2010 ▶). Of these, the least studied members are the ancient conserved domain proteins (ACDPs), which were first described by Wang et al. (2003 ▶) in an effort to identify the gene(s) responsible for a rare genetic disorder known as urofacial syndrome (UFS) or Ochoa syndrome (Wang et al., 2003 ▶; Ochoa, 2004 ▶). Their name is based on the fact that they all share an ‘ancient conserved domain’ (ACD) that is present in a large number of species from bacteria to zebrafish to man, suggesting that they may be essential genes (Wang et al., 2003 ▶). In humans the ACDP family includes four members, ACDP1–4, whereas it appears to be a single-copy gene in lower organisms such as Caenorhabditis elegans, yeasts and bacteria (Wang et al., 2004 ▶). The ACDP proteins are evolutionarily expressed throughout development and adult tissues, except for ACDP1, which is mainly expressed in the brain, and all show very strong homology to the bacterial CorC and yeast Amip3 proteins, which are involved in magnesium and cobalt efflux and resistance to copper toxicity, respectively (Wang et al., 2004 ▶). In Saccharomyces cerevisiae, Mam3p, an orthologue of ACDP4, is implicated in manganese toxicity (Yang et al., 2005 ▶). The presence of a cyclin box-like motif, the predicted helical architecture (typical of cyclins) and their location in the plasma membrane has led to the suggestion that ADCPs might be involved in cell-cycle regulation (Wang et al., 2003 ▶). Accordingly, ACDPs are also known as cyclins M1–4 or CNNM1–4, although these proteins do not appear to possess a cyclin function in vivo. CNNM1 appears to act as a cytosolic copper chaperone (Alderton et al., 2007 ▶) and has been associated with urofacial syndrome (UFS; Ochoa, 2004 ▶), whereas CNNM2 has been identified as a magnesium transporter (Goytain & Quamme, 2005 ▶). CNNM4 shares 68% similarity (56% homology) to CNNM2 and is also implicated in magnesium transport and biomineralization (Wang et al., 2003 ▶; Guo et al., 2005 ▶; Polok et al., 2009 ▶; Quamme, 2010 ▶). Interestingly, mutations in the CNNM4 gene have clinical consequences that are limited to retinal function and biomineralization and are considered to be the cause of Jalili syndrome, which consists of autosomal recessive cone-rod dystrophy (CRD) and amelogenesis imperfecta (AI) (Parry et al., 2009 ▶; Polok et al., 2009 ▶).
Structurally, CNNM4 (UniProtKB/Swiss-Prot code Q6P4Q7) is a multidomain protein formed of (i) a DUF21 domain (residues 184–358) that includes a leucine-zipper pattern (residues 188–209) and four transmembrane helices (residues 19–39, 179–199, 241–261, 265–285 and 294–316), (ii) a cyclin-box motif (residues 548–578), (iii) a cyclic nucleotide monophosphate (cNMP)-binding domain (residues 575–695) similar to that usually present in ion channels and cNMP-dependent kinases (Shabb & Corbin, 1992 ▶) and (iv) two consecutive cystathionine β-synthase (CBS) domains (residues 377–438 and 445–511; Bateman, 1997 ▶; Wang et al., 2003 ▶; Fig. 1 ▶) that presumably exert a regulatory role on its biological activity. In the case of CNNM4, neither the structure nor the ligands of its CBS pair are known. CBS domains are considered to be energy-sensing modules that bind adenosine ligands with different affinities (Scott et al., 2004 ▶), although their exact functions are still poorly understood. For instance, CBS domains are involved in the gating of osmoregulatory proteins (Biemans-Oldehinkel et al., 2006 ▶), in the transport and binding of Mg2+ (Ishitani et al., 2008 ▶), in the modulation of intracellular trafficking of chloride channels (Carr et al., 2003 ▶), in nitrate transport (De Angeli et al., 2009 ▶) and as internal inhibitors of pyrophosphatase activity (Jämsen et al., 2010 ▶; Tuominen et al., 2010 ▶). Despite their low degree of sequence similarity, CBS motifs share common structural properties, either isolated or as domains of larger proteins. Interestingly, mutations in the CBS domains of several human proteins have been associated with several human diseases (OMIM 146690, MIM 602643, MIM 180105 and MIM 600858, where OMIM refers to the online version of Mendelian Inheritance in Man at http://www.ncbi.nlm.nih.gov/omim and MIM refers to the 1998 edition of the printed version; McKusick, 1998 ▶; Bowne et al., 2002 ▶; Scott et al., 2004 ▶; Kemp, 2004 ▶). With the aim of providing detailed information that will help in understanding the involvement of CNNM4 in the development of Jalili syndrome and in unravelling the underlying molecular mechanism, we initiated crystallographic studies of both the full-length protein and its individual domains. Here, we describe the success obtained in the purification, crystallization and preliminary analysis of the CBS pair of human CNNM4 as a first step towards elucidating the three-dimensional structure of the whole protein, for which no information is available.
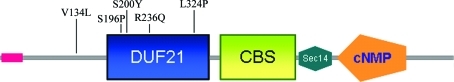
Figure 1.
Domain distribution in the human CNNM4 metal transporter. DUF21 (blue; residues 184–358; Pfam code PF01595) is a transmembrane region of unknown function that is usually found in the N-terminus of the proteins adjacent to two intracellular CBS domains. In CNNM4, it contains four transmembrane (TM) helices including residues 182–204 (TM1), 239–261 (TM2), 265–287 (TM3) and 294–316 (TM4). TM prediction was performed with the TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The cystathionine β-synthase (CBS) pair (light green) includes two CBS motifs (residues 377–438 and 445–511; Pfam code PF00571). The cyclic nucleotide monophosphate (cNMP)-binding domain (orange; Pfam code PF00027; residues 575–695) is a motif that is usually present in ion channels and cNMP-dependent kinases. The positions of known pathological mutations are indicated at the top. According to a BLAST search carried out against the Schnipsel database (http://smart.embl-heidelberg.de/help/smart_glossary.shtml#schnipsel), the region comprising residues 516–555 shares significant similarity to the Sec14 motif (dark green; InterPro code IPR001251), which is a domain found in homologues of an S. cerevisiae phosphatidylinositol-transfer protein (Sec14p) and in RhoGAPs, RhoGEFs and the RasGAP neurofibromin (NF1). The cyclin-box motif of CNNM4 (residues 548–578) is located within this Sec14-like region.
2. Materials and methods
2.1. Cloning, expression and purification of hCNNM4_CBS
Three different constructs, C1 (residues 359–511; MW = 17 752 Da; pI = 4.38), C2 (residues 369–511; MW = 16 668 Da; pI = 4.41) and C3 (residues 369–507; MW = 16 198 Da; pI = 4.48), encoding the CBS-pair region of human CNNM4 (referred to in the following as hCNNM4_CBS) were used in this study. An initial selection of constructs with potential crystallizability was made on the basis of multiple alignments against CBS domain-containing proteins of known three-dimensional structure. The three final constructs C1, C2 and C3 were chosen after careful analysis of all of the candidates using the XtalPred crystallizability prediction server (http://ffas.burnham.org/XtalPred-cgi/xtal.pl; Slabinski et al., 2007 ▶). The cloned cDNA of human CNNM4 was purchased from OriGene (Maryland, USA; catalogue No. SC113208). This cDNA was used as a template in order to obtain C1, C2 and C3, which were amplified by PCR using the primers 5′-CACCCATATGCTCAATATGATCCAGGGTGC (forward) and 3′-GGATCCTCACTCGTCCAGGATCTCCGA (reverse) for C1, 5′-CACCATGCGGACCAAAACTGTAGAGG (forward) and 3′-GGATCCTCACTCGTCCAGGATCTCCGA (reverse) for C2 and 5′-CACCATGCGGACCAAAACTGTAGAGG (forward) and 3′-TCACTCCGACTTGATGATCTCCTCG (reverse) for C3. The amplified DNA was cloned into vector pET101/ D-TOPO (Invitrogen). The plasmids were transformed into chemically competent Escherichia coli strain BL21 (DE3) Star One Shot (Invitrogen). Starter cultures were grown in Luria–Bertani (LB) medium containing 100 mg ml−1 ampicillin overnight at 310 K. The starters were diluted in 2 l Luria–Bertani medium containing 100 mg ml−1 ampicillin. Overexpression of the target proteins was induced by addition of IPTG to a final concentration of 0.5 mM when the optical density of the culture at 600 nm reached 0.5. Expression was allowed to proceed for 6 h. The cells were harvested by centrifugation at 4000g for 15 min at 277 K. The cell pellet was resuspended in 20 ml lysis buffer (50 mM HEPES pH 7.0, 1 mM EDTA, 1 mM β-mercaptoethanol, 1 mM benzamidine, 0.1 mM PMSF) and lysed by sonication in a Labsonic P sonicator (Sartorius) for 10–12 s at 90% amplitude, keeping the cells on ice to prevent overheating. The cell debris was separated by ultracentrifugation at 120 000g in a 70 Ti rotor (Beckman) for 25 min at 277 K. The first purification step consisted of anion-exchange chromatography. The supernatant was filtered through a 0.45 µm filter, injected into a pre-equilibrated 5 ml HiTrap Q column (GE Healthcare) at a flow rate of 0.5 ml min−1, washed with 10 column volumes (CV) of buffer A 1 (50 mM HEPES pH 7.0, 1 mM EDTA, 1 mM β-mercaptoethanol) and eluted with a continuous gradient of buffer B 1 (50 mM HEPES pH 7.0, 1 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol) over 30 CV. Fractions containing the protein of interest were pooled and dialyzed overnight against 1000 volumes of buffer A 2 (25 mM MES pH 6.0, 1 mM EDTA, 1 mM β-mercaptoethanol). The sample was centrifuged again as described above and filtered through a 0.22 µm filter before being injected onto a 1 ml MonoQ 5/50 GL column (GE Healthcare) at a flow rate of 0.5 ml min−1. The column was washed with buffer A 2 as described previously and eluted with a 0–100% gradient of buffer B 2 (25 mM MES pH 7.0, 1 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol) over 30 CV. The fractions of interest were combined and concentrated using Vivaspin centrifugal concentrators (5000 Da molecular-weight cutoff) to a volume of approximately 1 ml. A final gel-filtration step was carried out on a HiLoad Superdex 75 16/60 Prep Grade column (GE Healthcare) equilibrated in buffer A 3 (50 mM HEPES pH 7.0, 200 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol). The chromatography was performed at a flow rate of 0.5 ml min−1, collecting 0.5 ml fractions. The concentrations of the purified C1, C2 and C3 proteins were determined by UV absorption at 280 nm using the theoretical extinction coefficient computed from their amino-acid sequences (∊280 = 5960 M −1 cm−1 for all three proteins). Fractions containing pure hCNNM4_CBS were pooled and concentrated using Vivaspin concentrators (5000 Da molecular-weight cutoff) to a final concentration of 50 mg ml−1 for crystallization trials. Pure proteins were frozen in liquid nitrogen and stored at 193 K. SDS–PAGE (Laemmli, 1970 ▶) was used to analyze the protein purity. The typical yield from expression was approximately 10 mg purified protein per litre of original cell culture. The identity of hCNNM4_CBS was confirmed by mass spectrometry.
2.2. Mass-spectrometric analysis
SDS–PAGE gel bands containing the corresponding hCNNM4_CBS constructs were subjected to in-gel tryptic digestion according to Shevchenko et al. (1996 ▶) with minor modifications. The gel piece was swollen in a digestion buffer consisting of 50 mM NH4HCO3 and 12.5 ng µl−1 trypsin (Roche Diagnostics) in an ice bath. After 30 min the supernatant was removed and discarded, 20 µl 50 mM NH4HCO3 was added to the gel piece and digestion was allowed to proceed at 310 K overnight. Prior to MS analysis, the sample was acidified by adding 5 µl 0.5% TFA. 0.5 µl of the digested sample was directly spotted onto the MALDI target and was then mixed with 0.5 µl α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution [20 µg µl−1 in 70:30(v:v) ACN/0.1% TFA]. Peptide mass fingerprinting was performed on a Bruker Autoflex III mass spectrometer (Bruker Daltonics, Bremen, Germany). Positively charged ions were analyzed in reflector mode using delayed extraction. The spectra were obtained by randomly scanning the sample surface. About 600–800 spectra were averaged in order to improve the signal-to-noise ratio. The spectra were externally calibrated; the mass accuracy was <50 p.p.m. when external calibration was performed and typically <20 p.p.m. in the case of internal calibration. Protein identification was performed by searching a nonredundant protein database (NCBI) using the Mascot search engine (http://matrixscience.com). The following parameters were used for database searches: missed cleavages, 1; allowed modifications, carbamidomethylation of cysteine (complete) and oxidation of methionine (partial). Ultimately, the hCNNM4_CBS expressed in E. coli was the wild type and did not contain any mutations in the amino-acid sequence according to the Homo sapiens genome-sequence database.
2.3. Crystallization
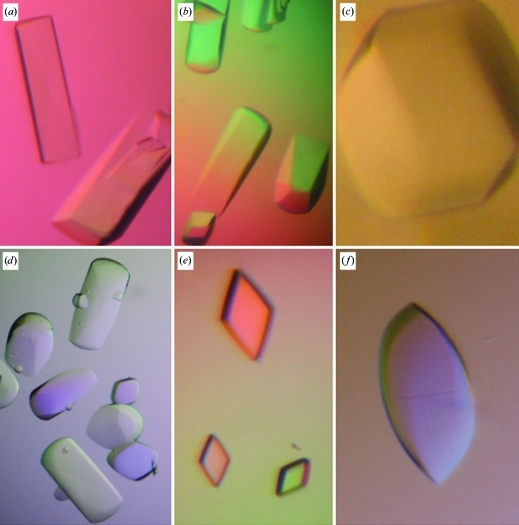
Dynamic light scattering indicated the presence of essentially monodisperse solutions of dimeric C1 species (data not shown) that were likely to correspond to CBS modules, which are the most frequently found associations in CBS-domain-containing proteins (Lucas et al., 2010 ▶; Gómez-García et al., 2010 ▶). Initial screening for crystals was performed with a variety of commercial screens (Crystal Screen, Crystal Screen 2, JCSG Core Suites I–IV, JCSG+ Suite, Index and PACT from Hampton Research, Molecular Dimensions and Qiagen) using the sitting-drop vapour-diffusion technique in 96-well plates at the High-Throughput Crystallization facility of CIC bioGUNE. Drops consisted of 0.1 µl protein solution at 60 mg ml−1 (in 100 mM HEPES pH 7.5, 1 mM β-mercaptoethanol, 1 mM EDTA) and 0.1 µl reservoir solution and were equilibrated against a reservoir volume of 70 µl at a constant temperature of 291 K. Initial experiments yielded square prisms that grew in 4.3 M NaCl, 100 mM HEPES pH 7.5 in 2–4 d but diffracted poorly to only 10 Å resolution. After extensive optimization, crystals with six different habits (trigonal prisms, flint axes, polyhedra, rounded-edge prisms, rhombic prisms and spears) were obtained (Fig. 2 ▶). Very often several habits grew within the same crystallization drop. The best diffraction-quality crystals (rounded-edge prisms) were grown at 295 K by mixing equal amounts (1 µl) of protein and reservoir solution (100 mM HEPES pH 7.5 and 3.5–4.1 M NaCl) in a 1:1 ratio; the crystals appeared in 2–4 d and grew to maximum dimensions of about 0.5 × 0.5 × 0.7 mm within two weeks. The use of detergents and/or additives (using screens from Hampton Research) did not improve the quality of the crystals. The C1 protein crystallized using a wide range of concentrations (20–60 mg ml−1), with higher concentrations giving increased nucleation and numerous but smaller crystals. Finding optimal cryogenic conditions for freezing C1 crystals was not straightforward and required significant experimental effort. The crystals often fractured after brief exposure (1–2 s) to various cryoprotectants, including MPD, low-molecular-weight PEGs, glycerol and ethylene glycol. The best results were obtained by soaking the crystals in crystallization solution containing a final sucrose concentration of 20% and a slight increase (5%) in the precipitant concentration. Dehydration of the crystals by slowly increasing the NaCl concentration (in steps of 5%) in the reservoir solution for varying times (from 1 h to 3 d) prior to flash-cooling in liquid nitrogen and/or to the addition of the cryoprotectants was also tested, with no further success. To check whether the cryoprotectant (sucrose) might be responsible for the limited resolution of the data sets, we analyzed the diffraction images obtained at room temperature and after freezing the crystals. Similar results were obtained, suggesting that the resolution limit might be intrinsic to the crystals despite their apparent external beauty.
Figure 2.
Crystals of C1 hCNNM4 showing six different habits: (a) trigonal prisms, (b) flint axes, (c) polyhedra, (d) rounded-edge prisms, (e) rhombic prisms and (f) spears. The highest resolution (3.6 Å) was obtained from the rounded-edge prisms (d).
2.4. Preliminary crystallographic analysis
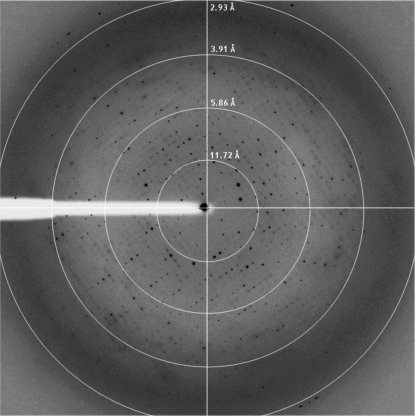
Prior to data collection, the crystals were transferred into crystallization buffer containing 20% sucrose as a cryoprotectant for a few seconds before being flash-cooled by direct immersion into liquid nitrogen at 93 K. Crystals were mounted for X-ray data collection using either CryoLoops (Hampton Research) or MicroMounts loops (MiTeGen). Data sets were collected in-house using a CCD detector mounted on a Microstar-H rotating-anode X-ray generator (Bruker) operated at 60 kV and 100 mA with Helios optics and a copper target (Cu Kα; λ = 1.542 Å) and on beamlines ID14.1 and ID23.2 at the ESRF synchrotron (Grenoble, France). Diffraction data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Preliminary analysis of the data sets was performed using the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). Native crystals of C1 hCNNM4 diffracted to 3.6 Å resolution (Fig. 3 ▶) and belonged to space group C222, with unit-cell parameters a = 90.9, b = 141.4, c = 87.6 Å. The presence of two, three or four molecules within the asymmetric unit gave Matthews coefficients of 3.97, 2.64 and 1.98 Å3 Da−1, respectively (Matthews, 1968 ▶), with solvent contents of 69, 54 and 38%. Considering the fragility of the hCNNM4_CBS crystals and their limited diffraction power, we estimated that two molecules per asymmetric unit (a dimer of the CBS pair of hCNNM4) was the most probable value. Such a dimer was likely to correspond to a CBS module, which is the most commonly observed association in CBS-domain-containing proteins (Lucas et al., 2010 ▶; Gómez-García et al., 2010 ▶). Data-collection statistics are summarized in Table 1 ▶.
Figure 3.
Representative X-ray diffraction image from a native C1 hCNNM4 crystal. The dimensions of the native crystals were 0.4 × 0.2 × 0.7 mm and they were exposed for 2 s over a 1° oscillation range. Resolution circles are included for clarity.
Table 1. Data-processing statistics for native C1 hCNNM4.
Values in parentheses are for the outer resolution shell.
| Beamline | ID14-1, ESRF |
| Wavelength (Å) | 0.934 |
| Total No. of reflections | 252964 |
| No. of unique reflections | 7697 |
| Resolution range (Å) | 50–3.6 |
| Space group | C222 |
| Unit-cell parameters (Å) | |
| a (Å) | 90.9 |
| b (Å) | 141.4 |
| c (Å) | 87.6 |
| Mosaicity (°) | 0.8 |
| Completeness (%) | 100 (100) |
| Multiplicity | 9.9 (10.3) |
| Rmerge† (%) | 8.1 (51.8) |
| Mean I/σ(I) | 26.1 (4.8) |
R
merge = 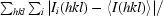
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
Acknowledgments
We thank the staff of ESRF beamlines ID14.1 and ID23.2 for support during synchrotron data collection. We also thank Dr Felix Elortza of the Proteomics Service at CIC bioGUNE for mass-spectrometric analysis and Dr Adriana Rojas for maintenance of the in-house X-ray equipment. This work was supported by grants from the Departamento de Educación, Universidades e Investigación del Gobierno Vasco (PI2010-17), the Basque Government (ETORTEK IE05-147, IE07-202), Diputación Foral de Bizkaia (Exp. 7/13/08/2006/11 and 7/13/08/2005/14), Spanish Ministerio de Ciencia e Innovación (MICINN; BFU2010-17857) and the MICINN CONSOLIDER-INGENIO 2010 Program (CSD2008-00005).
References
- Agus, M. S. & Agus, Z. S. (2001). Crit. Care Clin. 17, 175–186. [DOI] [PubMed]
- Agus, Z. S. (1999). J. Am. Soc. Nephrol. 10, 616–622.
- Alderton, A., Davies, P., Illman, K. & Brown, D. R. (2007). J. Neurochem. 103, 312–321. [DOI] [PubMed]
- Bateman, A. (1997). Trends Biochem. Sci. 22, 12–13. [DOI] [PubMed]
- Biemans-Oldehinkel, E., Mahmood, N. A. & Poolman, B. (2006). Proc. Natl Acad. Sci. USA, 103, 10624–10629. [DOI] [PMC free article] [PubMed]
- Bowne, S. J., Sullivan, L. S., Blanton, S. H., Cepko, C. L., Blackshaw, S., Birch, D. G., Hughbanks-Wheaton, D., Heckenlively, J. R. & Daiger, S. P. (2002). Hum. Mol. Genet. 11, 559–568. [DOI] [PMC free article] [PubMed]
- Carr, G., Simmons, N. & Sayer, J. (2003). Biochem. Biophys. Res. Commun. 310, 600–605. [DOI] [PubMed]
- Cefaratti, C., Romani, A. & Scarpa, A. (2000). J. Biol. Chem. 275, 3772–3780. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- De Angeli, A., Moran, O., Wege, S., Filleur, S., Ephritikhine, G., Thomine, S., Barbier-Brygoo, H. & Gambale, F. (2009). J. Biol. Chem. 284, 26526–26532. [DOI] [PMC free article] [PubMed]
- Eshaghi, S., Niegowski, D., Kohl, A., Martinez Molina, D., Lesley, S. A. & Nordlund, P. (2006). Science, 313, 354–357. [DOI] [PubMed]
- Gómez-García, I., Oyenarte, I. & Martínez-Cruz, L. A. (2010). J. Mol. Biol. 399, 53–70. [DOI] [PubMed]
- Goytain, A. & Quamme, G. A. (2005). Physiol. Genomics, 22, 382–389. [DOI] [PubMed]
- Günther, T. (1993). Miner. Electrolyte Metab. 19, 259–265. [PubMed]
- Guo, D., Ling, J., Wang, M.-H., She, J.-X., Gu, J. & Wang, C.-Y. (2005). Mol. Pain, 1, 15. [DOI] [PMC free article] [PubMed]
- Hattori, M., Tanaka, Y., Fukai, S., Ishitani, R. & Nureki, O. (2007). Nature (London), 448, 1072–1075. [DOI] [PubMed]
- Ishitani, R., Sugita, Y., Dohmae, N., Furuya, N., Hattori, M. & Nureki, O. (2008). Proc. Natl Acad. Sci. USA, 105, 15393–15398. [DOI] [PMC free article] [PubMed]
- Jämsen, J., Baykov, A. A. & Lahti, R. (2010). Biochemistry, 49, 1005–1013. [DOI] [PubMed]
- Kemp, B. E. (2004). J. Clin. Invest. 113, 182–184. [DOI] [PMC free article] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Lucas, M., Encinar, J. A., Arribas, E. A., Oyenarte, I., García, I. G., Kortazar, D., Fernández, J. A., Mato, J. M., Martínez-Chantar, M. L. & Martínez-Cruz, L. A. (2010). J. Mol. Biol. 396, 800–820. [DOI] [PubMed]
- Lunin, V. V., Dobrovetsky, E., Khutoreskaya, G., Zhang, R., Joachimiak, A., Doyle, D. A., Bochkarev, A., Maguire, M. E., Edwards, A. M. & Koth, C. M. (2006). Nature (London), 440, 833–837. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McKusick, V. A. (1998). Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders, 12th ed. Baltimore: Johns Hopkins University Press.
- Ochoa, B. (2004). Pediatr. Nephrol. 19, 6–12. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parry, D. A. et al. (2009). Am. J. Hum. Genet. 84, 266–273. [DOI] [PMC free article] [PubMed]
- Polok, B., Escher, P., Ambresin, A., Chouery, E., Bolay, S., Meunier, I., Nan, F., Hamel, C., Munier, F. L., Thilo, B., Mégarbané, A. & Schorderet, D. F. (2009). Am. J. Hum. Genet. 84, 259–265. [DOI] [PMC free article] [PubMed]
- Quamme, G. A. (1997). Kidney Int. 52, 1180–1195. [DOI] [PubMed]
- Quamme, G. A. (2010). Am. J. Physiol. Cell Physiol. 298, C407–C429. [DOI] [PubMed]
- Romani, A. M. & Scarpa, A. (2000). Front. Biosci. 5, D270–D374. [DOI] [PubMed]
- Schweigel, M., Kolisek, M., Nikolic, Z. & Kuzinski, J. (2008). Magnes. Res. 21, 118–123. [PubMed]
- Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G. & Hardie, D. G. (2004). J. Clin. Invest. 113, 274–284. [DOI] [PMC free article] [PubMed]
- Shabb, J. B. & Corbin, J. D. (1992). J. Biol. Chem. 267, 5723–5726. [PubMed]
- Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996). Anal. Chem. 68, 850–858. [DOI] [PubMed]
- Slabinski, L., Jaroszewski, L., Rychlewski, L., Wilson, I. A., Lesley, S. A. & Godzik, A. (2007). Bioinformatics, 23, 3403–3405. [DOI] [PubMed]
- Tashiro, M., Konishi, M., Iwamoto, T., Shigekawa, M. & Kurihara, S. (2000). Pflugers Arch. Eur. J. Physiol. 440, 819–827. [DOI] [PubMed]
- Tuominen, H., Salminen, A., Oksanen, E., Jämsen, J., Heikkilä, O., Lehtiö, L., Magretova, N. N., Goldman, A., Baykov, A. A. & Lahti, R. (2010). J. Mol. Biol. 398, 400–413. [DOI] [PubMed]
- Wang, C.-Y., Shi, J.-D., Yang, P., Kumar, P. G., Li, Q.-Z., Run, Q.-G., Su, Y.-C., Scott, H. S., Kao, K.-J. & She, J.-X. (2003). Gene, 306, 37–44. [DOI] [PubMed]
- Wang, C.-Y., Yang, P., Shi, J.-D., Purohit, S., Guo, D., An, H., Gu, J.-G., Ling, J., Dong, Z. & She, J.-X. (2004). BMC Genomics, 5, 7. [DOI] [PMC free article] [PubMed]
- Watanabe, M., Konishi, M., Ohkido, I. & Matsufuji, S. (2005). Am. J. Physiol. Renal Physiol. 289, F742–F748. [DOI] [PubMed]
- Yang, M., Jensen, L. T., Gardner, A. J. & Culotta, V. C. (2005). Biochem. J. 386, 479–487. [DOI] [PMC free article] [PubMed]