Serine/threonine protein kinase SnRK2.6/OST1 (OPEN STOMATA 1) from A. thaliana has been crystallized by the hanging-drop vapour-diffusion method and a native data set has been collected to 2.8 Å resolution.
Keywords: SnRK2.6/OST1, serine/threonine protein kinases, Arabidopsis thaliana
Abstract
The SnRK2.6 (SNF1-related kinase 2.6) gene from Arabidopsis thaliana encodes the serine/threonine protein kinase SnRK2.6/OST1 (OPEN STOMATA 1). It plays a central role in the drought-tolerance mechanism. OST1 is in fact the main positive effector in the hydric stress response. The SnRK2.6 gene was cloned into the pGEX4T1 plasmid, mutated and expressed in Escherichia coli, allowing purification to homogeneity in two chromatographic steps. Various OST1 mutants yielded crystals using vapour-diffusion techniques, but only one mutant showed a good diffraction pattern. Its crystals diffracted to 2.8 Å resolution and belonged to space group P2221, with unit-cell parameters a = 77.7, b = 99.4, c = 108.4 Å. A promising molecular-replacement solution was found using the structure of the kinase domain of the yeast AMP-activated protein kinase SNF1 (PDB entry 3hyh) as the search model.
1. Introduction
The phosphorylation and dephosphorylation of proteins, which is carried out by protein kinases and protein phosphatases, respectively, is the main mechanism of signal transduction in eukaryotic cells (Johnson et al., 1996 ▶). Several studies have shown that a subgroup of protein kinases that belong to the SNF1-related protein kinase 2 family (SnRK2s) and a group of 2C-type protein phosphatases (PP2Cs) are involved in decoding environmental stimuli to elaborate the proper response to abiotic stress in plants (Ma et al., 2009 ▶; Park et al., 2009 ▶; Cutler et al., 2010 ▶). Additionally, the critical role of the phytohormone abscisic acid (ABA) as a second messenger in plant adaptation in response to stress has been widely described (Zhu, 2002 ▶; Hetherington, 2001 ▶).
It has been shown that the regulation of the SnRK2s/PP2Cs signal transduction pathway depends on the intracellular concentration of ABA, on the phosphorylation states of the protein components and finally on the interactions between them. In a no-stress situation the SnRK2s are kept dephosphorylated and inhibited by their interaction with the PP2Cs (Vlad et al., 2009 ▶; Lee et al., 2009 ▶). Under salt-stress or drought-stress situations, the intracellular concentration of ABA increases and binds the PYR/RCAR ABA receptor. This complex interacts with the active site of the PP2Cs and abolishes their phosphatase activity (Nishimura et al., 2009 ▶; Miyazono et al., 2009 ▶). This allows the autophosphorylation and activation of the SnRK2s, which in turn are able to phosphorylate their target proteins to trigger the proper cell response. The balance between active and inactive kinases and phosphatases tunes the response.
SnRK2.6/OST1 (OPEN STOMATA 1) is the main positive effector in the cell response to hydric stress (Mustilli et al., 2002 ▶; Yoshida et al., 2002 ▶). It carries out activation by phosphorylation of transcription factors that are involved in a transcriptional response associated with ABA (Fujii et al., 2009 ▶), ionic channels that stimulate stomata closure (Lee et al., 2009 ▶, Sato et al., 2009 ▶) and oxidases that are involved in the production of second messenger molecules (Hubbard et al., 2010 ▶).
The structural characterization of OST1 is central to understanding the regulation mechanism of the kinase. In this work, we describe the expression, purification, crystallization and preliminary crystallographic analysis of two OST1 mutants (OST1 K50N and OST1 D160A). The expression, purification and crystallization of eukaryotic proteins using bacterial expression systems is a bottleneck in many structural studies, especially when working with the components of signal transduction cascades. We overcame these problems by using Rosetta (Novagen), a bacterial strain that avoids codon-bias problems, and a GST-fusion protein to improve the protein solubility (Frangioni & Neel, 1993 ▶). Moreover, in order to prevent sample heterogeneity arising from kinase autophosphorylation we used OST1 K50N and OST1 D160A mutants, which yield inactive kinases.
2. Experimental
2.1. SnRK2.6 gene cloning and site-directed mutagenesis
The OST1 gene from Arabidopsis thaliana encoding full-length OST1 was cloned into a pGEX4T1 (GE Healthcare) expression plasmid between BamHI and EcoRI sites (pGEX4T1OST1, construction provided by Dr Zhu). OST1 K50N and OST D160A mutants were produced using standard site-directed mutagenesis techniques (Sambrook & Russell, 2001 ▶). The primers used in mutagenesis were K50N Forward (5′-G CTT GTT GCT GTT GAC TAT ATC GAG AGA GG-3′), K50N Reverse (5′-CC TCT CTC GAT ATA GTC AAC AGC AAC AAG-3′), D160A Forward (5′-CCT CGT CTA AAG ATA TGT GCT TTC GGA TAT TCT AAG-3′) and D160A Reverse (5′-CTT AGA ATA TCC GAA AGC ACA TAT CTT TAG ACG AGG-3′). These constructions render GST-fusion proteins that contain a total of five amino acids (GSPNS) between the thrombin cleavage site and the start of OST1.
2.2. Protein expression and purification
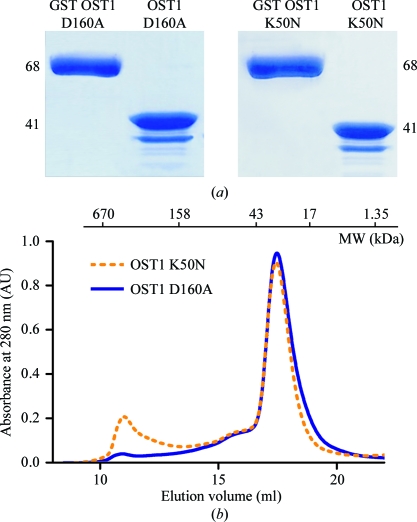
The pGEX4T1OST1 K50N and pGEX4T1OST1 D160A plasmids were transformed into Escherichia coli strain Rosetta (DE3) pLys (Novagen) for protein expression using standard protocols (Sambrook & Russell, 2001 ▶). The same expression and purification protocol was used for both mutants. A total of 5 ml of an overnight culture was subcultured into 500 ml fresh 2× TY broth (16 g Bacto tryptone, 10 g yeast extract and 5 g NaCl per litre of solution) containing ampicillin (100 µg ml−1) and chloramphenicol (20 µg ml−1). Transformed cells were grown at 310 K; when the OD at 600 nm reached 0.6–0.8 protein expression was induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside overnight at 289 K. Cells were harvested by centrifugation (15 min, 4500g), resuspended in 50 mM Tris–HCl pH 7.5, 50 mM NaCl and disrupted by sonication. After centrifugation (40 min, 40 000g) at 277 K, the clear supernatant was filtered (pore diameter 0.45 µm; Millipore, Bedford, Massachusetts, USA). The GST-tagged OST1 K50N and OST1 D160A mutants were purified using Glutathione Sepharose beads (GE Healthcare) according to the manufacturer’s instructions. The filtered supernatant was mixed with the previously equilibrated beads. After incubation, a washing step was performed with ten volumes of 50 mM Tris–HCl pH 7.5, 50 mM NaCl. SDS–PAGE analysis revealed that both constructs yielded 67.5 kDa products. The OST1 mutants were cleaved from GST using 7.5 units of thrombin protease (Novagen) per milligram of GST-tagged protein. The cleavage yielded protein fragments corresponding to the GST protein (26 kDa) and to the full-length OST1 mutants (41.5 kDa). The time courses of chromatography and enzymatic digestion were monitored by SDS–PAGE (Fig. 1 ▶ a). A final polishing step was performed using Superdex 200 16/60 (Amersham Biosciences; Fig. 1 ▶ b). OST1 K50N and OST1 D160A were concentrated to a final concentration of 12 mg ml−1 using a 10 kDa cutoff Amicon protein concentrator (YM-10; Millipore, Bedford, Massachusetts, USA). The final protein concentration was determined spectrophotometrically using the calculated molar absorption coefficient at 280 nm (30 370 M −1 cm−1; Gasteiger et al., 2005 ▶). The samples were kept at 277 K.
Figure 1.
(a) SDS–PAGE analysis of the purified OST1 D160A and OST1 K50N mutants. The lane on the left corresponds to the GST-fusion protein and the lane on the right to the product after thrombin cleavage. The expected molecular weights are indicated. (b) The size-exclusion chromatogram of OST1 K50N and OST1 D160A mutants. The lines show the absorbance recorded at 280 nm. Molecular-weight markers (Bio-Rad) are indicated in kDa.
2.3. Crystallization
Initial crystallization conditions for OST1 K50N and OST1 D160A were screened by high-throughput techniques with a NanoDrop robot (Innovadyne Technologies Inc.) using the commercial screens Crystal Screen and Crystal Screen 2 (Hampton Research), PACT Suite and JCSG Suite (Qiagen) and JBS Kinase (Jena Bioscience). Crystallization assays were carried out using the sitting-drop vapour-diffusion method at 291 K in 96-well plates (Innovaplate SD-2 microplates, Innovadyne Technologies Inc.).
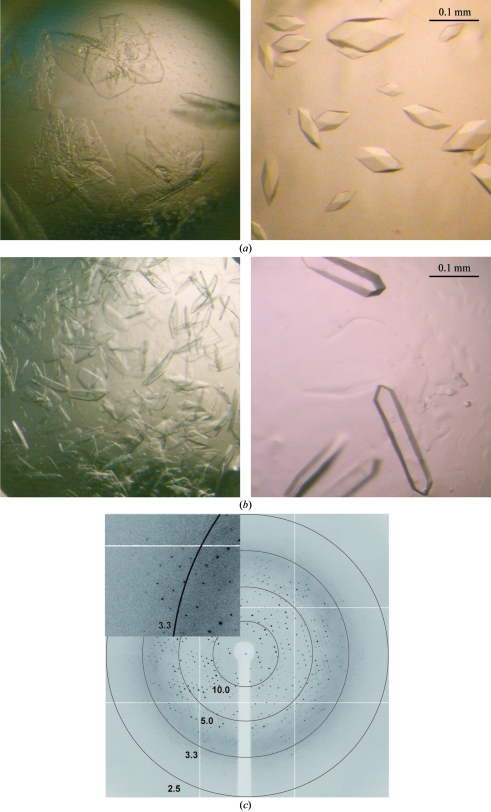
Prior to crystallization, the protein samples were incubated with the additives DTT, MgCl2 and AMPPNP at final concentrations of 1, 2 and 2 mM, respectively. Drops consisting of 250 nl protein at 12 mg ml−1 and 250 nl precipitant solution were mixed and equilibrated against 65 µl well solution. Crystals of both OST1 mutants were obtained using a precipitant solution consisting of 0.1 M HEPES pH 7.5, 10% PEG 10 000, 8% ethylene glycol (condition H7 of the JBS Kinase screen from Jena Bioscience; Figs. 2 ▶ a and 2 ▶ b). Several strategies were used to optimize these crystallization conditions, which included adjusting the protein-sample composition, the precipitant concentration and pH value, screening different additives (Additive Screen, Hampton Research) and detergents and streak-seeding. A sample composition containing DTT was essential in order to produce crystals and the use of MgCl2 and AMPPNP improved their overall shape. The final conditions were scaled up on 24-well plates (Linbro plates, Hampton Research) in hanging-drop experiments at 291 K. The OST1 K50N and OST1 D160A crystals used in our analysis were obtained from drops comprised of 2 µl protein solution and 1 µl precipitant solution consisting of 0.1 M HEPES pH 6.5, 12% PEG 10 000 and 12% ethylene glycol (Figs. 2 ▶ a and 2 ▶ b).
Figure 2.
Comparison of the initial and optimized crystals of OST1 K50N (a) and OST1 D160A (b). (c) X-ray diffraction pattern of an OST1 D160A crystal obtained using a synchrotron-radiation source. The inset shows a magnification of a sector of the diffraction frame. Numbers show the resolutions (in Å) of the circles.
Several cryoprotectants were tested, including glycerol, MPD and ethylene glycol. The best cryoprotectant solution resulted from increasing the ethylene glycol concentration in the crystal mother liquor from 12 to 18%.
2.4. Data collection and analysis
Crystals were mounted in a fibre loop, transferred into cryoprotectant solution and flash-frozen at 100 K in a nitrogen-gas stream. Preliminary diffraction data were collected on an in-house MAR345dtb imaging-plate detector (MAR Research) using Cu Kα X-rays generated by a rotating-anode generator (Microstar, Bruker) equipped with Helios mirrors (Bruker) and operated at 45 kV and 60 mA. The OST1 K50N crystals were not suitable for X-ray data analysis; their diffraction was very poor. However, the crystals of OST1 D160A showed good-quality diffraction patterns (Fig. 2 ▶ c). A complete data set was collected to 3.2 Å resolution using the in-house X-ray source. A diffraction data set was collected from another OST1 D160A crystal using an ADSC Q4 CCD detector on the ID14.2 beamline of the European Synchrotron Radiation Facility (Grenoble, France). Diffraction data were processed with XDS (Kabsch, 2010 ▶) and scaled with SCALA from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶). A summary of the diffraction protocol and data-collection statistics is given in Table 1 ▶.
Table 1. Diffraction protocol and data-collection statistics for the OST1 D160A crystal.
Values in parentheses are for the highest resolution shell.
| Crystal data | |
| Space group | P2221 |
| Unit-cell parameters (Å) | a = 77.7, b = 99.4, c = 108.4 |
| Z | 2 |
| VM (Å3 Da−1) | 2.54 |
| Solvent content (%) | 52 |
| Diffraction protocol | |
| Radiation source | Synchrotron radiation (ID14.2) |
| Wavelength (Å) | 0.93 |
| Detector type | ADSC Q4 CCD |
| X-ray beam size (µm) | 100 |
| Crystal-to-detector distance (mm) | 234.17 |
| Oscillation range (°) | 0.5 |
| Temperature (K) | 100 |
| Data-collection statistics | |
| Resolution (Å) | 50.0–2.80 (2.95–2.80) |
| No. of unique reflections | 12079 (2942) |
| No. of observed reflections | 81914 (20393) |
| Completeness (%) | 99.1 (99.5) |
| Multiplicity | 4.0 (4.1) |
| Mean I/σ(I) | 20.4 (2.3) |
| Rmerge† | 0.05 (0.50) |
| Rp.i.m.‡ | 0.03 (0.33) |
R
merge = 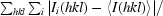
 , where Ii(hkl) is the ith observed amplitude of reflection hkl and 〈I(hkl)〉 is the mean amplitude for the measurements of reflection hkl.
, where Ii(hkl) is the ith observed amplitude of reflection hkl and 〈I(hkl)〉 is the mean amplitude for the measurements of reflection hkl.
R p.i.m. is the precision-indicating (multiplicity-weighted) R merge (Weiss, 2001 ▶).
3. Results and discussion
The K50N and D160A mutants of OST1 from A. thaliana were expressed as a GST-fusion proteins that allowed their purification from the soluble fraction under native conditions in two chromatographic steps. We select these inactive mutated versions of the protein to prevent heterogeneity arising from self-phosphorylation of the protein. The K50N mutant precludes an effective ATP-binding conformation, while the D160A mutant hinders the essential magnesium binding (Adams, 2001 ▶). The sample purity at the final purification step was at least 95% as monitored by SDS–PAGE (Fig. 1 ▶ a). The lanes corresponding to OST1 D160A and OST1 K50N after thrombin cleavage showed several bands resulting from partial protein degradation. Size-exclusion chromatography suggested that OST1 K50N and OST1 D160A are monomeric (Fig. 1 ▶ b).
Preliminary crystallization conditions led to crystal clusters of thin plates, always in the presence of PEG (0.1 M HEPES pH 7.5, 10% PEG 10 000, 8% ethylene glycol). After the refinement of several parameters, including a very fine tuning of the ethylene glycol concentration and the pH value, isolated prismatic and rod-shaped crystals were obtained. The best crystal forms of OST1 K50N and OST1 D160A both appeared in conditions containing PEG 10 000 as the precipitant, mainly at 0.1 M HEPES pH 6.5, 12% PEG 10 000, 12% ethylene glycol (Figs. 2 ▶ a and 2 ▶ b, respectively). In general, the crystals appeared in 3–4 d and continued to grow over the following week to maximum dimensions of 0.15 × 0.07 × 0.07 mm for OST1 K50N and 0.3 × 0.06 × 0.03 mm for OST1 D160A. Data collections were performed using ethylene glycol as a cryoprotectant.
OST1 D160A crystals displayed a good-quality diffraction pattern (Fig. 2 ▶ c), diffracted to 2.8 Å resolution and belonged to space group P2221, with unit-cell parameters a = 77.7, b = 99.4, c = 108.4 Å. In contrast, OST1 K50N crystals were not suitable for X-ray diffraction studies; they only diffracted to very low resolution even using synchrotron radiation. This is surprising since the mutant proteins showed similar behaviour in the SDS–PAGE gel analysis, similar profiles on size-exclusion chromatography (Figs. 1a and 1b ▶) and crystallized using similar conditions.
We investigated the local symmetry relating the units in the asymmetric unit using the CCP4 package program POLARRFN (Collaborative Computational Project, Number 4, 1994 ▶). Several self-rotation functions were computed in the resolution range 15–3 Å with Patterson vectors from 15 to 30 Å radius of integration. Analysis of self-rotation peaks did not reveal the presence of noncrystallographic symmetry. However, specific volume calculations (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) predicted the presence of two molecules of OST1 D160A in the asymmetric unit, with a V M of 2.54 Å3 Da−1 and an estimated solvent content of 52%. This suggests that either there is only one molecule in the asymmetric unit, there are two different conformers of the OST1 D160A molecule or there is a noncrystallographic twofold axis parallel to one of the crystallographic axes.
The strategy used to solve the OST1 D160A structure was molecular replacement with the program MOLREP (Vagin & Teplyakov, 2010 ▶) using the coordinates of the protein kinase domain of the yeast AMP-activated protein kinase SNF1 (PDB entry 3hyh; Rudolph et al., 2005 ▶) as a model. Although the model shows a sequence identity of 42% (BLAST; Altschul et al., 1990 ▶), it yielded a promising but inconclusive solution that contained two molecules in the asymmetric unit. The automated density-modification and model-building package implemented in PHENIX (Adams et al., 2010 ▶) automatically traced an initial model; however, the calculated electron-density map was noisy and strongly biased. We are currently refining this model and also preparing heavy-atom crystal derivatives to phase the structure using single-wavelength anomalous dispersion techniques.
Acknowledgments
The authors thank the ESRF for the access to the synchrotron-radiation source. This work was funded by grant BFU2008-00368/BMC and ‘Factoría de Cristalización’ Consolider-Ingenio 2010 of the Spanish ‘Plan Nacional’ (MICINN) to AA.
References
- Adams, J. A. (2001). Chem. Rev. 101, 2271–2290. [DOI] [PubMed]
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cutler, S. R., Rodríguez, P. L., Finkelstein, R. R. & Abrams, S. R. (2010). Annu. Rev. Plant Biol. 61, 651–679. [DOI] [PubMed]
- Frangioni, J. V. & Neel, B. G. (1993). Anal. Biochem. 210, 179–187. [DOI] [PubMed]
- Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S.-Y., Cutler, S. R., Sheen, J., Rodríguez, P. L. & Zhu, J.-K. (2009). Nature (London), 462, 660–664. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Hetherington, A. M. (2001). Cell, 107, 711–714. [DOI] [PubMed]
- Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D. & Schroeder, J. I. (2010). Genes Dev. 24, 1695–1708. [DOI] [PMC free article] [PubMed]
- Johnson, L. N., Noble, M. E. & Owen, D. J. (1996). Cell, 85, 149–158. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Lee, S. C., Lan, W., Buchanan, B. B. & Luan, S. (2009). Proc. Natl Acad. Sci. USA, 106, 21419–21424. [DOI] [PMC free article] [PubMed]
- Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A. & Grill, E. (2009). Science, 324, 1064–1068. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H.-J., Asano, A., Miyauchi, Y., Takahashi, M., Zhi, Y., Fujita, Y., Yoshida, T., Kodaira, K.-S., Yamaguchi-Shinozaki, K. & Tanokura, M. (2009). Nature (London), 462, 609–614. [DOI] [PubMed]
- Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F. & Giraudat, J. (2002). Plant Cell, 14, 3089–3099. [DOI] [PMC free article] [PubMed]
- Nishimura, N., Hitomi, K., Arvai, A. S., Rambo, R. P., Hitomi, C., Cutler, S. R., Schroeder, J. I. & Getzoff, E. D. (2009). Science, 326, 1373–1379. [DOI] [PMC free article] [PubMed]
- Park, S.-Y. et al. (2009). Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed]
- Rudolph, M. J., Amodeo, G. A., Bai, Y. & Tong, L. (2005). Biochem. Biophys. Res. Commun. 337, 1224–1228. [DOI] [PubMed]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
- Sato, A., Sato, Y., Fukao, Y., Fujiwara, M., Umezawa, T., Shinozaki, K., Hibi, T., Taniguchi, M., Miyake, H., Goto, D. B. & Uozumi, N. (2009). Biochem. J. 424, 439–448. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., Leung, J., Rodriguez, P. L., Laurière, C. & Merlot, S. (2009). Plant Cell, 21, 3170–3184. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst. 34, 130–135.
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., Ecker, J. R. & Shinozaki, K. (2002). Plant Cell Physiol. 43, 1473–1483. [DOI] [PubMed]
- Zhu, J.-K. (2002). Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed]