Crystals of S. aureus MazF belong to space group P212121, diffract to 2.1 Å resolution and contain two MazF dimers in the asymmetric unit.
Keywords: MazF, toxin–antitoxin modules, mRNA interferases, ribonucleases
Abstract
mazEF modules encode toxin–antitoxin pairs that are involved in the bacterial stress response through controlled and specific degradation of mRNA. Staphylococcus aureus MazF and MazE constitute a unique toxin–antitoxin module under regulation of the sigB operon. A MazF-type mRNA interferase is combined with an antitoxin of unknown fold. Crystals of S. aureus MazF (SaMazF) were grown in space group P212121. The crystals diffracted to 2.1 Å resolution and are likely to contain two SaMazF dimers in the asymmetric unit.
1. Introduction
Toxin–antitoxin (TA) modules are small operons encoding a growth inhibitor (the ‘toxin’) and a protein (the ‘antitoxin’) that regulates the activity of the toxin by direct inhibition and by acting as a transcription regulator (Buts et al., 2005 ▶; Gerdes et al., 2005 ▶). TA modules were initially discovered as ‘plasmid addiction modules’ on low-copy-number plasmids such as F or R1 and on plasmidic phages such as P1 (Ogura & Hiraga, 1983 ▶; Bravo et al., 1987 ▶; Lehnherr et al., 1993 ▶). They ensure plasmid maintenance by a mechanism of post-segregational killing that relies on a difference in the lifetime of the toxin and the antitoxin (Gerdes et al., 1986 ▶). However, most TA modules are encoded on chromosomes and their functional role is a matter of heavy debate. A number of different functions have been proposed, ranging from selfish genes through stabilization of specific chromosomal segments to roles in stress response and persister-cell formation (Magnuson, 2007 ▶; Van Melderen, 2010 ▶).
The mazEF family of TA modules constitutes one of the larger TA families and is found on chromosomes of both Gram-negative and Gram-positive bacteria (but not in archaea; Pandey & Gerdes, 2005 ▶). The toxin MazF is a ribonuclease that cuts mRNA at highly specific sites (Zhang et al., 2003 ▶), leading to a redirection of translation towards the synthesis of a few specific proteins (Amitai et al., 2009 ▶; Baik et al., 2009 ▶). Different family members show different cutting specificities which relate to the specific cellular functions of individual MazF-family members (Yamaguchi & Inouye, 2009 ▶). This property has been used to create a single protein-production system (Suzuki et al., 2005 ▶). In contrast to RelE (the mRNA-cleavage enzyme from the RelBE TA pair), mRNA cleavage by MazF generally occurs independently of translation, although for some specific RNAs translation significantly influences the observed cleavage rates (Christensen-Dalsgaard & Gerdes, 2008 ▶). Long-term activation of MazF leads to cell death, and MazF activation within an Escherichia coli population has been linked to quorum sensing (Amitai et al., 2004 ▶; Kolodkin-Gal et al., 2007 ▶).
While mazEF modules from Gram-negative bacteria, and especially E. coli and Mycobacterium tuberculosis, have been heavily studied (Inouye, 2006 ▶), less is known about mazEF modules in Gram-positive bacteria. The genome of Staphylococcus aureus contains a single mazEF module that is highly conserved in all Staphylococcus species, including MRSA and VRSA strains (Fu et al., 2007 ▶). In contrast to the typical situation in Gram-negative bacteria, the S. aureus mazEF module (SamazEF) does not form an isolated operon but belongs to the sigB operon that encodes the σB sigma factor and its control genes rsbU, rsbV and rsbW (Donegan & Cheung, 2009 ▶). SaMazE and SaMazF are co-transcribed with these four genes. While the SamazF gene encodes a protein that is weakly related to E. coli MazF (16% sequence identity), the 56-amino-acid SaMazE protein shows no detectable sequence identity to MazE proteins from Gram-negative bacteria. SaMazF was shown to cleave mRNA in vitro, although there is discussion about its sequence specificity (Fu et al., 2007 ▶; Zhu et al., 2009 ▶). Here, we present the crystallization and preliminary X-ray analysis of SaMazF.
2. Material and methods
2.1. Protein production
The SamazE (SACOL2059; UniProtKB entry Q5HED2) and SamazF (SACOL2058; UniProtKB entry Q5HED3) genes were cloned into pDuet1 as described previously (Fu et al., 2007 ▶), placing a His tag at the N-terminus of the SaMazF protein. The vector was transformed into E. coli BL21 (DE3) cells using the CaCl2 method. A 25 ml overnight preculture in LB medium supplemented with 100 µg ml−1 ampicillin was used to inoculate (1:100 dilution) 1 l LB medium supplemented with the same concentration of ampicillin and 0.2%(w/v) glucose. When the culture reached an OD600 of 0.6, expression of the proteins was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The induced culture was further incubated overnight at 310 K while stirring at 120 rev min−1. The cells were harvested by centrifugation (25 min at 11 000g using a Beckman JLA 8.1000 rotor at 5500 rev min−1) and the pellet was resuspended in 50 ml lysis buffer (100 mM Tris–HCl pH 8.0, 1 M NaCl, 20 mM imidazole, 0.1 mg ml−1 AESBF and 1 µg ml−1 leupeptin). The suspension was then passed twice through a French press and centrifuged at 43 500g for 30 min (using a Beckman JA-20 rotor at 18 000 rev min−1). The supernatant was filtered through a 0.45 µm Sartorius Minisart filter and loaded onto a pre-packed 1 ml Ni–NTA column (Qiagen) equilibrated in 50 mM Tris–HCl pH 7.0, 250 mM NaCl (washing buffer). The column was then washed with ten column volumes of washing buffer prior to elution with a gradient of 0–1 M imidazole in 50 mM Tris–HCl pH 7.0, 250 mM NaCl. The SaMazE–SaMazF complex thus obtained was dialyzed against 50 mM Tris–HCl pH 7.0, 250 mM NaCl and concentrated using a Vivaspin concentrator (10 000 Da molecular-weight cutoff). SaMazF and SaMazE were obtained by unfolding and refolding as described previously (Fu et al., 2007 ▶). The purity of the proteins was evaluated using SDS–PAGE.
2.2. Crystallization
Protein samples (SaMazE, SaMazF and their complex) were dialyzed against a suitable buffer (50 mM Tris pH 7.0, 250 mM NaCl; 20 mM Tris pH 7.5, 150 mM NaCl; 20 mM sodium phosphate pH 6.6) and concentrated to 5–20 mg ml−1. A complex between SaMazF and a 33-amino-acid peptide corresponding to the C-terminus of SaMazE (obtained from BioSynthesis, USA) was prepared under the same conditions by slowly adding a fivefold molar excess of the peptide to a concentrated SaMazF solution followed by concentration. Crystallization conditions were screened using the hanging-drop vapour-diffusion method in 48-well plates with drops consisting of 2 µl protein solution and 2 µl reservoir solution equilibrated against 200 µl reservoir solution. Various commercial screens from Hampton Research were used for screening.
2.3. Data collection and analysis
An SaMazF crystal was cryoprotected by direct transfer into the cryostream without the need for additional cryoprotectant. X-ray diffraction data were collected on the PROXIMA1 beamline at the SOLEIL synchrotron (Gif-Sur-Yvette, France) using a wavelength of 1.0394 Å. 150 useful images (out of 300) were collected on an ADSC Q315r CCD detector with a crystal-to-detector distance of 300 mm, 1.0° rotation per image and 1.0 s exposure time. A beam attenuation of 50% and a periodic translation of the crystal in the X-ray beam were used to minimize potential radiation damage. Data were indexed, integrated and scaled with DENZO and SCALEPACK from the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). Analysis of the unit-cell contents was performed with the program MATTHEWS_COEF and self-rotation functions were calculated with MOLREP, both of which are part of the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
An SaMazE–SaMazF (SaMazEF) complex was purified by Ni–NTA affinity chromatography and the individual SaMazE and SaMazF proteins were separated by treatment of the Ni–NTA-bound complex with GdHCl followed by refolding of SaMazF on the column. Both SaMazEF and SaMazE formed large aggregates as assessed by DLS and analytical size-exclusion chromatography and attempts at crystallization failed (data not shown). SaMazF, on the other hand, was monodisperse (2.1 nm, corresponding to a dimer, as is also the case for other MazF-family members) but still failed to crystallize in its free form. Screens set up with SaMazF (20 mM Tris–HCl pH 7.5, 150 mM NaCl) in the presence of a fivefold excess of the C-terminal domain of SaMazE (SaMazE23–56, residues Met23–Glu56) showed promising hits in a number of conditions. Well diffracting needle-shaped crystals were obtained in 0.2 M ammonium acetate, 0.1 M sodium acetate pH 4.6, 30%(w/v) polyethylene glycol 4000.
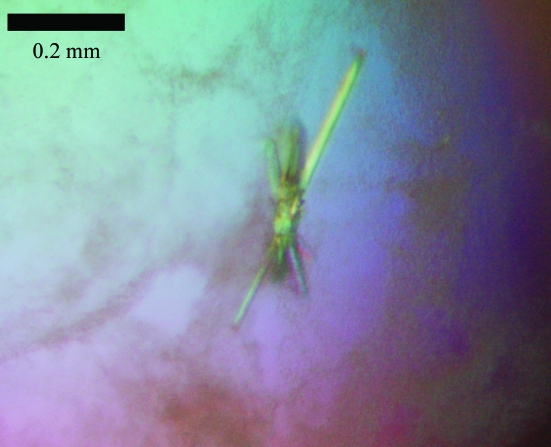
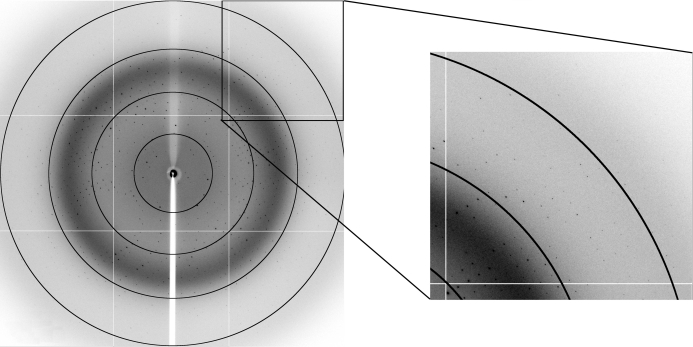
The crystals were needle-shaped and diffracted to about 2.1 Å resolution (Figs. 1 ▶ and 2 ▶). Despite their reasonable size (0.4 mm in their largest dimension) and the low X-ray dose used, they suffered from radiation damage during data collection. Therefore, a full data set required periodic translation of the crystal in the beam to expose fresh areas of the crystal. This involved three different areas of exposure, for each of which 100 images were collected. Only those images where radiation damage was not obvious (as monitored by the increase in B factor remaining below 5.0 Å2 during scaling of the frames) were retained. This corresponds to 150 images, leading to a data set with an overall redundancy of 5.4. Full data-collection statistics are given in Table 1 ▶.
Figure 1.
Typical crystals of SaMazF.
Figure 2.
Diffraction pattern of a crystal of SaMazF showing spots to 2.1 Å resolution. The image corresponds to 1.0 s exposure on the PROXIMA1 beamline of the SOLEIL synchrotron with the crystal rotated by 1°. Rings corresponding to resolutions of 8.7, 4.4, 2.9 and 2.2 Å are indicated. The inset shows an enlargement of the detector corner, showing spots to about 2.1 Å resolution.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | PROXIMA1 |
| Wavelength (Å) | 1.0394 |
| Detector | ADSC Q315r |
| Crystal-to-detector distance (mm) | 300 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 150 |
| Exposure time per image (s) | 1.0 |
| Resolution range (Å) | 56.34–2.10 (2.16–2.10) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 60.72, b = 65.36, c = 112.01 |
| Mosaicity (°) | 0.21–0.61 |
| Total no. of measured intensities | 144316 (9030) |
| Unique reflections | 26598 (2150) |
| Multiplicity | 5.4 (4.2) |
| Mean I/σ(I) | 14.0 (3.4) |
| Completeness (%) | 99.8 (98.8) |
| Rmerge† (%) | 10.4 (34.1) |
| Overall B factor from Wilson plot (Å2) | 30.3 |
R
merge = 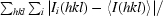
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
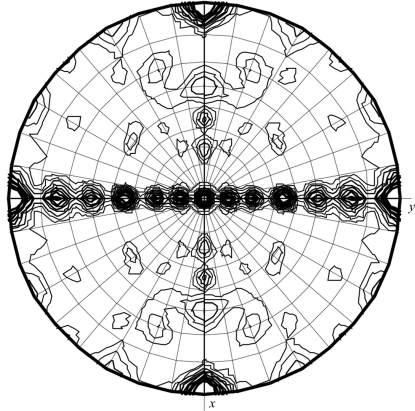
The unit-cell parameters were a = 60.72, b = 65.36, c = 112.01 Å and analysis of the systematic absences pinpointed the space group as P212121. MazF proteins are known to be obligate dimers. Therefore, owing to crystal symmetry constraints the asymmetric unit can only contain one or more complete dimers. If it is assumed that each SaMazF dimer has two SaMazE23–56 peptides bound, the resulting heterotetramer would have a molecular mass of 36 kDa. Matthews analysis then points to a single heterotetramer being present in the asymmetric unit, corresponding to a solvent content of 59%. However, analysis of the self-rotation function indicated a more complex situation, with several significant peaks (Fig. 3 ▶). This indicates the presence of more than one dimer, which would only be possible if MazF crystallized without the MazE-derived peptide. Indeed, SDS–PAGE analysis of a washed crystal revealed only a single band corresponding to the size of SaMazF. We thus conclude that the crystals are crystals of SaMazF, although the SaMazE-derived peptide apparently aided crystallization. It is possible that in the SamazEF system the C-terminus of the antitoxin may not be sufficient to recognize the toxin with high affinity as is the case for the E. coli mazEF module (Li et al., 2006 ▶). The unit cell is likely to contain two SaMazF dimers, corresponding to a solvent content of 34.1%. Structure determination is ongoing and is likely to increase our understanding of MazF-mediated mRNA degradation.
Figure 3.
Self-rotation function showing the presence of several peaks on the κ = 180° section. The self-rotation function was calculated using all data and a Patterson integration radius of 12 Å. Changing the resolution limits did not significantly affect the observed peaks, but their relative intensities varied when varying the integration radius between 8 and 14 Å.
Acknowledgments
This work was supported by NIH grant AI076298 and by grants from FWO, OZR-VUB and VIB. The authors thank Andrew Thompson for beamline support.
References
- Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi, M., Sacher, A. & Engelberg-Kulka, H. (2009). PLoS Genet. 5, e1000390. [DOI] [PMC free article] [PubMed]
- Amitai, S., Yassin, Y. & Engelberg-Kulka, H. (2004). J. Bacteriol. 186, 8295–8300. [DOI] [PMC free article] [PubMed]
- Baik, S., Inoue, K., Ouyang, M. & Inouye, M. (2009). J. Bacteriol. 191, 6157–6166. [DOI] [PMC free article] [PubMed]
- Bravo, A., de Torrontegui, G. & Díaz, R. (1987). Mol. Gen. Genet. 210, 101–110. [DOI] [PubMed]
- Buts, L., Lah, J., Dao-Thi, M.-H., Wyns, L. & Loris, R. (2005). Trends Biochem. Sci. 30, 672–679. [DOI] [PubMed]
- Christensen-Dalsgaard, M. & Gerdes, K. (2008). Nucleic Acids Res. 36, 6472–6481. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Donegan, N. P. & Cheung, A. L. (2009). J. Bacteriol. 191, 2795–2805. [DOI] [PMC free article] [PubMed]
- Fu, Z., Donegan, N. P., Memmi, G. & Cheung, A. L. (2007). J. Bacteriol. 189, 8871–8879. [DOI] [PMC free article] [PubMed]
- Gerdes, K., Christensen, S. K. & Løbner-Olesen, A. (2005). Nature Rev. Microbiol. 3, 371–382. [DOI] [PubMed]
- Gerdes, K., Rasmussen, P. B. & Molin, S. (1986). Proc. Natl Acad. Sci. USA, 83, 3116–3120. [DOI] [PMC free article] [PubMed]
- Inouye, M. (2006). J. Cell. Physiol. 209, 670–676. [DOI] [PubMed]
- Kolodkin-Gal, I., Hazan, R., Gaathon, A., Carmeli, S. & Engelberg-Kulka, H. (2007). Science, 318, 652–655. [DOI] [PubMed]
- Lehnherr, H., Maguin, E., Jafri, S. & Yarmolinsky, M. B. (1993). J. Mol. Biol. 233, 414–428. [DOI] [PubMed]
- Li, G.-Y., Zhang, Y., Chan, M. C., Mal, T. K., Hoeflich, K. P., Inouye, M. & Ikura, M. (2006). J. Mol. Biol. 357, 139–150. [DOI] [PubMed]
- Magnuson, R. D. (2007). J. Bacteriol. 189, 6089–6092. [DOI] [PMC free article] [PubMed]
- Ogura, T. & Hiraga, S. (1983). Proc. Natl Acad. Sci. USA, 80, 4784–4788. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pandey, D. P. & Gerdes, K. (2005). Nucleic Acids Res. 33, 966–976. [DOI] [PMC free article] [PubMed]
- Suzuki, M., Zhang, J., Liu, M., Woychik, N. A. & Inouye, M. (2005). Mol. Cell, 18, 253–261. [DOI] [PubMed]
- Van Melderen, L. (2010). Curr. Opin. Microbiol. 13, 781–785. [DOI] [PubMed]
- Yamaguchi, Y. & Inouye, M. (2009). Prog. Mol. Biol. Transl. Sci. 85, 467–500. [DOI] [PubMed]
- Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G. & Inouye, M. (2003). Mol. Cell, 12, 913–923. [DOI] [PubMed]
- Zhu, L., Inoue, K., Yoshizumi, S., Kobayashi, H., Zhang, Y., Ouyang, M., Kato, F., Sugai, M. & Inouye, M. (2009). J. Bacteriol. 191, 3248–3255. [DOI] [PMC free article] [PubMed]