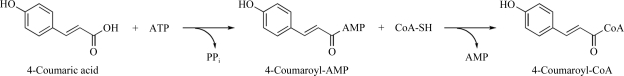
4-Coumarate:CoA ligase 2 from A. thaliana has been overexpressed in E. coli, purified and crystallized. Diffraction data were collected to a resolution of 1.6 Å.
Keywords: CoA ligases, ANL superfamily, phenylpropanoids, lignin biosynthesis
Abstract
4-Coumarate:CoA ligase 2 (4CL2) from Arabidopsis thaliana catalyzes the ATP-dependent formation of the 4-coumaroyl-CoA thioester through the formation of 4-coumarate-AMP. Recombinant 4CL2 protein was expressed in Escherichia coli and crystallized by the sitting-drop vapour-diffusion method. The crystals belonged to space group P21, with unit-cell parameters a = 91.6, b = 55.5, c = 124.4 Å, α = γ = 90.0, β = 111.1°.
1. Introduction
4-Coumarate:coenzyme A ligase (4CL; EC 6.2.1.12) plays a pivotal role in the biosynthesis of phenylpropanoids such as flavonoids and lignin in plants. The enzyme reaction proceeds by the initial adenylation of 4-coumaric acid by ATP and MgCl2, which is followed by a thioester bond-forming reaction with CoASH and the concomitant release of 4-coumaroyl-CoA and AMP (Fig. 1 ▶; Knobloch & Hahlbrock, 1975 ▶). The activation of a carboxylic acid substrate by ATP to form an acyl-AMP intermediate is a common mechanism for adenylate-forming enzymes that catalyze different overall reactions, such as fatty acyl-CoA and aryl-CoA ligases, firefly luciferase and the adenylation domains of the modular nonribosomal peptide synthetases (NRPSs). These enzymes, which share at least ∼20% overall amino-acid sequence identity and conserved ATP-binding motif sequences, are structurally homologous and form the ANL (Acyl-CoA ligases, NRPS adenylation domains and Luciferase enzymes) superfamily of adenylating enzymes (Gulick, 2009 ▶).
Figure 1.
Proposed mechanism for the formation of 4-coumaroyl-CoA by At4CL2.
4CL isoform 2 from Arabidopsis thaliana (At4CL2) is constitutively expressed in lignifying tissues and mainly contributes to lignin biosynthesis (Stuible & Kombrink, 2001 ▶; Schneider et al., 2003 ▶, 2005 ▶). The substrate specificity for the carboxylic acid starter molecule has been well studied and it has been reported that At4CL2 selectively accepts 4-coumaric acid and caffeic acid as starter acids to produce the corresponding CoA thioesters. However, the enzyme does not activate sinapic acid, benzoic acid, phenylacetic acid or phenylalanine (Schneider et al., 2005 ▶). Furthermore, site-directed mutagenesis studies of At4CL2 revealed the amino-acid residues that determine the starter acid specificity, which led to the generation of At4CL2 mutants that activate sinapic acid, a substrate that is normally not accepted by the wild-type At4CL2 enzyme (Schneider et al., 2003 ▶). Recently, the X-ray crystal structures of Populus tomentosa 4CL1, which shares 68.2% amino-acid sequence identity with At4CL2, was reported in the apo form and in forms complexed with AMP and the intermediate analogue 5′-[3-(4-hydroxyphenyl)propyl]phosphate (APP; Hu et al., 2010 ▶). To further clarify the structure and function of the 4CL enzymes, we expressed hexahistidine-fused recombinant At4CL2 protein in Escherichia coli and obtained good-quality crystals of the recombinant enzyme.
2. Experimental
2.1. Construction of the expression plasmid
Total RNA was obtained from three-week-old A. thaliana seedlings using an RNeasy Plant Mini Kit (Qiagen) and was reverse-transcribed using SuperScript II Reverse Transferase (Invitrogen) with an oligo-dT primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′). The first-strand cDNA mixture thus obtained was then used as the template to amplify the At1g02050 cDNA encoding full-length 4CL2 by PCR using 5′-ACGTACGCATGCATGACGACACAAGATGTGATAG-3′ as the sense primer, which introduces an SphI restriction site, and 5′-GGTCAGCTGCAGCTAGTTCATTAATCCATTTGCTAG-3′ as the antisense primer, which introduces a PstI restriction site. The amplified DNA fragment was digested with SphI/PstI and ligated into the SphI/PstI sites of the pQE80L vector (Qiagen) for expression as a fusion protein with a His6 tag at the N-terminus.
2.2. Expression and purification
After confirmation of the sequence, the pQE80L vector encoding full-length 4CL2 was transformed into E. coli M15. The cells harbouring the plasmid were cultured to an OD600 of 0.6 in LB medium containing 100 µg ml−1 ampicillin at 303 K. Isopropyl β-d-1-thiogalactopyranoside was then added to a final concentration of 1 mM to induce gene expression and the culture was incubated for a further 16 h at 303 K.
All of the following procedures were performed at 277 K. The E. coli cells were harvested by centrifugation at 5000g and resuspended in 50 mM Tris–HCl buffer pH 8.0 containing 0.2 M NaCl and 5%(v/v) glycerol (buffer A). The cells were disrupted by sonication and the lysate was centrifuged at 12 000g for 30 min. The supernatant was loaded onto an Ni Sepharose 6 Fast Flow column (GE Healthcare) equilibrated with buffer A. After washing the resin with 50 mM HEPES–NaOH buffer pH 7.0 containing 200 mM NaCl and 5%(v/v) glycerol (buffer B), the recombinant 4CL2 protein was subsequently eluted with buffer B containing 500 mM imidazole. The protein solution was then diluted fivefold with 50 mM HEPES–NaOH buffer pH 7.0 containing 5%(v/v) glycerol and 2 mM DTT (buffer C) and applied onto a Resource-Q column (GE Healthcare). The column was washed with buffer C containing 50 mM NaCl and the protein was subsequently eluted using a linear gradient of 50–300 mM NaCl. The protein solution was further purified to homogeneity by gel-filtration chromatography on Superdex 200 HR (10/300 GL; GE Healthcare) and concentrated to 10 mg ml−1 in 20 mM HEPES–NaOH pH 7.5 buffer containing 100 mM NaCl and 2 mM DTT.
A dynamic light-scattering (DLS) analysis was performed using a DynaPro-MSXTC molecular-sizing instrument (Protein Solutions Inc.). After centrifugation through an 0.22 µm Ultrafree-MC filter (Millipore) to remove particulate material from the protein solution, the solution properties of the purified protein were monitored. Data were acquired as 50 scattering measurements at 278 K and data from three sets of measurements were analyzed using the DYNAMICS software package (Protein Solutions Inc.) and averaged.
2.3. Crystallization and X-ray data collection
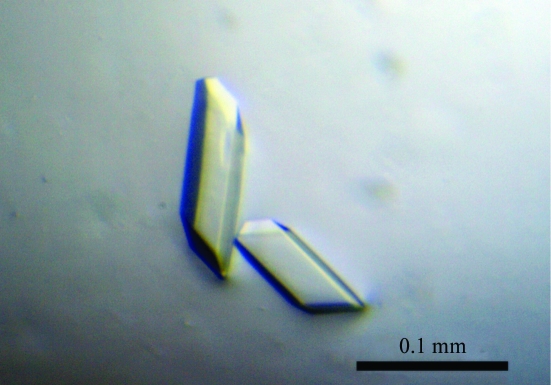
Initial crystallization attempts and optimization of the crystallization conditions were performed at 293 K using the sitting-drop vapour-diffusion method. 0.5 µl purified recombinant 4CL2 protein and an equal volume of reservoir solution were mixed and equilibrated against 100 µl reservoir solution using a 96-condition crystallization screen that was originally designed by Mitsubishi Chemical Corporation. Crystals were observed a few days later in a crystallization solution consisting of 100 mM MES–NaOH pH 6.5, 2 mM ATP and 24%(w/v) PEG 4000. Further crystallization was attempted by using Additive Screen (Hampton Research) at various pH values and by using 22%(w/v) PEG 4000 and 2 mM ATP as precipitants. Diffraction-quality crystals were finally obtained at 293 K in 100 mM MES–NaOH pH 6.0 containing 20% PEG 4000, 5 mM MgCl2, 0.5 mM glutathione (GSH) and 2 mM ATP using the sitting-drop vapour-diffusion method (Fig. 2 ▶).
Figure 2.
Crystal of 4CL2. The dimensions of the crystal were approximately 0.1 × 0.03 × 0.01 mm.
The crystals were transferred into a cryoprotectant consisting of 100 mM MES–NaOH pH 6.0, 20% PEG 4000, 5 mM MgCl2, 0.5 mM GSH, 2 mM ATP and 20% glycerol. After a few seconds, the crystals were picked up in a nylon loop and then flash-cooled at 100 K in a nitrogen-gas stream. X-ray diffraction data sets were collected on beamline NW12 of Photon Factory-AR (wavelength 1.00000 Å) using an ADSC Quantum 210 CCD detector, with a distance of 130 mm between the crystal and the detector. A total of 180 frames were recorded with a 1° oscillation angle and 1 s exposure time. The data were indexed, integrated and scaled with the HKL-2000 program package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The recombinant 4CL2 protein was expressed in E. coli. The purified His6-tag-fused 4CL2 protein migrated as a single band with a molecular mass of 61 kDa on SDS–PAGE, which agrees well with the calculated value of 62.4 kDa. In contrast, gel-filtration analysis of the recombinant 4CL2 protein yielded a molecular mass of 65.9 kDa, suggesting that the recombinant 4CL2 protein exists as a monomeric enzyme. Furthermore, DLS analysis of the recombinant 4CL2 protein after gel filtration revealed a monomodal distribution, with a polydispersity value of 26.6% and an estimated molecular mass of 52 kDa, also suggesting that the recombinant enzyme is monomeric.
Crystals appeared reproducibly within 3 d and the largest crystal grew to dimensions of approximately 0.1 × 0.03 × 0.01 mm (Fig. 2 ▶). A complete data set was collected to 1.6 Å resolution. Detailed data-processing statistics are shown in Table 1 ▶. Based on the diffraction data, the space group was determined to be P21, with unit-cell parameters a = 91.6, b = 55.5, c = 124.4 Å, α = γ = 90.0, β = 111.1°. With two monomers in the asymmetric unit, the Matthews volume (V M; Matthews, 1968 ▶) was calculated to be 2.4 Å3 Da−1 and the estimated solvent content was 47.0%, which is in the range normally observed for protein crystals. A self-rotation function analysis using the CNS program (Brünger et al., 1998 ▶) indicated that the twofold-symmetry axes are parallel to the crystallographic axes. Furthermore, a native Patterson function revealed the presence of a non-origin peak, which supports this conclusion. Structure determination by the molecular-replacement method is now under way using the MOLREP program (Vagin & Teplyakov, 2010 ▶) with the A. thaliana 4CL2 structure model generated by the SWISS-MODEL package (http://swissmodel.expasy.org/) based on the crystal structure of P. tomentosa 4CL (PDB code 3a9v; Hu et al., 2010 ▶), which shares 68% identity with A. thaliana 4CL2, as a search model. Simultaneously, we are also attempting to crystallize the 4CL2 protein complexed with substrate and product analogues. These structural analyses will provide insights into not only the structure–-function relationship of 4CL2 but also the functional diversity of the ANL superfamily enzymes.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Beamline | NW12, Photon Factory-AR |
| Wavelength (Å) | 1.000 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 210 CCD |
| Crystal-to-detector distance (mm) | 130 |
| Oscillation angle (°) | 1 |
| No. of frames | 180 |
| Exposure time (s) | 1 |
| No. of crystals | 1 |
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 91.6 |
| b (Å) | 55.5 |
| c (Å) | 124.4 |
| β (°) | 111.1 |
| Resolution (Å) | 50.0–1.6 (1.63–1.60) |
| Total reflections | 555709 |
| Unique reflections | 151778 |
| Mosaicity (°) | 0.45 |
| Multiplicity | 3.7 (3.6) |
| Completeness (%) | 98.6 (98.5) |
| 〈I/σ(I)〉 | 40.4 (9.2) |
| Rmerge† (%) | 3.4 (17.2) |
R
merge = 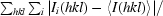
 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl,  is the sum over all reflections and
is the sum over all reflections and 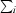 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to IA).
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Gulick, A. M. (2009). ACS Chem. Biol. 4, 811–827. [DOI] [PMC free article] [PubMed]
- Hu, Y., Gai, Y., Yin, L., Wang, X., Feng, C., Feng, L., Li, D., Jiang, X.-N. & Wang, D.-C. (2010). Plant Cell, 22, 3093–3104. [DOI] [PMC free article] [PubMed]
- Knobloch, K. H. & Hahlbrock, K. (1975). Eur. J. Biochem. 52, 311–320. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Schneider, K., Hövel, K., Witzel, K., Hamberger, B., Schomburg, D., Kombrink, E. & Stuible, H. P. (2003). Proc. Natl Acad. Sci. USA, 100, 8601–8606. [DOI] [PMC free article] [PubMed]
- Schneider, K., Kienow, L., Schmelzer, E., Colby, T., Bartsch, M., Miersch, O., Wasternack, C., Kombrink, E. & Stuible, H.-P. (2005). J. Biol. Chem. 280, 13962–13972. [DOI] [PubMed]
- Stuible, H.-P. & Kombrink, E. (2001). J. Biol. Chem. 276, 26893–26897. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]