Crystals of a geranyl pyrophosphate methyltransferase in the biosynthetic pathway of the off-flavor terpenoid alcohol, 2-methylisoborneol were obtained in the absence and presence of cofactor, cofactor analog and substrate.
Keywords: geranyl pyrophosphate methyltransferase, 2-methylisoborneol biosynthesis, Streptomyces lasaliensis
Abstract
The biosynthetic pathway of the off-flavour terpenoid alcohol 2-methylisoborneol (2-MIB) requires geranyl pyrophosphate methyltransferase (GPPMT) to methylate GPP before the cyclization reaction. GPPMT is the first example of an S-adenosyl-l-methionine-dependent methyltransferase that acts on general intermediates such as geranyl pyrophosphate and farnesyl pyrophosphate in isoprenoid biosynthetic pathways. In this study, recombinant GPPMT was overproduced, purified and crystallized in the absence and presence of cofactor, cofactor analogue and substrate. Well diffracting crystals of apo GPPMT containing one molecule in the asymmetric unit were obtained and the structure of this form was solved by the molecular-replacement method. Two crystal forms of the tertiary complex with GPP and sinefungin were also obtained. Structure analysis of these crystals is currently under way in order to understand the enzyme reaction mechanism.
1. Introduction
Isoprenoids form a major group of natural products that are found ubiquitously in all living organisms and serve as visual pigments, reproductive hormones, defensive agents and membrane constituents (Sacchettini & Poulter, 1997 ▶). Their molecular diversity accounts for their various functions in eukaryotes as well as prokaryotes. Despite their extraordinary diversity, all isoprenoids originate from only two isoprene units, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). These two five-carbon isoprene units undergo a condensation reaction to give three linear polyprenyl compounds, geranyl diphosphate (or geranyl pyrophosphate; GPP), farnesyl pyrophosphate (FPP) and geranylgeranyl diphosphate (GGDP), which are the substrates of various enzymes involved in isoprenoid synthesis. It is interesting to study the enzymes that determine the diversity of isoprenoid products. In the biosynthesis of most isoprenoids, a terpene cyclase is the first enzyme to act on GPP, GGDP and/or FPP (Daum et al., 2009 ▶), followed by other modification enzymes. The biosynthesis of the isoprenoid 2-methylisoborneol (2-MIB) is an exception. In this pathway, it was reported that the precursor GPP undergoes a methylation reaction before cyclization and that the reaction involves a unique methyltransferase, GPP methyltransferase (GPPMT; Fig. 1 ▶; Komatsu et al., 2008 ▶). Although methyltransferases that catalyze the methylation of steroid or triterpenoid side chains have been reported, there have been no reports of examples that are involved in catalyzing the methylation of universal pyrophosphate intermediates, which may increase the risk of malfunction of living cells. To our knowledge, this GPP methyltransferase is the first enzyme reported to catalyze this unusual methylation. The enzyme requires the universal methyl donor S-adenosyl-l-methionine (SAM) as a cofactor to produce 2-methyl GPP, which in turn undergoes a cyclization reaction to form the final product 2-MIB (Fig. 1 ▶).
Figure 1.
Biosynthetic pathway of 2-MIB from GPP and the chemical structures of SAM and sinefungin.
2-MIB is responsible for an undesirable earthy odour in water supplies and resists conventional water treatment. The odorous threshold of 2-MIB is as low as 15 ng l−1 (Gagné et al., 1999 ▶). It is a natural isoprenoid product that is commonly found in cyanobacteria and actinomycetes, including many Streptomyces spp. (Juttner & Watson, 2007 ▶). To date, genes encoding GPPMT have been reported in a number of Streptomyces spp. and the GPP biosynthetic pathway has been identified in S. coelicolor and S. lasaliensis (Komatsu et al., 2008 ▶; Wang & Cane, 2008 ▶). However, the details of the molecular mechanism of catalysis and the properties of GPPMT itself are not known. To understand how GPPMT is able to selectively catalyze an atypical reaction in isoprenoid synthesis, the present study was undertaken in order to determine the catalytic mechanism of GPPMT at the atomic level.
In this report, we present the crystallization of recombinant GPPMT from S. lasaliensis. To analyze how GPPMT recognizes the substrate/cofactor and how the reaction proceeds, the molecule was also cocrystallized with SAM, as well as with the substrate GPP and sinefungin (SFG), which is used as a mimic of SAM (Fig. 1 ▶). All of the crystals diffracted sufficiently well for analysis of the substrate-recognition mechanism of GPPMT.
2. Materials and methods
2.1. Expression and purification
The gene encoding GPPMT was cloned into the expression plasmid pET-26b(+) (Stratagene) in fusion with an N-terminal His6 tag. The protein was overproduced in Escherichia coli strain BL21 (DE3) RIL. Single colonies were picked and allowed to grow overnight at 310 K in preculture medium consisting of Luria broth with 25 µg ml−1 kanamycin and 34 µg ml−1 chloramphenicol. The precultures were then transferred into flasks containing 1 l Luria broth with 25 µg ml−1 kanamycin and 34 µg ml−1 chloramphenicol. When the cell density reached an OD600 of 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium to a final concentration of 0.5 mM for induction. The cells were cultured further overnight at 289 K and were then harvested by centrifugation at 4500g for 30 min at 277 K and washed with 50 mM sodium phosphate buffer pH 7.5, 300 mM NaCl, 10% glycerol before being stored at 203 K until use.
The cultured cell pellets were resuspended in a buffer consisting of 50 mM sodium phosphate pH 7.5, 300 mM NaCl, 10% glycerol and disrupted by sonication. The lysates were centrifuged for 60 min at 40 000g and 277 K. The supernatants were collected and filtered through 0.22 µm pore-size filters (Sterivex, Millipore). Crude protein samples were purified at 277 K on an ÄKTApurifier system (GE Healthcare) using an Ni2+-chelating column (HiTrap Chelating HP, GE Healthcare). Nonspecifically bound proteins were removed by successive washes with a buffer consisting of 50 mM sodium phosphate pH 7.5, 300 mM NaCl, 10% glycerol. GPPMT was eluted with a linear gradient to 500 mM imidazole and was further purified by size-exclusion chromatography using a 16/60 Superdex 200 column (GE Healthcare) with buffer consisting of 50 mM sodium phosphate pH 7.5, 300 mM NaCl, 500 mM imidazole and 30% glycerol. The purity of the protein was assessed on 12.5% SDS–PAGE, which showed a band around 40 kDa corresponding to the molecular mass of GPPMT. All purification steps were carried out at 277 K or on ice. If not being used immediately, the purified protein was stored at 193 K until crystallization trials. To perform crystallization, the frozen protein was thawed on ice and the buffer was changed using a concentrating filter (Amicon Ultra-15, Millipore) to a final buffer consisting of 50 mM sodium phosphate pH 7.5, 300 mM NaCl and 10% glycerol.
2.2. Crystallization
Prior to crystallization trials, GPPMT was concentrated to a final concentration of 5 mg ml−1 in a buffer consisting of 50 mM sodium phosphate pH 7.5, 300 mM NaCl and 10% glycerol. Concentration was carried out using a Millipore centrifugal filter device (Amicon Ultra-4, 10 kDa cutoff, Millipore). To cocrystallize GPPMT with the cofactor SAM, the protein sample was mixed with SAM at a protein:SAM molar ratio of 1:7 and left at 277 K for 30 min before setting up crystallization drops. Cocrystallization screening of GPPMT with the substrate GPP was also attempted in the presence and the absence of the SAM analogue sinefungin (SFG); the protein:GPP:SFG molar concentration ratio used was 1:21:21.
Screening for the crystallization of apo GPPMT, GPPMT–SAM and GPPMT–GPP–SFG was performed using Wizard Screens I and II (Emerald BioStructures), Crystal Screen, Crystal Screen 2, Index Screen, Crystal Screen Lite, Crystal Screen Cryo and PEG/Ion Screen (Hampton Research) using the sitting-drop vapour-diffusion method in 96-well plates. Drops were made up of 1 µl sample and an equal volume of reservoir solution and were equilibrated against 0.1 ml reservoir solution at 293 K. Additional screening kits (Nucleix Suite, Protein Complex Suite, Anions and Cations Suite and AmSO4 Suite; Qiagen) were also used for screening of GPPMT–GPP–SFG.
The initial crystallization conditions for the apoprotein and the complex with SAM were adopted from Crystal Screen 2 condition No. 42 [1.5 M ammonium sulfate, 0.1 M Tris pH 8.5, 12%(v/v) glycerol]. Crystals from the mixture with GPP were obtained using Wizard I condition No. 9 [0.1 M sodium acetate pH 4.5, 1 M (NH4)2HPO4]. Crystals of the ternary complex with GPP and SFG were obtained using Nucleix Suite condition No. 73 (0.05 M sodium cacodylate pH 6.0, 5.0 mM spermidine, 15 mM magnesium chloride, 2.0 M lithium sulfate).
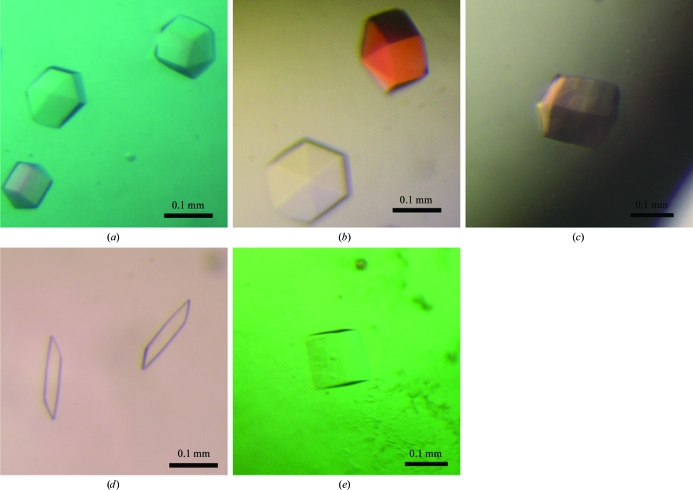
Several conditions were further screened by the hanging-drop method using 24-well VDX plates (Hampton Research) with drops composed of 1.5 µl protein solution and 1.5 µl reservoir solution equilibrated against 0.5 ml reservoir solution at 293 K. Five types of crystal were obtained as shown in Fig. 2 ▶. The apoprotein crystals and the crystals obtained in the presence of SAM and of GPP shared a similar hexagonal shape, while the GPP–SFG cocrystals had two different shapes. The best conditions for the apoprotein crystals were 0.1 M Tris pH 7.75, 1 M ammonium sulfate and 8% glycerol. GPPMT–SAM crystals were obtained under similar conditions (0.1 M Tris pH 8.75, 1 M ammonium sulfate, 20% glycerol). Optimized crystals of GPPMT in the presence of GPP were best crystallized in the presence of 0.1 M sodium acetate pH 4.5, 0.75 M (NH4)2HPO4. Two sets of conditions optimized from Nucleix Suite condition No. 73 gave different forms of GPPMT–GPP–SPG crystals (Figs. 2 ▶ d and 2 ▶ e): 50 mM sodium cacodylate pH 6.5, 1.4 M lithium sulfate, 30 mM MgCl2 and 1 mM spermine for GPPMT–GPP–SFG form I and 50 mM sodium cacodylate pH 6.7, 1.3 M lithium sulfate, 30 mM MgCl2 and 1 mM spermine for GPPMT–GPP–SFG form II.
Figure 2.
Crystals of GPPMT from S. lasaliensis. (a) GPPMT crystals without any ligand obtained using 0.1 M Tris pH 7.75, 1 M ammonium sulfate, 8% glycerol. (b) GPPMT–SAM crystals obtained using 0.1 M Tris pH 8.75, 1 M ammonium sulfate, 20% glycerol. (c) GPPMT crystals obtained in the presence of GPP using 0.1 M acetate pH 4.5, 1 M (NH4)2HPO4. (d) GPPMT–GPP–SFG form I crystals obtained using 50 mM sodium cacodylate pH 6.5, 1.4 M lithium sulfate, 30 mM magnesium chloride, 1 mM spermine. (e) GPPMT–GPP–SFG form II crystals obtained using 50 mM sodium cacodylate pH 6.7, 1.3 M lithium sulfate, 30 mM magnesium chloride, 1 mM spermine.
2.3. Data collection and reduction
All data sets were collected on beamline BL41XU of SPring-8 using an ADSC Q315 CCD detector. Prior to diffraction data collection, the crystals were cryoprotected by transfer into a solution containing 25%(v/v) glycerol for a few seconds and flash-cooled. The data sets were integrated, merged and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The apoprotein crystals belonged to space group P6322, with unit-cell parameters a = b = 143.7, c = 66.8 Å, as did the SAM cocrystals, with unit-cell parameters a = b = 147.9, c = 66.5 Å, and the crystals obtained in the presence of GPP, with unit-cell parameters a = b = 143.9, c = 66.7 Å, whereas cocrystallization with both SFG and GPP gave two types of crystal forms, both of which belonged to space group P1, with unit-cell parameters a = 83.1, b = 91.8, c = 335.9 Å, α = 85.19, β = 83.85, γ = 89.91° for form I and a = 78.9, b = 87.7, c = 160.2 Å, α = 100.01, β = 96.65, γ = 90.89° for form II. Based on the values of the Matthews coefficient (V M; Matthews, 1968 ▶), it was estimated that there was one molecule in the asymmetric unit of the apo GPPMT crystals, the GPPMT–SAM crystals and the crystals of GPPMT obtained in the presence of GPP, with V M values of 2.76 Å3 Da−1 (V solv = 55.4%), 2.91 Å3 Da−1 (V solv = 57.7%) and 2.76 Å3 Da−1 (V solv = 55.4%), respectively. Between 20 and 41 molecules in the asymmetric unit gives a reasonable V M value range (between 1.7 and 3.5 Å3 Da−1) for the form I crystal, whereas between nine and 17 molecules are possible in the form II crystal. Details of the data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| GPPMT–GPP–SFG | ||||
|---|---|---|---|---|
| GPPMT-apo (GPP present during crystallization) | GPPMT–SAM | Form I | Form II | |
| X-ray source | SPring-8 BL41XU | SPring-8 BL41XU | SPring-8 BL41XU | SPring-8 BL41XU |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 | 1.000 |
| Space group | P6322 | P6322 | P1 | P1 |
| Unit-cell parameters | ||||
| a (Å) | 143.9 | 147.9 | 83.1 | 78.9 |
| b (Å) | 143.9 | 147.9 | 91.8 | 87.7 |
| c (Å) | 66.7 | 66.5 | 335.9 | 160.2 |
| α (°) | 90 | 90 | 85.2 | 100.0 |
| β (°) | 90 | 90 | 83.9 | 96.6 |
| γ (°) | 120 | 120 | 89.9 | 90.9 |
| Resolution range (Å) | 50–2.20 (2.28–2.20) | 50–2.20 (2.28–2.20) | 50–3.20 (3.26–3.20) | 50–3.00 (3.05–3.00) |
| Total No. of observations | 317808 | 161781 | 425655 | 290328 |
| Unique reflections | 21196 (2059) | 22017 (2174) | 151410 (5056) | 82669 (4153) |
| Multiplicity | 15.0 (12.7) | 7.4 (7.3) | 2.8 (2.1) | 3.5 (3.5) |
| Completeness (%) | 99.9 (100) | 98.8 (99.6) | 94.9 (63.3) | 96.3 (95.8) |
| Mean I/σ(I) | 42.0 (9.1) | 25.6 (3.6) | 7.1 (3.8) | 8.0 (2.9) |
| Rmerge† | 0.07 (0.42) | 0.06 (0.42) | 0.24 (0.64) | 0.12 (0.36) |
R
merge = 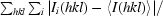
 , where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections.
, where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections.
3. Results and discussion
The biological significance of methylation varies widely and most methylation reactions are catalyzed by the S-adenosyl-l-methionine-dependent methyltransferases (SAM-MTs). SAM-MTs can be classified into five distinct classes (classes I–V) based on their fold; the largest group of SAM-MTs possess the class I fold (Schubert et al., 2003 ▶). We focused on GPPMT, the substrate of which is GPP, which is widely used in isoprenoid biosynthetic pathways. As GPP contains a pyrophosphate moiety, a feature that is also shared with other common building blocks for isoprenoid biosynthesis such as IPP, FPP and GGDP, we thought that it would be possible to discover a structural basis that is shared throughout this family of SAM-MTs in this study. We attempted to crystallize the apoenzyme and to cocrystallize GPPMT with three different ligands in order to analyze its mechanisms of ligand recognition and catalysis. A molecular-replacement solution for the apoprotein crystal was found using the structure of the SAM-dependent rebeccamycin sugar 4′-O-methyltransferase RebM (Singh et al., 2008 ▶; PDB entry 3bus) as a search model. The sequence identity between these proteins is 22%, with 15% strong similarity. The molecular replacement was carried out using MOLREP (Vagin & Teplyakov, 2010 ▶). The arrangement of the molecule in the asymmetric unit in the crystal of GPPMT–SAM was essentially identical to that of apo GPPMT. Preliminary structure refinement of these models was performed with REFMAC5 (Murshudov et al., 1997 ▶). We confirmed that a bound SAM molecule was present in the active site of GPPMT cocrystallized with SAM, whereas no residual electron density was found around the active site of the crystal structure obtained by mixing GPPMT with only GPP. Therefore, we regarded these data as the apo form in this paper. We also found the locations of the protein chains in the crystal structure of GPPMT–GPP–SFG in both form I and form II by the molecular-replacement procedure using MOLREP (Vagin & Teplyakov, 2010 ▶). There are 24 molecules in the form I unit cell and 12 molecules in the form II unit cell and structure refinement of each form is currently under way in our laboratory.
Acknowledgments
We are grateful to the beamline staff of SPring-8 (Hyogo, Japan) for technical help during data collection. This project was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. OA was supported by a scholarship from MEXT.
References
- Daum, M., Herrmann, S., Wilkinson, B. & Bechthold, A. (2009). Curr. Opin. Chem. Biol. 13, 180–188. [DOI] [PubMed]
- Gagné, F., Ridal, J., Blaise, C. & Brownlee, B. (1999). Bull. Environ. Contam. Toxicol. 63, 174–180. [DOI] [PubMed]
- Juttner, F. & Watson, S. B. (2007). Appl. Environ. Microbiol. 73, 4395–4406. [DOI] [PMC free article] [PubMed]
- Komatsu, M., Tsuda, M., Omura, S., Oikawa, H. & Ikeda, H. (2008). Proc. Natl Acad. Sci. USA, 105, 7422–7427. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sacchettini, J. C. & Poulter, C. D. (1997). Science, 277, 1788–1789. [DOI] [PubMed]
- Schubert, H. L., Blumenthal, R. M. & Cheng, X. (2003). Trends Biochem. Sci. 28, 329–335. [DOI] [PMC free article] [PubMed]
- Singh, S., McCoy, J. G., Zhang, C., Bingman, C. A., Phillips, G. N. Jr & Thorson, J. S. (2008). J. Biol. Chem. 283, 22628–22636. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, C. M. & Cane, D. E. (2008). J. Am. Chem. Soc. 130, 8908–8909. [DOI] [PMC free article] [PubMed]