To investigate the pathogenesis of the RNA splicing factor forms of RP, the authors generated and characterized the retinal phenotypes of Prpf3-T494M, Prpf8-H2309P knockin mice, and evaluated the retinal ultrastructure of Prpf31-knockout mice. All three mouse models demonstrate degenerative changes in the RPE with age, suggesting that the RPE may be the primary cell type affected in the RNA splicing factor forms of RP.
Abstract
Purpose.
Mutations in genes that produce proteins involved in mRNA splicing, including pre-mRNA processing factors 3, 8, and 31 (PRPF3, 8, and 31), RP9, and SNRNP200 are common causes of the late-onset inherited blinding disorder retinitis pigmentosa (RP). It is not known how mutations in these ubiquitously expressed genes lead to retina-specific disease. To investigate the pathogenesis of the RNA splicing factor forms of RP, the authors generated and characterized the retinal phenotypes of Prpf3-T494M, Prpf8-H2309P knockin mice. The retinal ultrastructure of Prpf31-knockout mice was also investigated.
Methods.
The knockin mice have single codon alterations in their endogenous Prpf3 and Prpf8 genes that mimic the most common disease causing mutations in human PRPF3 and PRPF8. The Prpf31-knockout mice mimic the null alleles that result from the majority of mutations identified in PRPF31 patients. The retinal phenotypes of the gene targeted mice were evaluated by electroretinography (ERG), light, and electron microscopy.
Results.
The RPE cells of heterozygous Prpf3+/T494M and Prpf8+/H2309P knockin mice exhibited loss of the basal infoldings and vacuolization, with accumulation of amorphous deposits between the RPE and Bruch[b]'s membrane at age two years. These changes were more severe in the homozygous mice, and were associated with decreased rod function in the Prpf3-T494M mice. Similar degenerative changes in the RPE were detected in Prpf31± mice at one year of age.
Conclusions.
The finding of similar degenerative changes in RPE cells of all three mouse models suggests that the RPE may be the primary cell type affected in the RNA splicing factor forms of RP. The relatively late-onset phenotype observed in these mice is consistent with the typical adult onset of disease in patients with RP.
Pre-mRNA splicing is an essential step in the expression of most eukaryotic transcripts. This ubiquitous process takes place in a large ribonucleoprotein complex, the spliceosome, which in addition to the pre-mRNA substrate is made up of five core small nuclear ribonucleoprotein complexes (snRNPs), and a host of accessory proteins which assemble in a stepwise fashion on each intron to carry out the splicing reaction (for reviews see Refs. 1, 2). Mutations in five protein factors integral to this process have been implicated in the inherited blinding disorder retinitis pigmentosa (RP).3–7 These factors are ubiquitously expressed and are common to the U4/U6/U5 tri-snRNP complex. The pre-mRNA processing factor 3 (PRPF3) protein is associated with the U4/U6 snRNP complex, and is necessary for the integrity of a spliceosomal precursor, the U4/U6/U5 tri-snRNP complex, without which splicing cannot occur.8,9 PRPF8 is a U5 snRNP and U4/U6/U5 tri-snRNP component and is thought to be an assembly platform at the core of the activated spliceosome (for review see Ref. 10). PRPF31 is also a component of the U4/U6/U5 tri-snRNP and is involved in maintaining the stability of the complex.11,12 RP9 and SNRNP200 interact with PRPF3 and PRPF8, respectively, and are also components of the U4/U6.U5-tri-snRNP complex.7,13,14 Due to the ubiquitous nature of RNA splicing, there is much debate regarding the underlying disease mechanism by which mutations in the RNA splicing factors lead to photoreceptor specific disease.
RP is the most common inherited form of blindness, with an estimated prevalence of approximately 1:1000 to 4000 people.15–18 This translates into ∼1.5 to 6 million individuals affected with RP worldwide. RP patients typically present in early adulthood with progressive night blindness and loss of peripheral visual field due to the loss of rod photoreceptor cells of the retina. The cone photoreceptors die after the rods, leading to a further loss of daytime vision, which gradually progresses to complete blindness in later life.19 RP displays all three modes of Mendelian inheritance, and most of the 37 genes implicated in non-syndromic RP are expressed specifically in either photoreceptor cells or the retina20 (for a current listing of retinal disease genes see RetNet: http://www.sph.uth.tmc.edu/Retnet/). As a group, the five pre-mRNA processing factor forms of dominant RP constitute a leading cause of dominant RP, second only to mutations in rhodopsin.21,22 For most RP genes, the mechanisms by which the mutations lead to disease are not understood, but since most of the disease genes are photoreceptor specific, it follows naturally that a photoreceptor-specific disease results. The pre-RNA splicing factor forms of RP are different in that they are due to mutations in ubiquitous proteins involved in RNA splicing in all cells and tissues of the body, and yet still lead to retina-specific disease, suggesting a novel pathway to retinal degeneration.
There are several potential explanations for the retinal degeneration in splicing-factor RP. Splicing-factor RP could result from haploinsufficiency of functional splicing factors. Sub-optimal levels of pre-mRNA processing factors could lead to retinal damage because the retina is among the most biosynthetically active tissues in the body. A second hypothesis is that spliceosomes containing the mutant factors are competent to splice most introns correctly, but when assembled on introns that, even under normal circumstances, splice relatively inefficiently, the mutant proteins compromise the correct splicing of those introns. The correlate to this hypothesis is that one or more vital retinal transcripts contain these introns, which then lead to RP when mis-spliced. The third hypothesis is that the mutations result in a gain of function to these factors which is toxic to the retina (for review see Ref. 23).
Our laboratories have produced mouse models to study the effects of mutations in PRPF3, PRPF8 and PRPF31 on vision. A recent report from one of our groups described Prpf3 knockout mice, in which heterozygous animals had no evident retinal phenotype at two years of age, arguing against the first hypothesis of haploinsufficiency.24 In contrast, homozygosity of the Prpf31-A216P knockin allele caused early embryonic lethality, consistent with the idea that a majority of mutations in PRPF31 result in null alleles and the disease is caused by haploinsufficiency of this splicing factor.4,25–33 Heterozygous Prpf31+/A216P and Prpf31+/− mice did not develop photoreceptor dysfunction at ages up to 18 months.33 Here we describe generation and characterization of Prpf3 knockin mice bearing the p.T494M missense mutation found in RP18 patients, and Prpf8 knockin mice with the p.H2309P missense mutation found in RP13.3,5 Additionally, we report ultrastructural characterization of the heterozygous Prpf31 knockout (Prpf31+/−) mouse model.33 We find that at aged time points, all three animal models display degenerative changes in the retinal pigment epithelium (RPE). The RPE degeneration is associated with decreased retinal function in the Prpf3-T494M mice.
Materials and Methods
Animals
Animal research was performed under protocols approved by the Institutional Animal Care and Use Committees at the University of Pennsylvania and the Centre de Recherche Institut de la Vision, Université Pierre et Marie Curie-Paris, and conforms to the ARVO Statement for Use of Animals in Ophthalmic and Vision Research. Wild-Type C57BL/6J mice and FLPe deleter mice were obtained from Jackson Laboratories, (Bar Harbor, ME).34
Production of Targeting Vectors
Prpf3-T494M.
We used homologous recombination to screen a lambda phage mouse genomic library as in Zhang et al.35 Using a floxed tetracycline resistance cassette flanked by regions of homology to the 10th intron of the Prpf3 gene, we isolated five clones spanning different regions of the C terminus of Prpf3. Of these clones, one contained a 10 kb insert spanning exons 10–16. This region of Prpf3 is the most highly conserved and contains exon 11, which is the location of the p.T494M mutation found in RP18 patients. To make the knockin vector (See Fig. 1) we transformed this clone into bacteria expressing cre-recombinase to remove the tetracycline cassette.35 We then used oligonucleotide-directed mutagenesis to mutate the 11th exon, resulting in the p.T494M mutation (QuikChange, Stratagene, LaJolla, CA). A silent base change which eliminates an XmnI digest site was also included for rapid genotyping purposes. Next, another round of homologous recombination was performed to insert the positive selection cassette for targeting in ES cells using short regions of homology to the 10th intron to create the final targeting vector for the knockin36 (Fig. 1B).
Figure 1.
Targeting of Prpf3-T494M and Prpf8-H2309P knockin mice. (A) Exons 9 to 16 of the Prpf3 locus with restriction sites indicated. (B) Knockin vector with T494M mutation, and positive and negative selection cassettes. (C) Targeted Prpf3-Neo-T494M locus. (D) Final targeted Prpf3-T494M knockin allele. (E) Southern blot analysis of wild-type and Prpf3-Neo-T494M shown in (C) using indicated 5′ and 3′ probes for SpeI and BstXI digests, respectively. (F) Genomic sequence data from wild-type, heterozygous, and homozygous Prpf3-T494M mice. (G) Exons 25 to 42 of the Prpf8 locus. (H) Prpf8 Knockin vector containing exons 30–42, the H2309P mutation, and selection cassette for ES targeting. (I) Targeted Prpf8-Neo-T494M locus. (J) Final targeted Prpf8-H2309P knockin allele. (K) Southern blot analysis of targeted Prpf8 knockin mice detects the shorter NotI fragment in targeted mice. (L) Sequence analysis of wild-type and targeted Prpf8 genomic DNA showing replacement of the endogenous codon with the mutant codon.
Prpf8-H2309P.
We created the Prpf8-H2309P targeting vector using a similar approach to that described above, starting with a BAC clone which contained the Prpf8 gene. We again used recombineering techniques to isolate a 16 kb portion of the Prpf8 gene containing exons 30 through 42 (Fig. 1H). We added appropriate selection cassettes, and mutated exon 42 to introduce the p.H2309P substitution (Figs. 1H–I).
Transfection of ES Cells, Verification of Targeting, and Production of Mice
After sequence verification that the knock-in constructs contained the desired mutation and that the rest of the exons were correct, the linearized vectors were electroporated into 129SvEvTac mouse embryonic stem (ES) cells.37 Embryonic stem cells were cultured in embryonic stem cell medium (Dulbecco[b]'s Modified Eagle[b]'s Medium (DMEM; GIBCO; Gaithersburg, MD) with 15% fetal bovine serum (Hyclone Laboratories, Logan, UT), 1% non essential amino acids (GIBCO), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO), and 1250 U/mL leukemia inhibitory factor (Chemicon International, Temecula, CA) on a primary mouse embryo fibroblast monolayer (Chemicon). G418- and ganciclovir-resistant clones were isolated and expanded for verification of targeting by Southern blot analysis using probes in the 5′ and 3′ surrounding region outside the targeting vector. The presence of the T494M and H2309P mutations was verified by PCR followed by sequencing. Two correctly targeted clones for each gene were injected into C57BL/6J blastocysts to produce chimeric mice at the Transgenic and Chimeric Mouse Core Facility at the University of Pennsylvania.38 Highly chimeric founder mice for each targeted allele were backcrossed with C57BL/6J mice. Agouti F1 progeny were screened for the targeted Prpf3 and Prpf8 alleles by PCR and Southern blotting. We obtained germline transmission from at least two founder mice for both targeted alleles. The F1 Prpf3-Neo-T494M and Prpf8-Neo-H2309P mice were crossed with FLPeR mice to remove the neomycin selection cassettes.34 Excised Prpf3-T494M and Prpf8-H2309P mice were backcrossed with C57BL/6J mice to generate heterozygous F1 mice. Male and female F1 Prpf3-T494M or Prpf8-H2309P mice were inter-crossed to generate homozygous F2 mice. Expected Mendelian ratios were obtained from these crosses, and the resultant mice were healthy and fertile, with normal lifespan. The Prpf31+/− mice were generated as described previously.33
Genotyping of Mice
Genotyping of Prpf3-T494M and Prpf8-H2309P knockin mice and Prpf31 knockout mice was performed by Southern blotting and PCR. For Prpf3 Southern blotting, 2 probes were used. Probe 1 was amplified from genomic DNA 5′ to the targeting vector using primers: forward 5′-CATTCTTAGCATTAAAGCAACAAAATC-3′ and reverse 5′-ACAGACCCAATCAGCTAAACCACCTATC-3′. It detects a wild-type SpeI fragment of 8 kb and a targeted 6 kb fragment. Probe 2 was amplified 3′ to the targeting vector using primers: forward 5′-GCCCATGACCCGTACGGTGTGTGTATC-3′ and reverse 5′-TGCTTCACGTTCCCTGCATGTTCCATCC-3′. It detects a 16.9 kb wild-type BstXI fragment versus a 19.5 kb targeted BstXI fragment. For Prpf8 Southern blotting Probe1 was amplified 5′ to the vector using primers: forward 5′-CCCTTCAGCTCGTCCCTGGTTTGAG-3′ and reverse 5′-GATCTAGAAAATATCCAACTCCACAACTTACTGC-3′. It detects a wild-type NotI fragment of 17 kb and a 10 kb targeted fragment.
For genotyping by PCR, the following primer sets were used. For Prpf3-T494M mice, amplification with forward primer 5′-CTCATTCATTGGCATTAAAAATAAATAAACTCC-3′ and reverse primer 5′-CGTTGGCCTCTTCATGCGCTCTGTCGTGAC-3′, followed by digestion of the PCR products with Xmn1 was used. The presence of a band at 649 bp signifies the mutant allele. Prpf8 mice were genotyped using primers located in the 3′ UTR (forward primer 5′-CCAGACAAGCTGCTGACATTCAGCAGTC-3′ and reverse primer: 5′-TAGAGCTGTAAGTGGTCACTTCAGGC-3′). The mutant allele is 400 bp larger than the wild-type. Prpf31+/− mice were identified by PCR using primers in exons 4 and 8 (forward primer: 5′-CTCCTGAGTACCGAGTCATTGTGGATGC; reverse primer: 5′-GTAGAAGAGAAGCCAGACAGGGTCTTGC). The mutant allele was 897 bp shorter than the wild-type.
Northern Blotting
Four or more retinas of 4-week-old mice of the indicated genotype were pooled and RNA isolated using reagent (Trizol; Invitrogen, Carlsbad, CA). Fifteen to 20 micrograms of total RNA were loaded per lane on a denaturing 0.8% agarose gel, transferred overnight to a nylon membrane (Schleicher & Schuell, Keene, NH), cross-linked and stored or hybridized according to standard protocols.39 A 652-bp radiolabeled probe against the mouse Prpf3 transcript was amplified from cDNA using primers: forward 5′-CAGATGATGGAAGCAGCAACACGAC-3′ and reverse 5′-TTCTAGCAGCTTGTGAAATCTCT-3′. This probe spans exons 5 to 8 of the Prpf3 transcript. To detect the mouse Prpf8 transcript, a 1300-bp probe was amplified using primers: forward 5′-GCTGCCGGATTATATGTCAGAGGAGAAGC-3′ and reverse 5′-GTACTTAAGCAGCTTCTGATAAGAGACTCG-3′. This probe spans exons 1 to 9 of the Prpf8 transcript. To assess total RNA per lane, a probe against the housekeeping gene acidic ribosomal phosphoprotein P0 (36B4) was used.40 Probes were hybridized to membranes overnight at 65°C, washed, and exposed to phosphor screens for detection. Phosphor screens were scanned using a phosphorimager (Storm Phosphorimager, GE Healthcare, Piscataway, NJ) and quantified using imaging software (ImageQuant 5.2, GE Healthcare).
Western Blotting
Retinas were solubilized by sonication in LDS sample buffer (Invitrogen) and 100 micrograms of reduced protein were separated in each lane of 3–8% Tris-Acetate polyacrylamide gel (NuPage; Invitrogen). Proteins were transferred electrophoretically to polyvinylidene difluoride (PVDF) membrane (Invitrogen), and blocked in 10% nonfat dry milk solution for 1 hour at room temperature. Primary antibodies against Prpf3 protein were a generous gift from James Hu.41 Alkaline phosphatase conjugated anti-rabbit secondary antibodies (Vector Laboratories, Burlingame, CA) were used in conjunction with ECF reagent (Amersham Biosciences, Piscataway, NJ) and blots were scanned with an imager (Storm Phosphorimager or Typhoon Variable Mode Imager; GE Healthcare). Band intensities were quantified (ImageQuant 5.2 software; GE Healthcare).
ERG Analysis
Electroretinography was performed as previously described.42 Briefly, full-field ERG[b]'s were recorded in a Ganzfeld on dark-adapted, anesthetized mice taking care to maintain 37°C body temperature at all times. Pupils were dilated with 1% tropicamide. Retinal responses were detected with platinum electrodes embedded in contact lenses contacting the cornea and recorded using custom software.
Light and Electron Microscopy
Preparation of retinas for light and electron microscopy was performed as previously described.24,43 For histologic analysis of the retina animals were perfused with 4% paraformaldehyde in phosphate buffered saline (PBS; Electron Microscopy Sciences, Hatfield, PA), and eyecups were processed for cryosectioning. Ten-micron sections were cut, mounted onto slides (Superfrost Plus, Fisher Scientific, Pittsburgh, PA), and stained with alkaline toluidine blue for light microscopy. For co-localization studies, antibody Y12 (ab3138; Abcam, Cambridge, MA) was used for detection of snRNPs. For the measurement of ONL thickness, a section from 12 o[b]'clock, through the optic nerve head, and 6 o[b]'clock was used and measurements were taken at 10 intervals in both directions toward the periphery starting at the optic nerve head. For electron microscopy, perfused eyecups were transferred to 2% paraformaldehyde + 2% glutaraldehyde in 0.2 M sodium cacodylayte buffer (pH 7.4) and further processed for plastic sectioning in EMbed812 (Electron Microscopy Sciences). One micrometer sections were then cut and stained with alkaline toluidine blue for light microscopy and 60 to 80 nM ultrathin sections were stained with lead citrate /uranyl acetate and examined using a transmission electron microscope (FEI Tecnai, Hillsboro, OR).
Results
Generation of Prpf3-T494M and Prpf8-H2309P Mice.
We generated Prpf3-T494M and Prpf8-H2309P knockin mice using standard protocols as described in Materials and Methods (Fig. 1). After removal of the selection cassettes via crosses with FLPe deleter mice, heterozygous and homozygous knockin mice were generated for the experiments described. Both lines of homozygous knockin mice were healthy, and fertile. The generation of the Prpf31+/− mice has been described recently.33
Expression of Prpf3 and Prpf8.
Northern blot analysis and qRT-PCR analyses showed that the sizes and levels of the mutant Prpf3 and Prpf8 transcripts produced in the retinas of the homozygous Prpf3-T494M and Prpf8-H2309P mice were similar to that observed in controls (Figs. 2A, 2B, 2C). Similarly, Western blot analysis showed that the level of Prpf3 protein in the retinas of homozygous knockin mice was also normal (Fig. 2D). No suitable antibodies for mouse Prpf8 were available to perform this analysis in the Prpf8-H2309P line.
Figure 2.
Expression of Prpf3 and Prpf8 knockin transcripts and associated proteins. (A) Northern blot analysis of RNA from four wild-type and four homozygous Prpf3-T494M retinas. (B) Northern blot analysis of four wild-type and four homozygous Prpf8-H2309P retinas. (C) SYBR green qRT-PCR for Prpf3 and Prpf8 from wild-type versus knockin retina (n = 3). (D) Western blotting of Prpf3 from wild-type, heterozygous, and homozygous knockin retina. (E) Frozen sections of wild-type and Prpf3-T494M/T494M retinas stained with antibodies to Prpf3 (red). The Prpf3 protein is located in the inner segment, outer nuclear layer, and inner nuclear layer in both wild-type and homozygous knockin animals. (F) Confocal microscopy of frozen sections co-stained with Prpf3 (red) and antibody Y12 (green). Prpf3 localizes to nuclear speckles detected by Y12 in the periphery of photoreceptor nuclei (ONL panels) and throughout the nuclei of the inner nuclear layer (INL panels). The location of Prpf3 is the same in wild-type and Prpf3-T494M/T494M animals. IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer.
We asked whether the presence of the T494M mutation in the Prpf3 protein alters its subcellular localization, as has been reported following over-expression of mutant protein in cultured cells.44 Prpf3 was detected in photoreceptor inner segments, the outer nuclear layer, and in the inner nuclear layer of the retina, consistent with its ubiquitous nature. This localization was unchanged in the retinas of homozygous knockin mice (Fig. 2E). Furthermore, we performed a co-localization study with monoclonal antibody Y1245 which recognizes Sm proteins in snRNPs, to determine whether the mutation alters the inclusion of Prpf3 into nuclear speckles, which are sites of snRNP storage, assembly and modification (for review see Ref. 46). We found an interesting staining pattern in photoreceptor nuclei, for both Prpf3 and the Y12 antibody. The staining concentrates to nuclear speckles in the periphery of the nucleus in photoreceptors, but in non-photoreceptor cells displays the more typical pattern of speckles throughout the nuclei. In both cases we found that in wild-type versus homozygous knockin retina, the overlap of the two signals was unchanged, indicating that the mutant Prpf3 is able to enter the nucleus and incorporate into snRNPs (Fig. 2F).
Retinal Function.
Electroretinography was used to evaluate the retinal function of the Prpf3 and Prpf8 knockin mice.42 We measured scotopic a-waves and b-waves to analyze the rod photoreceptors and bipolar cells, respectively, as well as the photopic b-waves to assess cone function. We compared wild-type, heterozygous and homozygous knockin mice at various time points up to 24 months of age. At time points up to 18 months, no significant differences were found between the wild-type and knockin animals (data not shown). However, at the oldest time point of 24 months, the maximal rod a-wave of Prpf3 heterozygous and homozygous knockin mice was significantly decreased (Fig. 3A). Although the rod b-waves were decreased as well, this decrease did not reach statistical significance. No significant differences were found in the cone b-wave at this time point (Figs. 3B, 3C). No significant differences were found for Prpf8 knockin mice compared to Prpf8 wild-type mice.
Figure 3.
Retinal Function. (A) Maximum scotopic a-wave responses in 24-month-old mice. Significant decreases were found in Prpf3-Wt/T494M (n = 12, P = 0.0149 via two-tailed Student's t-test) and Prpf3-T494M/T494M (n = 11, P = 0.0135)) compared to wild-type (n = 9). No significant changes were found in Prpf8 mice (Wt/Wt, n = 12, Prpf8-Wt/H2309P, n = 6, Prpf8-H2309P/H2309P, n = 12). (B) Scotopic b-wave amplitudes as measured from baseline to b-wave peak were decreased in Prpf3 knockin mice relative to wild-type, but this change did not reach significance. (C) No significant differences were found in photopic b-wave responses. Error bars indicate SEM.
Retinal Morphology and Ultrastructure.
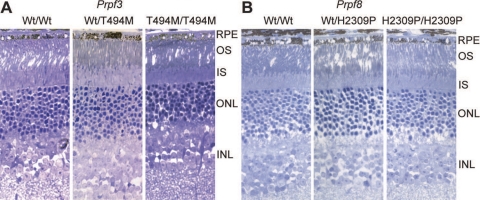
Light microscopy showed that there was no major photoreceptor degeneration at any age tested in Prpf3-T494M (Fig. 4A) or Prpf8-H2309P (Fig. 4B) mice. Similar results were also reported for 1-year-old Prpf31+/− mice.33 However, ultrastructural analyses did reveal significant differences between the wild-type and knockin animals in the retinal pigment epithelium (RPE) for the Prpf3 and Prpf8 mutant mouse lines at two years of age, and the Prpf31 mice at one year of age (Fig. 5). The wild type RPE appears normal, with long apical microvilli, interdigitating with the photoreceptor outer segments, and visible basal infoldings on the basal side of the RPE cells (Fig. 5). In contrast, the RPE cells of the heterozygous Prpf3-T494M mice and Prpf8-H2309P mice exhibit loss of the basal infoldings and accumulation of amorphous deposits between the RPE and Bruch[b]'s membrane (Figs. 5A, 5B). There are also vacuoles in the RPE cells of the mutant mice. These changes are more severe in the homozygous Prpf3-T494M and Prpf8-H2309P mice. Photoreceptor cells in the knockin mice appear to be normal in histologic and ultrastructural analyses. One-year-old Prpf31+/− mice also demonstrate degenerative changes in the RPE, with loss of the basal infoldings and accumulation of amorphous deposits between the RPE and Bruch's membrane (Fig. 5C). Analysis showed that all the mutant mice develop loss of basal infoldings and accumulation of deposits between the RPE and Bruch's membrane (n ≥ 3 per mutant line), compared to none of the control mice (n ≥ 3 per mutant line).
Figure 4.
Retinal Morphology. Thick plastic retinal sections stained with toluidine blue. (A) 24-month-old wild-type, Prpf3-Wt/T494M, and Prpf3-T494M/T494M mice. (B) 23-month-old wild-type, Prpf8-Wt/H2309P, and Prpf8-H2309P/H2309P mice. Normal ONL thickness and photoreceptor outer segment length indicates no major loss of photoreceptors at these time points.
Figure 5.
RPE Ultrastructure. Electron microscopy of retina and RPE from (A) two-year-old Prpf3-T494M and littermate control mice, (B) two-year-old Prpf8-H2309P and littermate control mice, and (C) one-year-old Prpf31 knockout and control mice. In all three mouse models, loss of basal infoldings and accumulation of amorphous material between Bruch's membrane and the RPE is evident (arrows). These changes are shown in more detail in the insets at the bottom of each image, with the deposits outlined. Vacuoles are also present in the RPE of the mutant mice. These changes were not detected in the control mice. All images were taken at the same magnification (×1100). Scale bars: 2 μm. BI, basal infoldings; BrM, Bruch's membrane; OS, outer segments; RPE, retinal pigment epithelium.
Discussion
The results presented above provide several important insights into the pathogenesis of RNA splicing factor RP. First, all three mouse models manifest degenerative changes in the RPE. The finding of a similar phenotype in all three models suggests that the RPE is the primary retinal cell type affected in these forms of RNA splicing factor RP, which in turn leads to photoreceptor degeneration over the long term. The RPE changes are associated with decreased photoreceptor function in the Prpf3-T494M mice, although overt photoreceptor degeneration was not observed in any of the models. The later-onset phenotype in these three mouse models is consistent with the adult-onset phenotype and vision loss observed in many patients with RNA splicing factor forms of RP.7,47,48 These three mouse models provide a platform for future comparative studies to elucidate the mechanism of pathogenesis of the RNA splicing factor forms of RP.
An interesting finding from this work is that all the Prpf mutant mice develop loss of RPE basal infoldings and sub-RPE deposits. The deposits share features with the basal deposits associated with macular degeneration, including membranous debris and vesicular structures.49 Both the heterozygous and homozygous mice accumulate these deposits, in agreement with the dominant inheritance of the human conditions. Ultrastructural analysis of the retina from one patient with RNA splicing factor RP has been reported. In this sample, from a patient with a mutation in PRPF8, the RPE was clearly abnormal with loss of basal infoldings, but since the patient had end-stage RP it is difficult to determine whether the RPE abnormalities were a cause or consequence of photoreceptor loss.50,51 While other forms of RP are not known to develop sub-RPE deposits, the RPE and photoreceptors have an intimate relationship and there are several types of retinal degeneration caused by mutations in genes required for RPE function. Some examples are LCA caused by mutations in LRAT and RPE65 and RP caused by MERTK and RGR.52–55 Further, RPE phagocytic dysfunction in β5 integrin-deficient mice leads to photoreceptor dysfunction, and accumulation of lipofuscin in RPE cells.56
The RPE is also a major part of the blood-retinal barrier, and therefore regulates the extracellular environment of photoreceptors. One recent study reported that mice which lack the RPE monocarboxylic acid transporter 3 have an altered pH in the sub-retinal space, and that this leads to altered photoreceptor function.57 One possible explanation for the decreased a-wave ERG observed in the older Prpf3-T494M mice is that the RPE defects observed lead to a similar alteration in the extracellular milieu of photoreceptor outer segments. In short, the RPE and the retina are integrally related, and problems in one can have consequences for the other. Additional studies of the effects of mutations in the three RNA splicing factors on RPE function and its relationship to photoreceptor degeneration are warranted.
The homozygous Prpf3-T494M and Prpf8-H2309P knockin mouse lines are viable, demonstrating that the Prpf3-T494M and Prpf8-H2309P mutations do not create null alleles, or the animals would have died embryonically, as was recently reported for Prpf3 knockout animals, Prpf8 knockout animals, and Prpf31 knockin and knockout animals (Deramaudt BM, et al. IOVS 2005;46:ARVO E-Abstract 5263).24,33 The use of gene-targeted knockin mice for these studies provided several important advantages over other methods that have been used to study the RNA splicing factor forms of RP to date,44,58 including the ability to study the Prpf3-T494M protein and the Prpf8-H2309P protein in the absence of wild-type Prpf3 or Prpf8 in vivo, which cannot be readily accomplished in cell culture. We have observed that in neither case do the expression levels of the mutant Prpf3 or Prpf8 transcripts change, nor does the size of the transcript, ruling out the possibility of the mutation altering a splice signal within the Prpf3 or Prpf8 transcripts themselves. Furthermore, the levels and location of Prpf3-T494M protein within the retina are normal, and no nuclear aggregates were seen, in contrast to a recent study in which aggregation of mutant PRPF3 was seen after over-expression of the protein.44 These findings are also consistent with the hypothesis that mutations in these genes produce disease via a dominant mechanism (dominant-negative or gain-of-function), rather than haploinsufficiency.
The location of the Prpf3 and Sm proteins in the periphery of the nuclei of photoreceptor cells is distinct from the typical staining pattern for splicing proteins, which localize to speckles and Cajal bodies throughout the nucleus but exclude nucleoli. The location of Prpf3 and Sm proteins in the mouse is still speckled but peripheral, consistent with recent findings that nocturnal mammal photoreceptor nuclei have a unique inverted pattern with a heterochromatin center surrounded by euchromatin, nascent transcripts as well as the splicing machinery.59 However, the mutant Prpf3-T494M location does not differ from that of the wild-type, implying no nuclear import defects or defects incorporating into the snRNPs.
The findings described here provide a new model for the pathogenesis of RNA splicing RP. Although the characteristic loss of photoreceptors is the defining feature of RP, our mouse models suggest that photoreceptor dysfunction in RNA splicing factor RP may arise secondary to an unhealthy RPE. Given these findings, we hypothesize that production of an aberrantly spliced transcript or group of transcripts in RPE and/or photoreceptor cells is responsible for the retinal degeneration phenotype observed in patients with RNA splicing factor RP. We believe analyses of the transcriptomes of the RPE and retinas of the Prpf3-T494M, Prpf8-H2309P, and Prpf31-knockout mice will help identify the pathogenic splice alterations responsible for retinal disease. This is a worthwhile endeavor, because identification of the altered transcripts that cause retinal degeneration will open the path to the development of therapies for these blinding disorders. Indeed, there are now several promising examples of the use of antisense oligonucleotides to correct splicing errors caused by mutations in vivo, including a recent report of efficacy in a small clinical trial.60,61
Acknowledgments
The authors thank Qin Liu (F.M. Kirby Center) and Christina Zeitz (Institut de la Vision) for research advice; and Marian Humphries and Peter Humphries (Trinity College, Dublin) for their help with creating Prpf31+/− line and animal husbandry.
Footnotes
Supported by The Foundation Fighting Blindness, Research to Prevent Blindness, the Zeigler Foundation, the F. M. Kirby Foundation, the Mackall Foundation Trust, and the National Eye Institute (Grant T32-EY-007035; JJG), Institut National de la Santé et de la Recherche Médicale (INSERM), Université Pierre et Marie Curie-Paris 6, Centre National de la Recherche Scientifique (CNRS), Départment de Paris, Marie Curie Actions (European Reintegration Grant (PERG04-GA-2008-231125); KB), Fondation Voir et Entendre and Fondation Bettencourt Schueller (Young Investigator Grants; EFN), and Agence Nationale de la Recherche (Chaire d[b]'Excellence; SSB).
Disclosure: J.J. Graziotto, None; M.H. Farkas, None; K. Bujakowska, None; B.M. Deramaudt, None; Q. Zhang, None; E.F. Nandrot, None; C.F. Inglehearn, None; S.S. Bhattacharya, None; E.A. Pierce, None
References
- 1. Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14 [DOI] [PubMed] [Google Scholar]
- 2. Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718 [DOI] [PubMed] [Google Scholar]
- 3. Chakarova CF, Hims MM, Bolz H, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:87–92 [DOI] [PubMed] [Google Scholar]
- 4. Vithana EN, Abu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001;8:375–381 [DOI] [PubMed] [Google Scholar]
- 5. McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet. 2001;10:1555–1562 [DOI] [PubMed] [Google Scholar]
- 6. Maita H, Kitaura H, Keen TJ, Inglehearn CF, Ariga H, Iguchi-Ariga SM. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp Cell Res. 2004;300:283–296 [DOI] [PubMed] [Google Scholar]
- 7. Zhao C, Bellur DL, Lu S, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauber J, Plessel G, Prehn S, et al. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941 [PMC free article] [PubMed] [Google Scholar]
- 9. Anthony JG, Weidenhammer EM, Woolford JL., Jr The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA. 1997;3:1143–1152 [PMC free article] [PubMed] [Google Scholar]
- 10. Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp Cell Res. 2005;302:61–68 [DOI] [PubMed] [Google Scholar]
- 14. van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–365 [DOI] [PubMed] [Google Scholar]
- 16. Xu L, Hu L, Ma K, Li J, Jonas JB. Prevalence of retinitis pigmentosa in urban and rural adult Chinese: The Beijing Eye Study. Eur J Ophthalmol. 2006;16:865–866 [DOI] [PubMed] [Google Scholar]
- 17. Grondahl J. Estimation of prognosis and prevalence of retinitis pigmentosa and Usher syndrome in Norway. Clin Genet. 1987;31:255–264 [DOI] [PubMed] [Google Scholar]
- 18. Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002:1–34 [DOI] [PubMed] [Google Scholar]
- 19. Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–1676 [PubMed] [Google Scholar]
- 20. Pierce EA. Pathways to photoreceptor cell death in inherited retinal degenerations. Bioessays. 2001;23:605–618 [DOI] [PubMed] [Google Scholar]
- 21. Sullivan LS, Bowne SJ, Birch DG, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809 [DOI] [PubMed] [Google Scholar]
- 23. Mordes D, Luo X, Kar A, et al. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis. 2006;12:1259–1271 [PMC free article] [PubMed] [Google Scholar]
- 24. Graziotto JJ, Inglehearn CF, Pack MA, Pierce EA. Decreased levels of the RNA splicing factor Prpf3 in mice and zebrafish do not cause photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2008;49:3830–3838 [DOI] [PubMed] [Google Scholar]
- 25. Wang L, Ribaudo M, Zhao K, et al. Novel deletion in the pre-mRNA splicing gene PRPF31 causes autosomal dominant retinitis pigmentosa in a large Chinese family. Am J Med Genet A. 2003;121A:235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia K, Zheng D, Pan Q, et al. A novel PRPF31 splice-site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol Vis. 2004;10:361–365 [PubMed] [Google Scholar]
- 27. Sato H, Wada Y, Itabashi T, Nakamura M, Kawamura M, Tamai M. Mutations in the pre-mRNA splicing gene, PRPF31, in Japanese families with autosomal dominant retinitis pigmentosa. Am J Ophthalmol. 2005;140:537–540 [DOI] [PubMed] [Google Scholar]
- 28. Abu-Safieh L, Vithana EN, Mantel I, et al. A large deletion in the adRP gene PRPF31: evidence that haploinsufficiency is the cause of disease. Mol Vis. 2006;12:384–388 [PubMed] [Google Scholar]
- 29. Rivolta C, McGee TL, Rio FT, Jensen RV, Berson EL, Dryja TP. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 2006;27:644–653 [DOI] [PubMed] [Google Scholar]
- 30. Sullivan LS, Bowne SJ, Seaman CR, et al. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:4579–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waseem NH, Vaclavik V, Webster A, Jenkins SA, Bird AC, Bhattacharya SS. Mutations in the gene coding for the pre-mRNA splicing factor, PRPF31, in patients with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2007;48:1330–1334 [DOI] [PubMed] [Google Scholar]
- 32. Rio FT, Wade NM, Ransijn A, Berson EL, Beckmann JS, Rivolta C. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest. 2008;118:1519–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bujakowska KM, Maubaret C, Chakarova CF, et al. Study of gene-targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP). Invest Ophthalmol Vis Sci. 2009;50:5927–5933 [DOI] [PubMed] [Google Scholar]
- 34. Rodriguez CI, Buchholz F, Galloway J, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140 [DOI] [PubMed] [Google Scholar]
- 35. Zhang P, Li MZ, Elledge SJ. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat Genet. 2002;30:31–39 [DOI] [PubMed] [Google Scholar]
- 36. Lee EC, Yu D, Martinez de Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65 [DOI] [PubMed] [Google Scholar]
- 37. Matise MP, Auerbach W, Joyner AL. Production of targeted embryonic stem cell lines. In: Joyner AL. ed. Gene Targeting: A Practical Approach, 2nd ed. Oxford, UK: Oxford University Press; 2000:101–132 [Google Scholar]
- 38. Nagy A, Gertsenstein M, Vintersten K, Behringer RR. Manipulating the Mouse Embryo, 3rd ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 2003 [Google Scholar]
- 39. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989 [Google Scholar]
- 40. Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nuc Acid Res. 1991;19:3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gonzalez-Santos JM, Wang A, Jones J, Ushida C, Liu J, Hu J. Central region of the human splicing factor Hprp3p interacts with Hprp4p. J Biol Chem. 2002;277:23764–23772 [DOI] [PubMed] [Google Scholar]
- 42. Lyubarsky AL, Falsini B, Pennesi ME, Valentini P, Pugh EN., Jr UV- and midwave-sensitive cone-driven retinal responses of the mouse: a possible phenotype for coexpression of cone photopigments. J Neurosci. 1999;19:442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Q, Lyubarsky A, Skalet JH, Pugh EN, Jr, Pierce EA. RP1 is required for the correct stacking of outer segment discs. Invest Ophthalmol Vis Sci. 2003;44:4171–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Comitato A, Spampanato C, Chakarova C, Sanges D, Bhattacharya SS, Marigo V. Mutations in splicing factor PRPF3, causing retinal degeneration, form detrimental aggregates in photoreceptor cells. Hum Mol Genet. 2007;16:1699–1707 [DOI] [PubMed] [Google Scholar]
- 45. Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981;78:2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612 [DOI] [PubMed] [Google Scholar]
- 47. Towns KV, Kipioti A, Long V, et al. Prognosis for splicing factor PRPF8 retinitis pigmentosa, novel mutations and correlation between human and yeast phenotypes. Hum Mutat. 2010 [DOI] [PubMed] [Google Scholar]
- 48. Vaclavik V, Gaillard MC, Tiab L, Schorderet DF, Munier FL. Variable phenotypic expressivity in a Swiss family with autosomal dominant retinitis pigmentosa due to a T494M mutation in the PRPF3 gene. Mol Vis. 2010;16:467–475 [PMC free article] [PubMed] [Google Scholar]
- 49. Curcio CA, Millican CL. Basal linear deposit and large drusen are specific for early age-related maculopathy. Arch Ophthalmol. 1999;117:329–339 [DOI] [PubMed] [Google Scholar]
- 50. Szamier RB, Berson EL. Retinal ultrastructure in advanced retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1977;16:947–962 [PubMed] [Google Scholar]
- 51. To K, Adamian M, Berson EL. Histologic study of retinitis pigmentosa due to a mutation in the RP13 gene (PRPC8): comparison with rhodopsin Pro23His, Cys110Arg, and Glu181Lys. Am J Ophthalmol. 2004;137:946–948 [DOI] [PubMed] [Google Scholar]
- 52. den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419 [DOI] [PubMed] [Google Scholar]
- 53. Morimura H, Saindelle-Ribeaudeau F, Berson EL, Dryja TP. Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa [Letter]. Nature Genet. 1999;23:393–394 [DOI] [PubMed] [Google Scholar]
- 54. Gal A, Li Y, Thompson DA, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–271 [DOI] [PubMed] [Google Scholar]
- 55. Nandrot E, Dufour EM, Provost AC, et al. Homozygous deletion in the coding sequence of the c-mer gene in RCS rats unravels general mechanisms of physiological cell adhesion and apoptosis. Neurobiol Dis. 2000;7:586–599 [DOI] [PubMed] [Google Scholar]
- 56. Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004;200:1539–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daniele LL, Sauer B, Gallagher SM, Pugh EN, Jr, Philp NJ. Altered visual function in monocarboxylate transporter 3 (Slc16a8) knockout mice. Am J Physiol Cell Physiol. 2008;295:C451–C457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yuan L, Kawada M, Havlioglu N, Tang H, Wu JY. Mutations in PRPF31 inhibit pre-mRNA splicing of rhodopsin gene and cause apoptosis of retinal cells. J Neurosci. 2005;25:748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Solovei I, Kreysing M, Lanctot C, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368 [DOI] [PubMed] [Google Scholar]
- 60. Aartsma-Rus A, Janson AA, van Ommen GJ, van Deutekom JC. Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med Genet. 2007;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686 [DOI] [PubMed] [Google Scholar]