In this study, high-throughput, next-generation sequencing was used to identify disease-causing mutations in a large set of unrelated families with autosomal dominant retinitis pigmentosa (adRP), a highly heterogeneous inherited disease. This is one of the first reports of the application of the technology to a large set of adRP families. Next-generation sequencing of a large set of candidate genes identified mutations in 24% of the families tested, bringing the mutation identification rate in this adRP cohort to 65%.
Abstract
Purpose.
To determine whether massively parallel next-generation DNA sequencing offers rapid and efficient detection of disease-causing mutations in patients with monogenic inherited diseases. Retinitis pigmentosa (RP) is a challenging application for this technology because it is a monogenic disease in individuals and families but is highly heterogeneous in patient populations. RP has multiple patterns of inheritance, with mutations in many genes for each inheritance pattern and numerous, distinct, disease-causing mutations at each locus; further, many RP genes have not been identified yet.
Methods.
Next-generation sequencing was used to identify mutations in pairs of affected individuals from 21 families with autosomal dominant RP, selected from a cohort of families without mutations in “common” RP genes. One thousand amplicons targeting 249,267 unique bases of 46 candidate genes were sequenced with the 454GS FLX Titanium (Roche Diagnostics, Indianapolis, IN) and GAIIx (Illumina/Solexa, San Diego, CA) platforms.
Results.
An average sequence depth of 70× and 125× was obtained for the 454GS FLX and GAIIx platforms, respectively. More than 9000 sequence variants were identified and analyzed, to assess the likelihood of pathogenicity. One hundred twelve of these were selected as likely candidates and tested for segregation with traditional di-deoxy capillary electrophoresis sequencing of additional family members and control subjects. Five disease-causing mutations (24%) were identified in the 21 families.
Conclusion.
This project demonstrates that next-generation sequencing is an effective approach for detecting novel, rare mutations causing heterogeneous monogenic disorders such as RP. With the addition of this technology, disease-causing mutations can now be identified in 65% of autosomal dominant RP cases.
Massively parallel next-generation sequencing has revolutionized the speed and cost associated with generating large quantities of sequence data, making it a promising technology for detecting disease-causing mutations associated with monogenic diseases.1–5 The low-cost, high-throughput attributes of next-generation sequencing make it particularly attractive for use in highly heterogeneous monogenic diseases such as retinitis pigmentosa (RP) where the number of potential disease-causing genes and mutations is high, and many are still unknown.
RP is a multifaceted Mendelian form of inherited photoreceptor degeneration that is monogenic in most individuals and families, but extremely heterogeneous in patient populations. RP affects approximately 1 in 4000 individuals in the United States, Europe, and Japan. This translates into approximately 1.5 million people affected with RP worldwide.6–10 Data from the Beijing Eye Study suggest that the prevalence of RP in China may be even higher (approximately 1 in 1000).11 Studies in Japan, Denmark, and Kuwait show that RP is among the leading cause of blindness or visual impairment worldwide, accounting for 25% to 29% of cases in the working-age group (21–60 years).12–15
RP can be inherited in an autosomal dominant (adRP), autosomal recessive (arRP), or X-linked (XlRP) manner with rare mitochondrial and digenic forms also reported.16,17 Several syndromic disorders, such as Bardet-Biedl and Usher syndrome, have RP associated with them.8,17 To further complicate matters, there are several other forms of inherited retinal degeneration—Leber's congenital amaurosis and cone–rod dystrophy to name a few—which have overlapping phenotypes, and to some extent, overlapping genes and mutations.16,18,19
Significant progress has been made in determining the molecular causes of RP, but much remains to be done. To date, research has identified 20 adRP, 35 arRP, and 6 XlRP loci, several overlapping with each other (RetNet, http://www.sph.uth.tmc.edu/RetNet; provided in the public domain by the University of Texas Houston Health Science Center, Houston, TX). Most of the genes have been identified for these loci, (17, 18, and 2 respectively), but mutations still cannot be found in a large fraction of individuals with RP, indicating that many of the causative genes and mutations have not been identified yet. For example, after testing for mutations in the known adRP genes, we can identify mutations in only approximately 60% of a clearly defined cohort of patients with adRP.17,20–24 In populations other than those of Western European origin, the mutation detection rate is even lower.25–29
To determine whether massively parallel next-generation sequencing is an option for adRP mutation identification and discovery, we sequenced pairs of affected individuals from 21 autosomal dominant families without mutations in known adRP genes, by using a combination of two platforms: 454GS FLX Titanium (Roche Diagnostics, Indianapolis, IN); and GAIIx (Illumina/Solexa, San Diego, CA). One thousand amplicons corresponding to the coding sequences and intron–exon junctions of 46 candidate genes were sequenced as part of this pilot project. Variants were assessed for potential pathogenicity using bioinformatic annotation, dbSNP, and manual review. One hundred twelve of the most likely variants were validated and subjected to additional segregation and population analysis using conventional di-deoxy sequencing. Five definitive mutations were identified in the 21 families proving that massively parallel next-generation sequencing is an effective approach for determining the genes and mutations associated with RP.
Materials and Methods
Samples
A subset of 21 families was selected from a cohort of 230 adRP families that has been described previously (Bowne SJ, et al. IOVS 2007;48:ARVO E-Abstract 2334).20,21,23,24 Families without mutations were selected from the adRP cohort based on pedigree analysis and the availability of DNA (Table 1). Pairs of affected individuals with the lowest kinship coefficient within the family were selected from each pedigree. Six additional positive control samples were selected from our diagnostic laboratory for analysis (Table 2).
Table 1.
Twenty-One Families Selected from the AdRP Cohort
| Family ID | Ethnicity | Generations | Affected Individuals | Male-to-Male Transmission | Lod ad:ar | Lod ad:XI | Kinship Coefficient |
|---|---|---|---|---|---|---|---|
| VCH007 | Black | 5 | 9 | Yes | 14 | 1 | 1/32 |
| VCH008 | Cauc | 4 | 10 | No | 14 | −1 | 1/8 |
| VCH009 | Cauc | 4 | 10 | Yes | 34 | 14 | 1/8 |
| VCH010 | Cauc | 4 | 8 | Yes | 8 | 4 | 1/8 |
| VCH011 | Cauc | 4 | 6 | No | 15 | 0 | 1/16 |
| VCH012 | Cauc | 5 | 15 | Yes | 21 | 8 | 1/8 |
| VCH013 | Cauc | 4 | 10 | Yes | 14 | 8 | 1/8 |
| VCH014 | Hisp | 5 | 4 | No | 5 | −1 | 1/4 |
| VCH015 | Cauc | 3 | 4 | Yes | 7 | 4 | 1/8 |
| VCH016 | Black | 6 | 13 | Yes | 35 | 10 | 1/16 |
| VCH017 | Cauc | 3 | 7 | No | 11 | 0 | 1/4 |
| VCH018 | Cauc | 6 | 11 | No | 12 | −2 | 1/4 |
| VCH019 | Cauc | 6 | 17 | Yes | 44 | 18 | 1/512 |
| VCH020 | Cauc | 4 | 7 | No | 14 | 6 | 1/16 |
| VCH021 | Cauc | 5 | 10 | Yes | 17 | 6 | 1/16 |
| VCH022 | Hisp | 4 | 8 | Yes | 13 | 7 | 1/8 |
| VCH023 | Asian | 4 | 21 | Yes | 26 | 11 | 1/32 |
| VCH024 | Cauc | 4 | 9 | Yes | 22 | 13 | 1/8 |
| VCH025 | Cauc | 5 | 13 | Yes | 24 | 8 | 1/4 |
| VCH026 | Cauc | 4 | 18 | Yes | 19 | 9 | 1/32 |
| VCH004 | Cauc | 5 | 12 | No | 16 | 5 | 1/16 |
Table 2.
Positive Control Samples
| Sample ID | Gene | Mutation | Protein | Locus | Genomic Variant |
|---|---|---|---|---|---|
| VCH001–01 | PRPF31 | c.del636 | Frame shift | 19q13 | del59319048 (G) |
| VCH002–01 | RHO | c.68 C>A | p.Pro23His | 3q22 | 130730334 C>A |
| VCH005–01 | RHO | [c.404 G>T; c.405 G>T] | p.Arg135Leu | 3q22 | [130732451 G>T; 130732452 G>T] |
| VCH027–01 | PRPH2 | IVS2+3 A>T | Unknown | 6p21 | 42780078 A>T |
| VCH028–01 | RP1 | del2280–2284 | Unknown | 8q12 | del55701275–55701279 |
| VCH029–01 | PRPF31 | deletion of entire gene + flanking | None | 19q13 | Minimum is del59283753–59328550 |
The research adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from each of the individuals tested. This study was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston and by the respective human subjects' review boards at each of the participating institutions.
Population Samples
Lymphoblast DNAs from four human population control collections (CEPH, Han people of Los Angeles, Mexican-American community of Los Angeles, and African American) were obtained from the Coriell Institute for Medical Research (Camden, NJ) or from the Centre d'Etude du Polymophisme (Paris, France).
Target Selection and PCR Assay Design
Genes targeted for sequencing were (1) known causes of autosomal dominant RP, (2) known causes of other forms of retinal degeneration with overlapping phenotypes, or (3) potential disease-causing candidate genes selected from sensory cilium proteome studies, EyeSAGE data, and other retinal expression and protein interaction studies (Table 3) (Liu O, et al. IOVS 2006;47:ARVO E-Abstract 3725).30–33
Table 3.
Targeted Candidate Genes
| Gene | Locus | Reason |
|---|---|---|
| AIPL1 | 17p13 | Other retinal degeneration |
| BEST1 | 11q12 | Other retinal degeneration |
| C1orf142 | 1q42 | Candidate gene |
| C1QTNF5 | 11q23 | Other retinal degeneration |
| CA4 | 17q23 | adRP |
| CKB | 1q32 | Candidate gene |
| CLN8 | 8p23 | Candidate gene |
| CORO1C | 12q24 | Candidate gene |
| CRB1 | 1q31 | Other retinal degeneration |
| CRX | 19q13 | adRP |
| FLT3 | 13q12 | Candidate gene |
| FSCN2 | 17q25 | adRP |
| GNAT1 | 3p21 | Other retinal degeneration |
| GUCA1A | 6p21 | Other retinal degeneration |
| GUCA1B | 6p21 | adRP |
| GUCY2D | 17p13 | Other retinal degeneration |
| IMPDH1 | 7q32 | adRP |
| KIAA0090 | 1p36 | Candidate gene |
| KLHL7 | 7p15 | adRP |
| LCA5 | 6q14 | Other retinal degeneration |
| MGC42105 | 5p12 | Candidate gene |
| NR2E3 | 15q23 | adRP |
| NRL | 14q11 | adRP |
| PAP1/RP9 | 7p14 | adRP |
| PDE6B | 4p16 | Other retinal degeneration |
| PITPNM3 | 17p13 | Other retinal degeneration |
| PROM1 | 4p15 | adRP |
| PRPF3 | 1q21 | adRP |
| PRPF31 | 19q13 | adRP |
| PRPF8 | 17p13 | adRP |
| PRPH2 | 6p21 | adRP |
| RDH12 | 14q24 | adRP |
| RTBDN | 19p13 | Candidate gene |
| RHO | 3q22 | adRP |
| RIMS1 | 6q13 | Other retinal degeneration |
| ROM1 | 11q12 | adRP |
| RP1 | 8q12 | adRP |
| RP1L1 | 8p23 | Candidate gene |
| RP2 | Xp11 | Other retinal degeneration |
| RPGR | Xp11 | Other retinal degeneration |
| RPGRIP1 | 14q11 | Other retinal degeneration |
| SEMA4A | 1q22 | adRP |
| TOPORS | 9p21 | adRP |
| TTC26 | 7q34 | Candidate gene |
| TUBB2C | 9q34 | Candidate gene |
| UNC119 | 17q11 | Candidate gene |
In-house amplimer design algorithms were used in conjunction with Primer3 primer-selection software (http://sourceforge.net/projects/primer3/develop)34 to design 1000 PCR amplicons (average size, 283 bp) targeting all coding and noncoding exonic sequences of the 46 genes. PCR primers were ordered from IDT (Coralville, IA) in four sets, each with an M13, MID1, MID2, or MID3 primer tail.
Polymerase Chain Reaction
Genomic DNA was subjected to whole-genome amplification (WGA; REPLI-g genome amplification service; Qiagen, Valencia, CA) before amplification of the targets. Only samples with an assessment rating indicating a >99.0% accuracy rate were used for the study. Each of the 1000 PCR amplicons was amplified individually with 10 ng of WGA DNA, PCR master mix (Amplitaq Gold Master Mix; Applied Biosystems, Inc., [ABI], Foster City, CA), 8% glycerol, and 2.4 picomoles of primer. Standard PCR cycling conditions were performed for 40 cycles with an annealing temperature of 60°C.
Amplicon Efficiency Pool
Before variant identification sequencing, WGA DNA from all 48 individuals was pooled and amplified to determine individual amplicon efficiencies. Each of the 1000 primer sets containing M13 tails was amplified independently. Equal volumes of PCR product from these amplimers (3 μL) were pooled and sequenced on one full 454/XLR plate (∼1,000× coverage per amplicon, or ∼20× per sample). Amplicons were classified into groups of high, medium, and low coverage based on the average 454GS FLX read depth as described in Table 4. Amplicon efficiency classifications and resulting input ratios were used for all subsequent sample sequencing library preparations.
Table 4.
Amplicon Efficiency Grouping and Library Input Ratios
| Group | Amplicons (n) | Total Read Count | Average Read Count | In Relation to Low | Volume to Add |
|---|---|---|---|---|---|
| Low | 384 | 79686.6 | 207.5 | 1.00 | 100 |
| Medium | 384 | 225675.8 | 587.7 | 0.35 | 35 |
| High | 232 | 227245.1 | 988.0 | 0.21 | 21 |
454GS FLX Library Construction and Sequencing
PCR products corresponding to four individuals (two individuals per family), were each amplified with a different primer tail. PCR products were pooled using the ratios established during the PCR efficiency run (Table 4). PCR product-pool libraries were created according to the protocol outlined in the manufacturer's instructions, with omission of the DNA fragmentation and size selection steps. Briefly, 2 μg of each pool was size purified (Agencourt AMPure; Beckman Coulter Genomics, Danvers, MA), according to the manufacturer's protocol. Fragments were end polished with T4 PNK and T4 DNA polymerase (both from Roche Diagnostics) and the adapters ligated at 25°C for 15 minutes. Fragments were immobilized, filled in, and denatured to construct the single-stranded DNA library. A library of positive controls was created using the same methodology but the pooled PCR product corresponding to the six mutation-positive DNAs was used without regard for the primer tail used in amplification.
Emulsion PCRs were performed according to the manufacturer's instructions (Roche Diagnostics). Briefly, library DNA fragments were captured on beads and then resuspended in amplification mix and prepared in oil (GS FLX Titanium emPCR kit; Roches Diagnostics). The emulsified beads were amplified and then recovered and washed (GS FLX Titanium emPCR Breaking Kit; Roche Diagnostics). Bead enrichment was performed via a biotinylated enrichment primer and streptavidin-coated magnetic beads. Sequencing primers were annealed to the bead-bound, single-stranded template DNA fragments and sequenced according to the manufacturer's protocol (Roche Diagnostics). The positive control library was run on one-half of a 454GS FLX XLR plate, while the remaining libraries were run on one fourth of a 454GS FLX XLR plate.
GAIIx Paired-End Library Construction and Sequencing
One paired-end GAIIx library containing PCR product from each of the 48 DNA samples was constructed. Excess primers and primer dimers were removed using PCR purification columns (QIAquick; Qiagen), according to the manufacturer's protocol. Ten micrograms of the PCR product was concatenated (Quick Ligase Kit; New England Biolabs, Ipswich, MA) with 7.2% PEG-8000. Concatenated DNA was then nebulized according to the manufacturer's protocol (Illumina/Solexa).
Sheared DNA was end repaired (DNA Terminator End Repair kit; Lucigen, Middleton, WI), purified (QIAquick columns; Qiagen), and verified on 1.2% gels (FlashGels; Lonza, Basel, Switzerland). An adenosine was added to the 3′ end of concatenated products (Klenow Fragment (3′→5′ exo−; New England Biolabs) at 37°C for 30 minutes. Adenosine-tailed DNA was column purified (MinElute; Qiagen) and ligated to an adapter-oligo mix at room temperature for 15 minutes. Ligated reactions were purified with the purification columns followed by gel purification using 1.2% agarose and extraction (MinElute Gel Extraction Kit; Qiagen). PCR enrichment was performed using DNA polymerase master mix (Phusion; New England Biolabs). Reactions were denatured at 98°C for 30 seconds followed by five cycles of 98°C for 10 seconds, 65°C for 30 seconds, and 72°C for 30 seconds with a final extension at 72°C for 5-minute PCR reactions were purified on the minicolumns, and the 300- to 400-bp library fragment was isolated by gel purification.
Cluster Generation Kit ver. 2 (Illumina/Solexa) and the manufacturer's protocol were used to generate paired ends, which were then sequenced on the GAIIx (Illumina/Solexa) platform (SBS Sequencing Kit, ver. 3; Illumina/Solexa) according to the manufacturer's protocol.
454GS FLX Alignments and Variant Detection
Sequencing data files (SFF format) were converted to FASTA format using sffinfo. Individual samples were separated using cross_match to match M13, MID1, MID2, and MID3 primer sequences with the first 20 bases of every read. Manifests of read names for each primer tail were composed, and sequences were extracted from the master SFF file for each individual.
Read sequences were extracted from individual SFF files using sffinfo and then aligned to the Hs36 reference sequence using BLAT (ver. 32 × 1) (http://genome.ucsc.edu/cgi-bin/hgBlat/ provided in the public domain by the University of California Santa Cruz).35 Alignments with <90% identity, a score of <25, or mapping to multiple locations in the genome (with the same score), were discarded. SNPs and indels were detected in the BLAT alignments using VarScan.36 A minimum of 10× coverage and at least 25% of reads supporting the variant allele was required for variant calling. Substitutions at sites with base quality <15 were discarded. Artifacts from regions homologous to MID primer tails were avoided by discarding variants called within 10 bp of the beginning or end of the read.
GAIIx Alignments and Variant Detection
GAIIx reads were aligned to the Hs36 reference sequence using Bowtie (ver. 0.9.8, http://bowtie-bio.sourceforge.net).37 Reads were required to have a single best alignment (−m 1) with no more than two high-quality mismatches to the reference sequence. Bowtie alignments were parsed to identify substitutions at positions with base quality of 15 or higher. The variant allele with the greatest read support was called for all positions at which variants were detected.
Indel Validation with Illumina/Solexa Data
To detect small indels, GAIIx reads were aligned to the Hs36 reference sequence (Novoalign, ver. 2.03.12; Novoalign, Selangor, Malaysia). VarScan36 was used to call indels based on unique read alignments. Indels with ambiguous positions or flanked by homopolymers were removed. Validation of 454GS FLX indels required that GAIIx indel calls be the same type, size (within 1 bp), and position (±5 bp).
Di-deoxy Sequencing of Potential Disease-Causing Variants
The 1000 amplicon PCR primers with M13 tails were used for all additional analyses of potential disease-causing variants. Genomic DNA was amplified using Taq master mix (AmpliTaq Gold 360 Master Mix; ABI) and standard amplification conditions. PCR product was treated with exonuclease I and shrimp alkaline phosphatase (ExoSapIt; USB, Cleveland, OH) before sequencing. Clean PCR product was sequenced as described previously with dye termination chemistry (BigDye Terminator ver. 1.1; ABI) and M13 primers.38 Sequence reactions were treated with a reaction cleanup kit (BigDye XTerminator; ABI), according to the manufacturer's protocol, and run on one of two automated capillary sequencers (3100 Avant or 3730XL; ABI). Sequence analysis was then performed with one of three commercial software programs (Sequencing Analysis, Variant Reporter, or SeqScape; ABI).
Results
Sample
The sample selected for this project included a subset of family members from our previously described AdRP Cohort (Bowne SJ, et al. IOVS 2007;48:ARVO E-Abstract 2334;).21,23,24,38,39 Families in the adRP cohort, based on pedigree analysis, have a high likelihood of having the autosomal dominant form of RP. Analysis of pedigrees for the likelihood of dominance versus X-linked or recessive inheritance did show some families with ad:Xl likelihood odds ratios of less than 0, indicating that the disease in a few families could be caused by mutations in an X-linked gene with clinical expression in carrier females (Table 1).
A proband from each family was tested previously for mutations in the complete coding regions of CA4, CRX, FSCN2, IMPDH1, NRL, PRPF31, RDS, RHO, ROM1, RP9, and TOPORS, and in mutation hot spots of RP1, PRPF3, PRPF8, NR2E3, and SNRNP200. Likely disease-causing mutations were identified in 141 of the 230 families. Only families without previously identified mutations were considered for this study.
Twenty-one families were selected for this project based on pedigree analysis and availability of family member DNAs. Two affected individuals from each family were selected to have the lowest kinship coefficient possible—that is, the most distant, available affected relatives with a common ancestor carrying a putative adRP mutation. This increased the probability that any shared variant identified in this project would be associated with disease, not just identical by chance. The demographic and pedigree characteristics of the families and the kinship coefficient of the two selected family members are shown in Table 1.
Six individuals with known mutations were also selected to use as positive controls in this study (Table 2). These individuals had a variety of mutation types in several different genes, thereby testing the identification rate of next-generation sequencing for different classes of DNA variants.
DNA Targets
Each of the 46 genes selected for this project was (1) a known cause of adRP, (2) a known cause of other forms of retinal degeneration with phenotypes overlapping adRP, or (3) a potential disease-causing candidate gene selected from sensory cilium proteome studies, EyeSAGE data, and other studies of retinal expression and protein interaction (Liu Q, et al. IOVS 2006;47:ARVO E-Abstract 3725).30–33 The genes selected, their chromosomal location, and the associated diseases, if any, are listed in Table 3.
PCR primers corresponding to 1000 amplicons were designed to amplify all the coding and noncoding exonic sequences for each of the 46 genes selected. PCR primers were manufactured in sets of four with each set containing the same genome-specific sequence and one of four different tail sequences (M13, MD1, MD2, and MD3). These four tail sequences allowed PCR product from four individuals to be combined after amplification, while retaining the ability to distinguish the four individuals on sequence assembly.
Target Amplification and Library Construction
Genomic DNA from each of the 48 individuals tested in this study was amplified by WGA before target amplification. A 454GS FLX amplicon efficiency test was performed on a pool of the DNAs to optimize product pooling such that each amplimer represented in the sequencing library was relatively equivalent (Table 4).
Eleven patient pool libraries were constructed for analysis on the 454GS FLX sequencing system. Each of these pools corresponded to two sets of affected family pairs that had been amplified individually with the four different, tailed target primers. The six positive control samples were pooled to form one library that was not sorted by MD tail.
One concatenated, paired-end library was constructed for analysis (PE sequencer; Illumina/Solexa). Concatenation and shearing of the PCR products before library construction enabled access to those regions that, due to the short read length of GAIIx sequencing, would otherwise not be sequenced, and also gives more random distribution of all positions in a particular sequence. This feature introduces less quality bias based on position of the variation within a given sequence. The paired-end library pooled all 42 unknown affected individuals and the six positive controls. Since this library was not sortable, it was used only for variant confirmation, not individual variant identification.
454GS FLX Analyses of Positive Controls
Sequence reads corresponding to the pooled positive control library were aligned to Hs36 and analyzed for the presence of each known mutation. Five of the six mutations were detected at a read frequency of 6% to 13%, which is in accordance with the predicted detection rate of 8% (Table 5). The sixth variant, a large 47-kb deletion of PRPF31 and several flanking genes present in VCH029, was not detected using this technology, as expected.
Table 5.
Detection of Positive Controls in Pooled 454 FLX Run
| Family | Locus | Position | Ref | Var | Read 1 | Read 2 | VarFreq (%) | P |
|---|---|---|---|---|---|---|---|---|
| VCH001 | 19q13 | 59319047 | T | G | 189 | 28 | 12.90 | 2.94E-09 |
| VCH002 | 3q22 | 130730334 | C | A | 225 | 31 | 12.11 | 3.54E-10 |
| VCH005 | 3q22 | 130732451 | G | T | 279 | 31 | 10.00 | 4.23E-10 |
| VCH027 | 6p21 | 42780078 | T | A | 375 | 24 | 6.02 | 8.35E-08 |
| VCH028 | 8q12 | 55701279 | T | -TAAAT | 456 | 36 | 7.32 | 1.50E-11 |
As expected, the large deletion present in family VCH029 was not detectable with this technology. 454GS FLX (Roche, Indianapolis, IN).
Sequence Alignment and SNP Variant Detection
454GS FLX Reads.
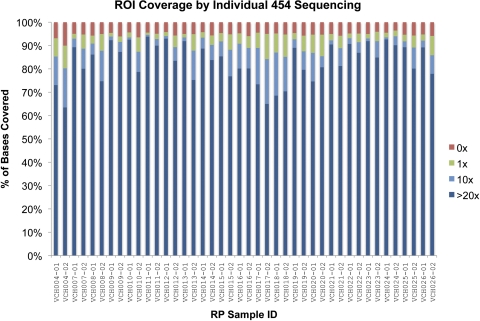
Approximately 1.5 million 454GS FLX sequence reads were separated by individual, by using primer tails with typically 90% to 95% of reads identified unambiguously. Alignment of the sequence reads to Hs36 resulted in an average sequence depth of 70×, with 93% of reads mapping to the 46 target genes and identification of more than 9000 variants (Fig. 1, Table 6).
Figure 1.
Coverage from 454GS FLX sequencing of individual samples (Roche Diagnostics, Indianapolis, IN). The fraction of targeted positions (∼250 kbp total) covered at 0× (red), 1× (green), 10× (light blue), and 20× (dark blue) are shown.
Table 6.
Sequence Data Generated on Next-Generation Platforms
| Regions for targeted sequencing | |
| Candidate genes | 46 |
| PCR amplicons | 1,000 |
| Total positions targeted (unique) | 249,267 |
| Individual 454 data* | |
| Number of samples | 48 |
| Total reads (∼230-bp XLR) | 1.46 million |
| Avg. sequence depth per sample | 70x |
| Pooled GAIIx data† | |
| Number of samples pooled | 48 |
| Total reads (36-bp frag) | 66.68 million |
| Avg. sequence depth per sample | 125x |
Roche Diagnostics, Indianapolis, IN.
Illumina/Solexa, San Diego, CA.
The list of unfiltered variants was compared with the nonpathogenic variants identified in positive controls on the assumption that variants other than the one pathogenic mutation would not be disease-causing in other individuals. All variants found in the positive controls were removed, as were any nonpathogenic variants found in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/ provided in the public domain by the National Institutes of Health, Bethesda, MD). Unfortunately, automated removal of variants in dbSNP was not possible, as a small portion of the variants in dbSNP are truly pathogenic.
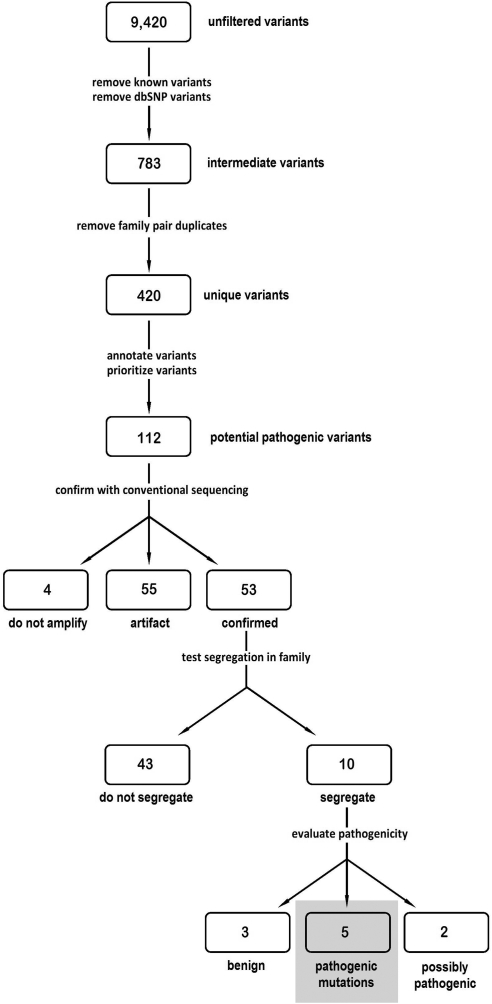
The remaining 783 intermediate variants were analyzed to remove duplicate variants found in the same family leaving 420 unique variants. These variants were then annotated and prioritized based on their location in a gene (exon, intron, and splice-site) and on the predicted transcript or protein alteration and by manual assessment of the potential of the affected gene to cause RP. The resulting 112 variants were classified as potentially pathogenic, thereby warranting additional analysis (Fig. 2).
Figure 2.
Flow chart of variant analysis.
GAIIx Reads.
The 66.7 million 36-bp reads from the pooled GAIIx library (Illumina/Solexa) were aligned to Hs36 with an average sequence depth of 125× (Table 6). Variants were called and compared with the list of the 454GS FLX variants. To confirm a 454GS FLX variant required GAIIx read coverage of at least 100× and at least two variant-supporting reads. These data were confirmatory but not used in the initial identification of the 112 potential pathogenic variants.
Evaluation of Potential Pathogenic Variants
The 112 potential pathogenic variants were subjected to a series of analyses to determine whether they were true variants, if they segregated with disease in the family in which they were identified, and whether they were present in unaffected controls.
Fluorescent di-deoxy capillary sequencing was used to determine whether the variants identified by next-generation sequencing were actually genomic variants. Genomic DNAs from the original affected family pair and from two additional family members (when available) were tested with the corresponding M13 tailed primers for the original 1000 amplimer amplifications. Traditional Sanger sequencing showed that 55 of the 112 potential pathogenic variants were artifacts of 454GS FLX sequencing. An additional four of the potential pathogenic variants did not amplify within the genomic regions specified for the variant and so were also assumed to be artifacts. With this strategy, 55 of the potential pathogenic variants were confirmed to be present in the identified individuals.
Once a variant was determined to be real, segregation analysis was used to assess its likelihood of it being disease-causing. If initial analyses showed correct segregation in the first set of four family members tested, then all available family DNAs were tested. Forty-three of the 53 confirmed variants did not segregate with disease in the family and hence were determined to be benign. Ten of the variants segregated with disease in all available family members (Table 7).
Table 7.
Ten Potential Disease-Causing Variants
| Family | Gene | Chr | Position | Ref | Var | Functional Class | Nucleotide | Protein | Reference sequence | Frequency in Controls (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| VCH010 | KLHL7 | 7 | 23146928 | C | T | Missense | c.458C>T | p.A153V | NM_001031710 | 0.0 |
| VCH017 | RPGR | X | 38030984 | G | T | Nonsense | c.2212G>T | p.G738* | NM_001034853 | 0.0 |
| VCH018 | RPGR | X | 38067103 | G | A | Missense | c.194G>A | p.G65D | NM_001034853 | 0.0 |
| VCH020 | PRPF31 | 19 | 59323259 | G | C | Splice-site | c.946–1 | Unknown | NM_015629 | 0.0 |
| VCH012 | GUCY2D | 17 | 7858743 | C | T | Missense | c.2512C>T | p.R838C | NM_000180 | 0.0 |
| VCH024 | PRPF8 | 17 | 1534891 | G | A | 5'UTR | c.1–51G>A | Unknown | NM_006445 | 2.0 |
| VCH013 | PITPNM3 | 17 | 6308263 | G | A | Missense | c.2108C>T | p.R703W | NM_031220 | 0.7 |
| VCH011 | TTC26 | 7 | 138502254 | G | T | Intronic | c.896+73G>T | Unknown | NM_024926 | 2.0 |
| VCH008 | PROM1 | 4 | 15619667 | C | T | Splice-site | c.1302+3C>T | Unknown | NM_006017 | 0.0 |
| VCH025 | MFRP | 11 | 118721331 | G | A | Splice-site | c.641+9G>A | Unknown | NM_031433 | 0.0 |
Three of the 10 segregating variants, KLHL7 p.A153V, RPGR p.G65D, and PRPF31 c.946–1, were identified and characterized in parallel laboratory testing and determined to be pathogenic.40,41 An additional variant in RPGR, p.G738*, was identified among the 10 segregating variants. Although not previously reported, RPGR p.G738*, like many other reported RPGR mutations, produces a premature termination codon in ORF15 and hence is most likely pathogenic. One additional segregating variant in GUCY2D, p.R838C, has also been reported to cause cone–rod dystrophy.42 No further testing was performed for these five disease-causing mutations (Fig. 3).
Figure 3.
Five families with identified pathogenic mutations. (A) VCH010. The p.A153V mutation in KLHL7 was present in all three affected family members tested. (B) VCH012. All five tested affected members of this family had the R838C mutation in GUCY2D (C) VCH017. Four affected members of this family were either heterozygous or hemizygous for the RPGR G738X mutation which was not present in the one unaffected family member tested. (D) VCH018. THE RPGR G65D mutation was present in seven affected members or female carriers in this family and absent from the one unaffected spouse tested. (E) VCH037. The c.946-1 splice site mutation in PRPF31 segregated with disease in the three family members tested. *Individuals tested in this study.
Ethnically matched control population DNAs were tested for the possible presence of the remaining five variants of unknown pathogenicity. Three of the variants, PRPF8 c.1–51G>A, PITPNM3 p.R703W, and TTC26 c.896+73G>T were found in control DNAs and hence are benign. The two remaining variants, PROM1 c.1302+3C>T and MRFP c.641+9G>A, were not found in the controls. The number of immediately available family member DNA samples was low (three and two, respectively) for the PROM1 and MRFP variants. Subsequent collection and testing of three additional VCH008 family members demonstrated that the PROM1 c.1302+3C>T variant does not segregate with disease. Collection and testing of three additional VCH025 family members also demonstrated that the MRFP variant does not segregate with disease. These data demonstrate that both the PROM1 and MRFP variants are benign.
Indels
Analysis of the individual reads from 454GS FLX sequencing identified 77 small, high-confidence indels ranging in size from 1 to 3 bp. Indels with ambiguous positions or flanked by homopolymers were removed and the remaining compared with GAIIx indel data. A total of 10 indels were identified with the GAIIx data (Table 8).
Table 8.
Indels present in 454 FLX and GAIIx Sequence Reads
| Family | Gene | Chr | Position | Type | Size (bp) |
|---|---|---|---|---|---|
| VCH021 | BEST1 | 11 | 61482241 | Deletion | 1 |
| VCH022 | |||||
| VCH026 | |||||
| VCH026 | ROM1 | 11 | 52138421 | Deletion | 1 |
| VCH015 | GUCY2D | 17 | 7847637 | Insertion | 1 |
| VCH014 | PROM1 | 4 | 15591264 | Insertion | 1 |
| VCH021 | PROM1 | 4 | 15604786 | Deletion | 1 |
| VCH022 | |||||
| VCH025 | |||||
| VCH026 | |||||
| VCH22 | LCA5 | 6 | 80259054 | Deletion | 1 |
| VCH25 | |||||
| VCH26 | |||||
| VCH019 | RIMS | 6 | 73159087 | Deletion | 1 |
| VCH013 | CRX | 19 | 53034641 | Deletion | 1 |
| VCH022 | RP1 | 8 | 55701373 | Deletion | 1 |
| VCH016 | RPGR | X | 38030864–38030866 | Deletion | 3 |
454GS FLX (Roche, Indianapolis, IN); GAIIx (Illumina/Solexa, San Diego, CA).
Indels were evaluated using the same fluorescent capillary sequencing strategy described above for the possibly pathogenic SNP variants. Traditional sequencing failed to confirm the presence of nine of the indels. The 10th indel, a 3-bp deletion in ORF15 of RPGR was not assessed, since RPGR is located on the X-chromosome and the family exhibited male-to-male transmission of RP. Furthermore, 3-bp deletions in ORF15 are common and usually benign.43–46
Discussion
An excess of 9000 variants was identified in the 21 families analyzed in this project. Most of these variants were classified as benign on the basis of their presence in controls with definitive disease-causing mutations distinct from the novel variants. This massive reduction in variants requiring follow-up analyses, over 8000, stresses the importance of running control individuals and multiple family members, if possible, when using next-generation sequencing to detect mutations in families with inherited diseases.
Additional laboratory analyses were performed for 112 possibly pathogenic variants found by 454GS FLX sequencing. The presence of 53 (47%) of these variants was confirmed by traditional sequencing, whereas the remaining 59 (53%) variants were found to be false positives. A large fraction of the false-positive variants occurred in a polynucleotide runs, which is a known limitation of 454 sequencing methodology. This result suggests that additional stringency should be used when identifying variants in polynucleotide runs to reduce the number of false positives.
The pooled GAIIx sequencing data were not used in the initial phases of the project, but were compared to the list of 112 variants to determine whether cross-platform comparisons might also reduce the number of false-positive variants. When compared with the pooled GAIIx data, 85% of the confirmed variants were present, but, 55% of the false positives were also seen in the GAIIx reads. This finding suggests that cross-platform comparisons may be useful for prioritizing variants for subsequent follow-up, but should not be used as an exclusive requirement for variant identification.
Next-generation sequencing of the 1000 amplicons corresponding to 46 candidate genes resulted in identification of five pathogenic mutations in the 21 families tested (Table 7, Fig. 3). As expected, three of these mutations are in genes reported to be associated with either autosomal dominant RP or autosomal dominant cone–rod dystrophy (Birch DG, et al. IOVS 2006;47:ARVO E-Abstract 1037).21,40–42 Somewhat surprising was the identification of two mutations in RPGR. It has been known for some time that mutations in RPGR cause X- linked RP, but the high frequency of symptomatic female carriers is just beginning to be appreciated.43,45,47
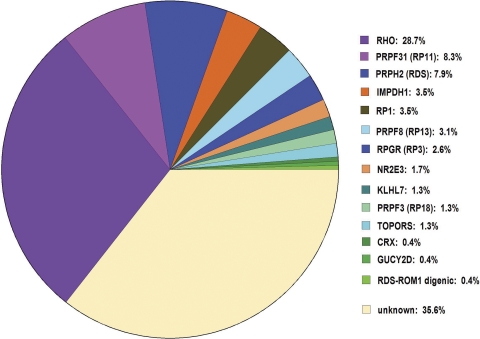
Identification of five additional mutations brings the known-mutation frequency of our AdRP cohort up to 64% (Fig. 4). That is, we can identify the disease-causing mutations in 148 of the 230 adRP cohort families.
Figure 4.
Prevalence of mutations in genes causing dominant RP. Pathogenic mutations have been identified in 148 of the 230 adRP cohort families including the five families reported in this study. The mutation remains to be identified in 82 (35%) of the families.
This project demonstrates that next-generation sequencing can be an effective tool for determining the pathogenic mutation in inherited disease families with highly heterogeneous causes. The large number of those variants proven to be artifacts identified in the limited region of the genome tested during this project raises concerns that the use of next-generation sequencing for larger genomic regions, such as a complete exome or genome, will be daunting. Coupled with the wide genetic variation known to exist in humans, this project makes it evident that, without the ability to perform segregation analysis, it will be extremely difficult to distinguish rare pathogenic variants from rare benign variants.
Acknowledgments
The authors thank Gita Dangol, Kaylie Webb, and Joe Ray for technical assistance.
Footnotes
Supported by the Foundation Fighting Blindness, National Institutes of Health Grants EY007142 and EY12910, and U54 HG003079 to the Center for Large Scale Genome Sequencing and Analysis.
Disclosure: S.J. Bowne, None; L.S. Sullivan, None; D.C. Koboldt, None; L. Ding, None; R. Fulton, None; R.M. Abbott, None; E.J. Sodergren, None; D.G. Birch, None; D.H. Wheaton, None; J.R. Heckenlively, None; Q. Liu, None; E.A. Pierce, None; G.M. Weinstock, None; S.P. Daiger, None
References
- 1. McKernan KJ, Peckham HE, Costa GL, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905 [DOI] [PubMed] [Google Scholar]
- 3. Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909 [DOI] [PubMed] [Google Scholar]
- 4. Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876 [DOI] [PubMed] [Google Scholar]
- 5. Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;233:1–34 [DOI] [PubMed] [Google Scholar]
- 7. Heckenlively JR. Retinitis Pigmentosa. Philadelphia: J. B. Lippincott; 1988 [Google Scholar]
- 8. Heckenlively JR, Daiger SP. Hereditary Retinal and Choroidal Degenerations. 5th ed. London: Churchill Livingston Elsevier; 2007 [Google Scholar]
- 9. Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–365 [DOI] [PubMed] [Google Scholar]
- 10. Grondahl J. Estimation of prognosis and prevalence of retinitis pigmentosa and Usher syndrome in Norway. Clin Genet. 1987;31:255–264 [DOI] [PubMed] [Google Scholar]
- 11. Xu L, Hu L, Ma K, Li J, Jonas JB. Prevalence of retinitis pigmentosa in urban and rural adult Chinese: The Beijing Eye Study. Eur J Ophthalmol. 2006;16:865–866 [DOI] [PubMed] [Google Scholar]
- 12. Buch H, Vinding T, La Cour M, Appleyard M, Jensen GB, Nielsen NV. Prevalence and causes of visual impairment and blindness among 9980 Scandinavian adults: the Copenhagen City Eye Study. Ophthalmology. 2004;111:53–61 [DOI] [PubMed] [Google Scholar]
- 13. Al-Merjan JI, Pandova MG, Al-Ghanim M, Al-Wayel A, Al-Mutairi S. Registered blindness and low vision in Kuwait. Ophthalmic Epidemiol. 2005;12:251–257 [DOI] [PubMed] [Google Scholar]
- 14. Hayakawa M, Fujiki K, Kanai A, et al. Multicenter genetic study of retinitis pigmentosa in Japan, II: prevalence of autosomal recessive retinitis pigmentosa. Jpn J Ophthalmol. 1997;41:7–11 [DOI] [PubMed] [Google Scholar]
- 15. Hayakawa M, Fujiki K, Kanai A, et al. Multicenter genetic study of retinitis pigmentosa in Japan, I: genetic heterogeneity in typical retinitis pigmentosa. Jpn J Ophthalmol. 1997;41:1–6 [DOI] [PubMed] [Google Scholar]
- 16. Daiger SP. Identifying retinal disease genes: how far have we come, how far do we have to go? Novartis Found Symp. 2004;255:17–27; discussion 27–36, 177–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419 [DOI] [PubMed] [Google Scholar]
- 19. Rivolta C, Sharon D, DeAngelis MM, Dryja TP. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum Mol Genet. 2002;11:1219–1227 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan LS, Bowne SJ, Seaman CR, et al. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:4579–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sullivan LS, Bowne SJ, Birch DG, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa (adRP): a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. RetNet. The Retinal Information Network, http://www.sph.uth.tmc.edu/RetNet/. Stephen P. Daiger, PhD, Administrator, The Univ. of Texas Health Science Center at Houston; 1996–present
- 23. Bowne SJ, Sullivan LS, Gire AI, et al. Mutations in the TOPORS gene cause 1% of autosomal dominant retinitis pigmentosa (adRP). Mol Vis. 2007;14:922–927 [PMC free article] [PubMed] [Google Scholar]
- 24. Gire A, Sullivan LS, Bowne SJ, et al. The Gly56Arg mutation in NR2E3 accounts for 1–2% of autosomal dominant retinitis pigmentosa. Mol Vis. 2007;13:1970–1975 [PubMed] [Google Scholar]
- 25. Ziviello C, Simonelli F, Testa F, et al. Molecular genetics of autosomal dominant retinitis pigmentosa (ADRP): a comprehensive study of 43 Italian families. J Med Genet. 2005;42:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato H, Wada Y, Itabashi T, Nakamura M, Kawamura M, Tamai M. Mutations in the pre-mRNA splicing gene, PRPF31, in Japanese families with autosomal dominant retinitis pigmentosa. Am J Ophthalmol. 2005;140:537–540 [DOI] [PubMed] [Google Scholar]
- 27. Wada Y, Sandberg MA, McGee TL, Stillberger MA, Berson EL, Dryja TP. Screen of the IMPDH1 gene among patients with dominant retinitis pigmentosa and clinical features associated with the most common mutation, Asp226Asn. Invest Ophthalmol Vis Sci. 2005;46:1735–1741 [DOI] [PubMed] [Google Scholar]
- 28. Wada Y, Tamai M. Molecular genetic analysis for Japanese patients with autosomal dominant retinitis pigmentosa. Nippon Ganka Gakkai Zasshi. 2003;107:687–694 [PubMed] [Google Scholar]
- 29. Zhang XL, Liu M, Meng XH, et al. Mutational analysis of the rhodopsin gene in Chinese ADRP families by conformation sensitive gel electrophoresis. Life Sci. 2006;78:1494–1498 [DOI] [PubMed] [Google Scholar]
- 30. Lord-Grignon J, Tetreault N, Mears AJ, Swaroop A, Bernier G. Characterization of new transcripts enriched in the mouse retina and identification of candidate retinal disease genes. Invest Ophthalmol Vis Sci. 2004;45:3313–3319 [DOI] [PubMed] [Google Scholar]
- 31. Bowes Rickman C, Ebright JN, Zavodni ZJ, et al. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wistow G, Bernstein SL, Wyatt MK, et al. Expressed sequence tag analysis of human retina for the NEIBank Project: retbindin, an abundant, novel retinal cDNA and alternative splicing of other retina-preferred gene transcripts. Mol Vis. 2002;8:196–204 [PubMed] [Google Scholar]
- 33. Mitton KP, Swain PK, Khanna H, Dowd M, Apel IJ, Swaroop A. Interaction of retinal bZIP transcription factor NRL with Flt3-interacting zinc-finger protein Fiz1: possible role of Fiz1 as a transcriptional repressor. Hum Mol Genet. 2003;12:365–3673 [DOI] [PubMed] [Google Scholar]
- 34. Steve Rozen HJS. Primer3. Code available at http://sourceforge.net/projects/primer3/develop 1996
- 35. Kent WJ. BLAT: the BLAST-like alignment tool. Genome Res. 2002;12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bowne SJ, Sullivan LS, Mortimer SE, et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47:34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao C, Bellur DL, Lu S, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friedman JS, Ray JW, Waseem N, et al. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2009;84:792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vithana EN, Abu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001;8:375–381 [DOI] [PubMed] [Google Scholar]
- 42. Kelsell RE, Gregory-Evans K, Payne AM, et al. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–1184 [DOI] [PubMed] [Google Scholar]
- 43. Pelletier V, Jambou M, Delphin N, et al. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype-phenotype correlations and impact on genetic counseling. Hum Mutat. 2007;28:81–91 [DOI] [PubMed] [Google Scholar]
- 44. Sharon D, Sandberg MA, Rabe VW, Stillberger M, Dryja TP, Berson EL. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003;73:1131–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vervoort R, Lennon A, Bird AC, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25:462–466 [DOI] [PubMed] [Google Scholar]
- 46. Shu X, McDowall E, Brown AF, Wright AF. The human retinitis pigmentosa GTPase regulator gene variant database. Hum Mutat. 2008;29:605–608 [DOI] [PubMed] [Google Scholar]
- 47. Rozet JM, Perrault I, Gigarel N, et al. Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J Med Genet. 2002;39:284–285 [DOI] [PMC free article] [PubMed] [Google Scholar]