Abstract
Heterozygous LIS1 mutations are the most common cause of human lissencephaly, a human neuronal migration defect, and DCX mutations are the most common cause of X-linked lissencephaly. LIS1 is part of a protein complex including NDEL1 and 14-3-3ε that regulates dynein motor function and microtubule dynamics, while DCX stabilizes microtubules and cooperates with LIS1 during neuronal migration and neurogenesis. Targeted gene mutations of Lis1, Dcx, Ywhae (coding for 14-3-3ε), and Ndel1 lead to neuronal migration defects in mouse and provide models of human lissencephaly, as well as aid the study of related neuro-developmental diseases. Here we investigated the developing brain of these four mutants and wild-type mice using expression microarrays, bioinformatic analyses, and in vivo/in vitro experiments to address whether mutations in different members of the LIS1 neuronal migration complex lead to similar and/or distinct global gene expression alterations. Consistent with the overall successful development of the mutant brains, unsupervised clustering and co-expression analysis suggested that cell cycle and synaptogenesis genes are similarly expressed and co-regulated in WT and mutant brains in a time-dependent fashion. By contrast, focused co-expression analysis in the Lis1 and Ndel1 mutants uncovered substantial differences in the correlation among pathways. Differential expression analysis revealed that cell cycle, cell adhesion, and cytoskeleton organization pathways are commonly altered in all mutants, while synaptogenesis, cell morphology, and inflammation/immune response are specifically altered in one or more mutants. We found several commonly dysregulated genes located within pathogenic deletion/duplication regions, which represent novel candidates of human mental retardation and neurocognitive disabilities. Our analysis suggests that gene expression and pathway analysis in mouse models of a similar disorder or within a common pathway can be used to define novel candidates for related human diseases.
Author Summary
Neuronal migration is a biological process that ensures proper organization of the cerebral cortex during development. Failure of this process leads to lissencephaly, a neuronal migration defect in humans and an important cause of mental retardation and intractable epilepsy. To study these defects, we generated mouse mutants by inactivating four genes (Lis1, Dcx, Ywhae, and Ndel1) that play a crucial role in neuronal migration. These genes are part of the same molecular complex (LIS1 complex) that we hypothesize have overlapping functions in neuronal migration and cell proliferation. To broaden our understanding of neuronal migration and to further test our hypothesis, we analyzed global gene expression in these mutants using informatic approaches, confirming some of them biologically. We found that several biological processes were commonly altered in all mutants, while others were altered only in specific mutants. Our results provide new insights into the pathways and biological processes that regulate normal brain development and that are altered in mouse mutants of human neuronal migration defects, and they suggest a genomic approach to use gene expression analysis of mouse models of human genetic disease to identify candidate genes for related disorders, such as mental retardation and epilepsy.
Introduction
Neuronal migration defects such as lissencephaly are an important cause of mental retardation and severe epilepsy in humans, and many of the genes responsible for these conditions have been identified, including LIS1 and DCX [1]–[4]. Heterozygous deletions or mutations of LIS1 are the most common cause of lissencephaly in humans [5]. LIS1 is part of a complex that includes NDEL1 [6], [7] and 14-3-3 epsilon [8]. This complex is essential for the regulation and localization of cytoplasmic dynein (light and heavy chains), centrosomal protein localization and function as well as microtubule dynamics (for review, see [9]–[11]). These interactions are critical for nuclear movements and neuronal migration. Similar to LIS1, mutations of DCX are the most common cause of X-linked lissencephaly in humans [5]. DCX is a microtubule binding protein that helps to stabilize and bundle microtubules in migrating neurons [12]. There is some evidence that DCX either directly interacts with the proteins of the LIS1 complex, and genetic interactions between Lis1 and Dcx have been demonstrated [13], [14].
Targeted gene mutations have been produced in the mouse to identify the molecular pathways and biological processes disrupted in lissencephaly and several studies demonstrate that Lis1 is essential for both for neuronal migration and neurogenesis [15]–[18]. These effects are mediated by the LIS1-complex components NDEL1 and 14-3-3ε, since disruption of Ndel1 and Ywhae (coding for 14-3-3ε) result in neuronal migration defects in the mouse [8], [18], [19]. Dcx mutant male mice or Dcx knockdown by RNAi display neuronal migration defects [14], [20], [21], and Dcx is essential for both neuronal migration and proliferation alone and in combination with Lis1 [14].
These studies support the hypothesis that Lis1, Ndel1, Ywhae and Dcx have overlapping functions in the regulation of neurogenesis and neuronal migration. However, it is unclear if there is substantial overlap in function among these genes, or whether these genes have significant independent functions. This question is of interest for Lis1, Ndel1 and Ywhae, which code for proteins in the same complex, and for Lis1 and Dcx, which have functional interactions. To address this important question, and to provide further information about normal brain development, we have analyzed the global gene expression differences in brains from WT and mutants for these four genes (Lis1, Ndel1, Ywhae and Dcx) at three developmental time points. Since their protein products are not transcription factors and hence do not directly regulate mRNA levels of target genes, we performed bioinformatic analyses to unveil the canonical pathways and biological processes that regulate normal brain development and are responsible for the pathophysiology of neuronal migration defects in these mutants. Normalized data were analyzed by several different statistical methods and the most differentially expressed genes were assessed by Gene Ontology (GO) enrichment. We provide evidence for surprising similarities and differences in the pathways and processes identified in the analyses of these mouse mutants as well as new potential candidate genes in neuronal migration and proliferation defects. Finally, we experimentally confirmed selected novel findings with additional functional assays. The results of our studies provide novel insights into genes whose expression levels mediate normal brain development and are altered in models of human neuronal migration defects.
Results
We investigated global gene expression of wild-type (WT) mice and four mouse models for human neuronal migration defects (Lis1, Dcx, Ywhae and Ndel1) at three developmental stages (E14, P0 and P14) following a three-step strategy (Figure S1) to define common and different gene expression signatures as well as dysregulated genes. The output of each analysis was transformed into GO enrichments to define and compare the canonical pathways and biological processes expressed at each time point in each mutant. Enrichment scores will be indicated as “Sc” and refer to the −Log (p-value) of enrichment values with FDR<0.05.
Cell cycle and developmental signaling pathways in wild-type embryonic brain development
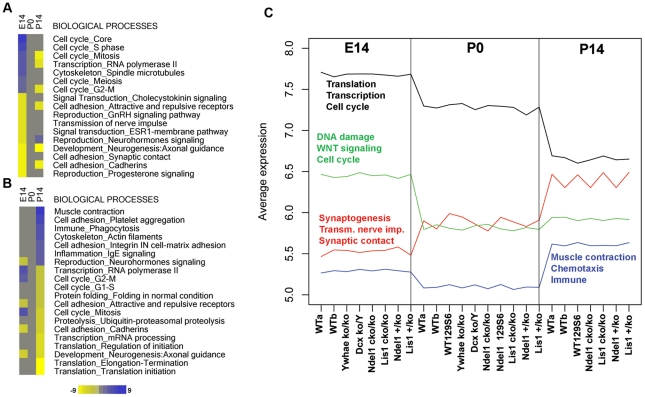
To set gene expression references for each time point, we analyzed differential expression data of WT samples by GO enrichment using Metacore and Ingenuity Pathway Analysis (IPA). We identified pathways and processes that were over- and under-represented at one developmental stage with respect to the other two stages (see methods). Gene expression at E14 was enriched for biological processes and canonical maps containing genes controlling cell cycle pathways (Figure 1A and Figure S2A). Consistently, DNA damage pathways were relatively highly expressed whereas cell adhesion (cadherins/synaptic contact processes), axonal guidance and axon growth repulsion pathways, were under-expressed (Figure 1A and Figure S2A). Positive enrichment in DNA damage checkpoint (ATM/ATR and BRCA1) and growth factor/proliferative (APC, IGF-R1, NOTCH, and VEGEF) signaling pathways supported the finding that proliferation is the main biological process important in the normal progression through these early stages of brain development (Figure 1A and Figure S2A). Surprisingly, there was no significant enrichment of pathways or processes at P0 with respect to the two other developmental time points, suggesting that gene expression at this time significantly overlaps each of the other two stages (Figure 1A, 1B and Figure S2). At P14, cell cycle pathways and processes as well as protein synthesis were down-regulated (Figure 1B and Figure S2B). Oxidative phosphorylation and other related pathways were up-regulated at this stage, indicating that energy metabolism and its regulation were important for later stages of brain development (Figure 1B and Figure S2B). Cytoskeleton processes were found to be up-regulated both at early and later stages of development, although different sets of genes were present. At E14, genes involved in spindle microtubule regulation were up-regulated, while actin filament pathways were up-regulated at P14 (Figure 1A, 1B). Similar results were found using the IPA software for GO analysis confirming that cell cycle and gene expression processes were positively enriched at E14, P0 had no characteristic gene expression signatures, and P14 was characterized by repression of protein synthesis (Figure S2C).
Figure 1. Cell cycle processes define early brain development in mouse, and unsupervised clustering revealed gene expression similarities in all genotypes at each developmental stage.
A,B) Differential expression analysis of WT brains followed by pathway analysis via Metacore software define biological processes that are up-/down-regulated at each developmental stage versus the other stages. A) Up-regulated (blue) and down-regulated processes (yellow) are sorted by intensity at E14. B) Similarly, up- and down-regulated processes are sorted by intensity at P14. No gene expression signature is characteristic of P0. Intensity values refer to enrichment values provided by Metacore and calculated as −Log(p-value) (FDR<0.05). C) Simultaneous analysis of gene expression data from all genotypes at all time points using the PAM algorithm, followed by gene enrichment (Metacore). PAM analysis revealed four patterns of gene expression that are similar across all genotypes, but different in a stage-dependent fashion. Black and green profiles represent genes highly expressed at E14, while red and blue represent genes with highest expression at P14. No particular expression pattern is characteristic of P0. Pathway analysis of the four patterns was consistent with the differential analysis results found in WT brains (see A,B).
Gene expression in mutant and wild-type brains is similar at each developmental stage
To determine whether mutant brains displayed similar or different gene expression patterns compared to WT, we performed unsupervised hierarchical clustering (PAM algorithm) using gene expression data sets from all WT and mutant time points. Due to the similar developmental outcome of all brains, similar expression patterns were more likely to be expected rather than different genotype-specific profiles. We used silhouette analysis to determine the optimal number of clusters (Figure S3A). Gene expression patterns fit the data best when distributed within four clusters (Figure 1C and Figure S3B). The four profiles distributed independently of genotypes along the x-axis, but cluster distribution was highly correlated with the E14 and P14 developmental stages (Figure 1C). The black and green clusters were expressed at higher average levels compared to red and blue. These two clusters peaked at E14 and declined at P0 and P14 (Figure 1C). The red and blue clusters displayed opposite profiles with lower levels at E14 and P0 that increased at P14 (Figure 1C). In agreement with the previous findings in WT brains (Figure 1A, 1B, and Figure S2), the clusters did not reveal a unique pattern of gene expression levels at P0 compared to E14 and P14 (Figure 1C), supporting the interpretation that gene expression at the P0 stage of brain development overlaps these other two stages.
GO analysis further supported the finding that critical biological processes are maintained during development independent of their genotype and background. Black and green clusters (enriched at E14) were highly enriched in genes coding for translation, transcription, cell cycle, DNA damage and WNT signaling (Sc = 40.4, Sc = 36.7, Sc = 19.3, Sc = 8.7, Sc = 8.5 respectively; Figure 1C and compare to Figure 1A and Figure S2), while red and blue clusters (highest at P14) were enriched in genes coding for synaptogenesis, transmission of nerve impulse (TNI), synaptic contact, muscle contraction and immune response (Sc = 21.7, Sc = 19.8, Sc = 12.3, Sc = 10.6, Sc = 4.2 respectively; Figure 1C and compare to Figure 1B). Consistent with the similar gross anatomical structures of all brains, unsupervised clustering of mutant brains displayed similar overall gene expression patterns to WT brains at each specific developmental stage, as described above.
To test the hypothesis that the genes within these pathways and processes are co-regulated during development, we analyzed the same dataset using WGCNA co-expression analysis (see methods). This analysis provides a clustering of genes based on the correlation of their expression levels and is a complementary method to the PAM analysis. Gene expression data were clustered into 25 modules (module = M in Figure S3C). We performed pathway analysis using Metacore to determine which biological processes were enriched in each module and compared them to the clusters identified by the PAM analysis. We found consistent results in which cell cycle processes constituted the largest cluster displaying the highest enrichment (M1-turquoise; Figure S3C and Table 1), followed by synaptogenesis/TNI (M3-brown) and translation/transcription (M4-yellow). The genes within the modules that contained at least 100 genes (M1-M18) were compared with the genes in the four PAM clusters. All 18 modules were included in the four PAM clusters and displayed a consistent enrichment (Figure S3D and Table 1). For instance, M1 (cell cycle) is composed of the black and green PAM clusters (cell cycle), which are enriched in genes displaying high levels of expression at E14. M3 (synaptogenesis) is entirely contained within the red PAM cluster (synaptogenesis), which is enriched in genes displaying high levels of expression at P14.
Table 1. Co-expression analysis of all datasets is consistent with the PAM unsupervised clustering.
| BIOLOGICAL PROCESSES | MODULE N° | −LOG(P-value) |
| Cell cycle | Module 1 | 18.39935 |
| Synaptogenesis/TNI | Module 3 | 14.01381 |
| Translation/Trancription | Module 4 | 11.70093 |
| Inflammation/Muscle contraction/Ossification | Module 2 | 5.817329 |
| DNA damage/Proteolysis/Cell cycle | Module 8 | 5.17276 |
| Translation in Mitocondria | Module 12 | 4.837137 |
| Cell cycle/Apoptosis/Synaptogenesis | Module 9 | 4.734231 |
| Inflammation/Muscle contraction | Module 6 | 4.009217 |
| Autophagy | Module 14 | 3.871601 |
| Immune phagocytosis/Cytoskeleton/Cell adhesion | Module 5 | 3.827582 |
| Axonal guidance/TNI/Cell adhesion | Module 11 | 3.631598 |
| Inflammation/Translation/Signal transduction | Module 19 | 3.383735 |
| Transcription/Apoptosis | Module 21 | 3.31721 |
| Neurogenesis in general/development | Module 22 | 3.209552 |
Metacore pathway analysis of the 25 modules (WGCNA) defined 14 modules with top enrichment values (FDR<0.05). Consistent with the PAM analysis, cell cycle, synaptogenesis and translation/transcription genes are highly co-expressed during brain development.
To further extend the analysis of gene co-regulation and to create a link with biological evidence of defective development we performed correlation analysis of the 25 modules (Figure S3C) with quantified phenotypes of neuronal migration and proliferation defects from each of the different mutants at E14 (Table 2). Several modules were significantly correlated with each phenotype and most of the modules correlated with spindle orientation and migration speed (Table 2). Metacore analysis of these modules indicated that the organization of cytoskeleton-actin filaments and regulation of cytoskeleton rearrangement were within the top three biological processes enriched in nearly all correlated modules (Table 2). This data confirmed the important role of cytoskeleton remodeling during brain development and its effects on neuronal migration and proliferation when altered in each of these mutants. Collectively, these analyses revealed gene expression patterns and clusters of co-expressed genes that reflected developmental processes in a stage-dependent fashion.
Table 2. Cytoskeleton processes are correlated with neuronal migration defects and proliferation phenotypes in the mutants.
| Phenotypes | N° of correlated modules | Correlation P-value | Biological processes | −LOG(P-value) |
| Length of leading process | 4 | <0.05 | Cytoskeleton_Actin filaments | 5.261537 |
| Immune_Phagocytosis | 3.789681 | |||
| Immune_Phagosome in antigen presentation | 3.643592 | |||
| Moving cells (%) | 3 | <0.05 | Immune_TCR signaling | 3.413638 |
| Cell adhesion_Integrin-mediated | 3.360514 | |||
| Cytoskeleton_Actin filaments | 3.257982 | |||
| Migration speed | 6 | <0.05 | Cytoskeleton_Actin filaments | 6.384787 |
| Inflammation_Protein C signaling | 3.970211 | |||
| Regulation of cytoskeleton rearrangement | 3.749092 | |||
| Spindle orientation | 9 | <0.01 | Cytoskeleton_Actin filaments | 7.448428 |
| Inflammation_Amphoterin signaling | 4.670399 | |||
| Regulation of cytoskeleton rearrangement | 4.600326 | |||
| BrdU-incorporation (%) | 5 | <0.05 | Cytoskeleton_Actin filaments | 6.297914 |
| Immune_Phagocytosis | 4.914353 | |||
| Muscle contraction | 4.242528 | |||
WGCNA defined modules of co-expressed gene that are correlated with quantified phenotypes in the mouse mutants. Pathways enrichment of these correlated modules revealed that cytoskeleton processes were within the top 3 processes, confirming their pivotal role during brain development.
Gene expression varies more from WT in Lis1cko/ko mutants than in Ndel1cko/ko mutants
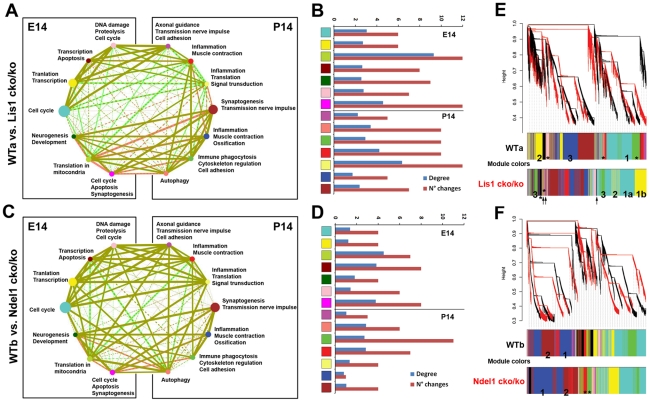
To investigate gene expression differences of Lis1 and Ndel1 mutants versus wild-type (WTa for Lis1, WTb for Ndel1), we used the lowest levels of LIS1 and NDEL1 protein (Lis1cko/ko and Ndel1cko/ko) and analyzed the interactions of the top 14 WGCNA co-expression modules (Table 1) to establish how gene expression correlates between pairs of modules (see methods). The correlation matrices were transformed into interaction networks with nodes representing the module enrichment of Table 1 and the edges representing the values of the correlation matrices (Figure 2A, 2C). We then represented this data as heat maps to visualize the differences between the mutants and WTs (Figure S4B). Overall, there were a higher number and degree of changes in biological processes in the Lis1cko/ko compared to the Ndel1cko/ko mutants (Figure 2B, 2D). The number of correlations that were unaltered or altered less than 10% was higher in Ndel1cko/ko mutants than in Lis1cko/ko mutants (Figure S4A). The number of correlations that mildly changed (greater than 10% but in the same direction) or severely changed (greater than 10% but in the opposite direction) was higher in Lis1cko/ko mutants than in Ndel1cko/ko mutants (Figure S4A). These results indicate that despite the gene expression similarities revealed by the PAM unsupervised clustering and co-expression analysis, mutant brains display substantial differences in the interactions among biological processes during brain development and these differences appear to be more severe in Lis1cko/ko than in Ndel1cko/ko mutants, consistent with the more severe brain phenotype of the Lis1 versus Ndel1 mutant [18].
Figure 2. Lis1cko/ko mutant is more dissimilar to WT than Ndel1cko/ko mutant.
Inter-genotype correlation matrix analysis of the top 14 biological processes (Table 1) and WGCNA of single WT and Lis1/Ndel1 mutants revealed more differences in the Lis1 genotype than the Ndel1 compared to WT. A,C) Correlation matrices are transformed into interaction networks with nodes representing the top 14 biological processes (Table 1) and edges represent the correlation values. The size of the nodes is proportional to the enrichment score of Table 1. The edges thickness is proportional to the strength of the correlation (thicker = more strongly correlated). The style of the edge lines represents the direction of the correlation (solid = direct correlation; dashed = inverse correlation). Nodes are color-coded according to the module colors of the WGCNA (Figure S3C). WT edges have been color-coded in red, while mutants' in green. The overlaps (Lis1 mutant over WTa, Ndel1 mutant over WTb) display the changes in the relationships among the processes. The preponderance of red (WT preponderance) or green (mutant preponderance), compared to no difference (mustard) reflects the underlying differences in the genotypes (WT versus mutant). B,D) Quantification of the number and degree of the changes between the compared genotypes revealed more severe alteration in the Lis1 mutant than Ndel1. E,F) WGCNA comparison of the same genotypes (WTa versus Lis1cko/ko; WTb versus Ndel1cko/ko). Modules with the same numbers in each comparison indicate similar GO enrichment, therefore representing similar modules. * and ↑ indicate that module enrichment is peculiar for that genotype and is not either found or significantly present in the compared genotype. Also this analysis revealed greater differences in the Lis1 mutant than Ndel1 mutant compared to WTs.
To further investigate gene expression differences in Lis1cko/ko and Ndel1cko/ko mutants, we analyzed the same four genotypes (WTa, WTb, Lis1cko/ko and Ndel1cko/ko) by WGCNA individually, using all three time points (E14, P0 and P14) and compared the different module assignments in terms of GO enrichment (Figure 2E, 2F). We identified several important differences in the co-expression of genes in the Lis1cko/ko mutant compared to WT (WTa), while only mild differences were present in the Ndel1cko/ko mutant compared to WT (WTb) (Table 3). The difference in number of modules and color assignment was consistent with major changes in clustering in the Lis1cko/ko mutant (Table 3). The co-expression of genes encoding three major pathways (cell cycle, synaptogenesis and translation) was altered more significantly in Lis1cko/ko than in Ndel1cko/ko mutants (Figure 2E and Table 3). Different modules were uniquely represented with significant enrichment in each genotype (see * in Figure 2E and Table 3). Of note, nine modules (for instance see ↑ in Figure 2E and Table 3) were enriched in cell adhesion processes in Lis1cko/ko mutants and not present in WT. We quantified the overall number of hits of cell adhesion processes with a Sc>3 in Lis1cko/ko mutant modules versus WT and found nearly three times greater representation in the mutant (data not shown). The clustering of Ndel1cko/ko mutants was overall more similar to WT than Lis1cko/ko with only translation co-expressed with cell cycle genes and two modules were uniquely enriched in the mutant (see * in Figure 2F and Table 3). These data indicate that there are significant changes in expression in Lis1 and Ndel1 mutants compared to WT, and the co-expression of genes during brain development in the Lis1cko/ko mutant are affected more than in the Ndel1cko/ko mutant, supporting the hypothesis that Lis1 mutants are more severely affected than in the Ndel1 mutants.
Table 3. Lis1 mutant is more dissimilar to WT than Ndel1 by WGCNA.
| Lis1cko/ko versus WTa | Ndel1cko/ko versus WTb | |||
| N° Modules | 23 | 16 | 10 | 10 |
| Color assignment | Different | Similar | ||
| Cell cycle | 2 modules(M1a-turquoise; M1b-yellow) | 1 module(M1-turquoise) | 1 module(M1-blue)similar to WT + translation | 1 module(M1-blue) |
| Synaptogenesis | is embedded within cell cycle | is opposite of cell cycle | Similar | |
| Translation | is small and split in 2 modules | is a large module | is co-expressed with cell cycle | is a distinct module |
| Unique enrichment(*) |
|
|
|
Not found |
Co-expression analysis of the individual genotypes (Lis1cko/ko, WTa, Ndel1cko/ko, WTb) revealed more severe differences in the clustering of Lis1 than Ndel1 mutants compared to WT. * and ↑ and colors refer to Figure 2E, 2F. In Lis1cko/ko: Module 1a (M1a) represents S-phase and Mitosis (Sc = 8.3 and Sc = 6.2); Module 1b (M1b) represents Mitosis and S-phase (Sc = 21.8 and Sc = 15.2); Synaptogenesis is represented by Module 2 (M2-light yellow, Sc>13.6); Translation is represented by two Modules 3 (M3-green, Sc = 13.3); 9 Modules with Sc>3 (Ms, see for examples ↑ in Figure 2E); Axonal guidance module is light green (Sc = 4.2); Inflammation module is black (Sc = 3.5). In WTa: Module 1 (M1) represents Mitosis and S-phase (Sc = 25.8 and Sc = 24.8); Synaptogenesis is represented by Module 2 (M2-yellow, Sc>10.5); Translation is represented by Module 3 (M3-blue, Sc = 21.1); Proteolysis module is green (Sc = 3.3); WNT signaling module is salmon (Sc = 3.4); Reproduction module is pink (Sc = 3.6). In Ndel1cko/ko: Cell cycle module (M1-blue) is similar to WTb and includes also translation processes (Sc = 32.1); Protein folding/Immune and Blood vessels processes are represented by module green and red (3>Sc<4); In WTb: Cell cycle module (M1-blue, Sc = 27.5) contains also DNA damage and transcription processes (Sc = 15.7, Sc = 13.7); Translation is represented by module 2 (M2-brown, Sc = 28.9).
Lis1 mutants reveal dosage-dependent patterns of gene expression
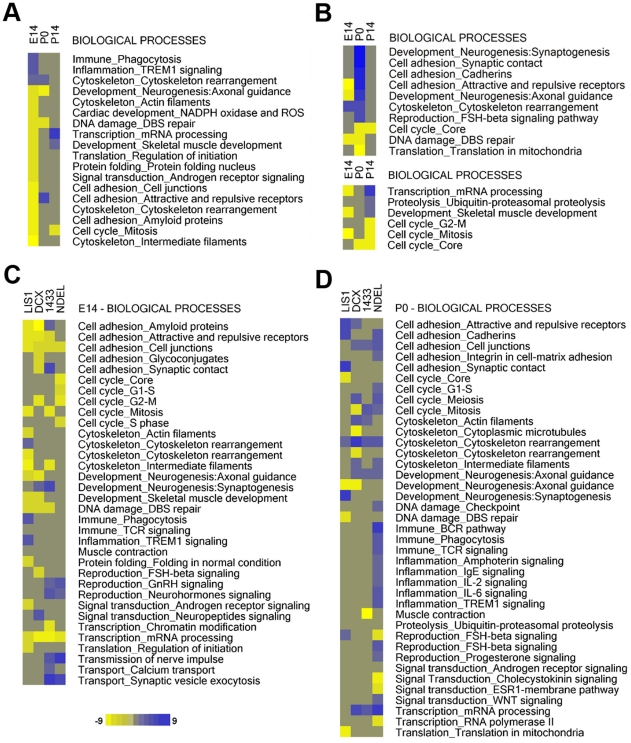
We next took advantage of an allelic series of Lis1 mutants to identify the gene expression changes that correlated with the effects of the different dosages of LIS1 (100%>50%>35%) [15]. In contrast to WT, brains with decreasing levels of LIS1 displayed a remarkable repression of cell cycle pathways and processes at E14 (Figure 3A), while transcription and DNA damage was also down-regulated at E14 and over-expressed at later stages. By contrast, immune phagocytosis and inflammation pathways were up-regulated. Cytoskeletal processes were found both up- and down-regulated suggesting that these processes were completely dysregulated in the Lis1 mutants. These findings are nearly opposite of the pattern of gene expression in WT E14 brains. Similarly, the expression pattern of P14 brains was consistently distinct from WT expression at P14. In Lis1 mutants, there was significant up-regulation of transcription, which was instead enriched at E14 in WT brains (Figure 3A). Gene expression was profoundly altered in the Lis1 mutants at P0 with over-representation of synaptogenesis and synaptic contact, suggesting a premature expression of pathways expressed in the WT at P14. This finding is consistent with the co-expression data in which synaptogenesis genes exhibited co-expression with earlier time point genes (cell cycle and transcription; Table 3). Additional cell adhesion pathways and processes were also abnormally enriched at this stage (Figure 3B). IPA analysis confirmed these findings and demonstrated that additional processes were altered in the Lis1 mutants (Figure S5). Cell cycle processes were found slightly enriched at E14. However, only a few genes (CDKN1A, CDKN1B, ID2, IGF1, TF and TGFA) were represented. Most of these genes are growth factors, not primary cell cycle regulatory genes, and this may reflect the use of cancer cell lines to define functional annotation classes in IPA. By contrast, primary cell cycle regulatory genes (Figure S2C) were present in the cell cycle progression processes of WT. Biological processes such as gene expression, cellular assembly and organization as well as cellular movement were down-regulated at E14 (Figure S5A), and the corresponding molecular functions associated with these processes (bundling of microtubules, transport of vesicles and migration of neurons) are surprisingly consistent with observed phenotypes of Lis1 mutants [13], [14], [18], . Axonal guidance and outgrowth were abnormally enriched at P0 compared to WT, together with other functions that indicated premature activity axonal related processes (Figure S5B). P0 was also characterized by a down-regulation of genes coding for RNA post-transcriptional modifications, which on the contrary were heavily activated at P14 in WT (Figure S5C), and cell cycle processes were found to be repressed at this stage. In summary, differential gene expression analysis of decreasing levels of LIS1 revealed opposite patterns of expression compared to WT at early and late developmental stages with a premature appearance of differentiation processes such as synaptogenesis at birth.
Figure 3. LIS1 protein reduction causes gene expression alterations of important biological processes, some of which are shared among the other mutants.
Differential expression analysis followed by pathway analysis (Metacore) revealed alterations in biological processes important for early and late brain development. Some of the processes are commonly up-/down-regulated in all mutants and some are specific for single mutants. A) up-regulated (blue) and down-regulated (yellow) processes are sorted by intensity at E14 in the Lis1 mutants with decreasing levels of LIS1 (100%>50%>35). B) Up-/down-regulated processes are sorted by intensity at P0 and P14 in the same Lis1 mutants. C) Up-/down-regulated processes are sorted by pathway category across the mutants at E14. D) Up-/down-regulated processes are sorted by pathway category across the mutants at P0. Intensity values refer to enrichment values provided by Metacore and calculated as −Log(p-value) (FDR<0.05).
Differential gene expression analysis in all mutants reveals common and distinct pathways of gene expression at E14 and P0
We next determined if there were gene expression signatures specific for each mutant or pathways commonly altered in all mutants compared with WT. The comparison of gene expression patterns in all neuronal migration mutants at E14 and P0 (Dcx and Ywhae mutants die soon after birth) revealed that several pathways were commonly affected (Figure 3C) and others were specific for individual genotypes. In E14 brains, cell cycle, transcription and distinct cell adhesion processes were commonly altered in all mutants with an opposite expression pattern compared to WT. Within cell cycle networks, brains from Lis1 and Ywhae mutants at E14 displayed down-regulation of mitosis, while brains from Dcx and Ndel1 mutants shared repression for G2-M genes (Figure 3C). Interestingly, only Ndel1 mutant brains displayed down-regulation of genes in almost all phases of cell cycle, while only Lis1 mutant brains displayed alteration of genes coding for cytoskeleton rearrangement and actin (Figure 3C). Reproduction process alterations (GnRH and neurohormone signaling) were characteristic of Ywhae and Ndel1, as was transmission of nerve impulse and vesicle exocytosis processes (Figure 3C). The latter two processes, together with synaptogenesis and synaptic contact processes, suggested that late stage processes (typical of P14 in WT) were prematurely expressed in Ywhae mutants at E14 (Figure 3C).
Gene expression patterns in brains from all mutants at P0 were clearly distinct, and different from WT, where no unique expression signatures were seen (Figure 3D). In general, over-expression was preponderant at this stage compared to E14 (Figure 3D and compare to Figure 1B). Cell adhesion processes were remarkably enriched at this stage in all mutant brains with a switch of expression compared to E14 (Figure 3C, 3D). Cell cycle processes displayed a switch of expression in Ndel1 and Ywhae mutants. Pathways for transcription of mRNA were up-regulated in all mutants, but not in Lis1 mutants (Figure 3D). Cytoskeleton rearrangement was commonly altered in all mutants and predominately affected in Dcxko/Y mutants. Gene expression in Ndel1 brains was characterized by a broad activation of immune and inflammation processes (Figure 3D). IPA analysis confirmed that processes for gene expression and cellular movement were down-regulated in all mutants at E14 (Figure S6A). In addition, cell morphology processes were widely and uniquely present in Dcxko/Y mutants (Figure S6A, 6B). Moreover, cell-to-cell signaling processes displayed an increasing gradient of expression across the mutants with maximum levels in Ywhae and Ndel1 mutants (Figure S6). In summary, this data suggest that, compared to WT, all mutants displayed common dysregulation for certain pathways (cell cycle and cell adhesion), while other pathways (synaptogenesis, cell morphology and immune response) were specifically altered in single mutants (Ywhae, Dcx and Ndel1 respectively).
Differential expression analysis in all mutants reveals common and distinct altered genes at E14 and P0
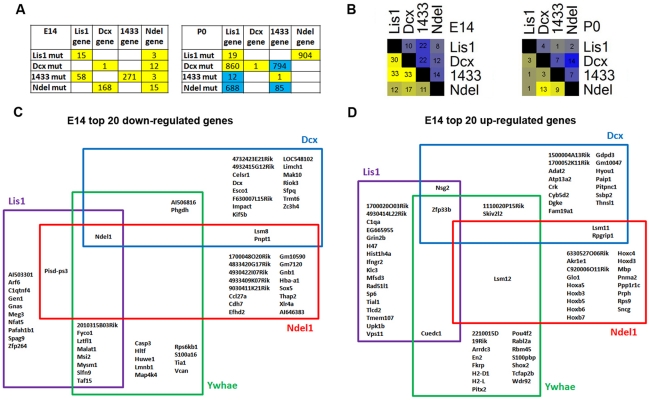
To better characterize the gene expression similarities and differences of our mutants, we directly compared the lists of up-/down-regulated genes in each genotype at E14 and P0. First, we addressed whether mutations of Lis1, Dcx, Ywhae and Ndel1 affected the expression of any of the other genes in the complex. We found that Lis1 was down-regulated in Lis1 and Ywhae mutants. Dcx was down-regulated in the Dcx and Ndel1 mutants. Ywhae was only down-regulated in Ywhae mutants. Surprisingly Ndel1 was down-regulated in all mutants, ranking within the top 15 dysregulated genes (Figure 4A,C). None of these genes were found up-regulated at E14, while Lis1 and Ywhae were both up- and down-regulated at P0 (Figure 4A). Two-way Venn analysis (data not shown) demonstrated that Lis1, Dcx, and Ywhae mutants shared a higher proportion of commonly down- and up-regulated genes at E14 compared to P0 and that Ndel1 differed the most with only about 10% of commonly altered genes between the other mutants (Figure 4B). Four-way Venn analysis of the top 20 differentially expressed genes at E14 supported the degree of similarities and differences noted in the previous section (Figure 4C). Examination of genes commonly down-regulated at E14 identified Ndel1, three un-characterized transcripts (ENSMUSG00000089788, ENSMUSG00000056919, ENSMUSG00000032425), and the Slfn9 gene, a gene with a role in the regulation of cell growth [24]. Overall, 45% of down-regulated genes in Lis1, Dcx and Ywhae mutants, were genotype specific. By contrast, 75% of down-regulated genes in Ndel1 mutants were specific to Ndel1, suggesting this mutant is the most divergent in terms of gene expression. We identified 10 genes that were commonly up-regulated at E14 in all mutants (Plek, Wdr92, Igf1, Hspa12a, C1qa, Nsg2, Cnp, Uqcr10, Rragd, Gpx3), and the proportion of genotype-specific up-regulated genes was more variable: 55% in Ywhae, 65% in Dcx, 75% in Lis1 and Ndel1 mutants. Of note, Ndel1 mutants displayed up-regulation of genes of “Hox family” (Figure 4D; 7 Hox genes in the top 20 and 10 Hox genes in the top 35).
Figure 4. Single genes are commonly and specifically altered in the mutants.
A) Ranking position of the four mutated genes (Lis1, Dcx, Ywhae and Ndel1) that were found differentially expressed in the mutants. Position number is in respect to lists of 1000 genes. Yellow represents down-regulation, blue represents up-regulation. B) Overlap of the top 1000 dysregulated genes across the mutants at E14 and P0. Numbers represent the percentage of genes that overlap in each pairwise comparison. Yellow represents down-regulation and blue represents up-regulation. Color intensity reflects the degree of overlap in which intense color indicates high degree of overlap. C,D) Four-way Venn analysis of the top 20 differentially expressed genes (down-regulated and up-regulated) at E14 in the mutants. The diagrams show that some of the genes are commonly altered in the mutants and others are genotype-specific.
From the surprisingly large proportion of down-regulated genes among Lis1, Dcx and Ywhae mutants at E14 (30%), we generated a list of potential candidates for neuronal migration and proliferation defects located within micro-deletion/duplication regions in human patients with mental retardation or neurocognitive disabilities (DECIPHER database; http://decipher.sanger.ac.uk/; see Table 4). In addition to NDEL1, we identified four genes that function directly in neuronal migration/proliferation (SYNE2, KIAA1009, TAOK1 and AP1S2 in Table 4). SYNE2 plays a role in nucleo-centrosome coupling that is critical during both neurogenesis and neuronal migration in the cortex [25]. KIAA1009 interacts with Cep120, which controls interkinetic nuclear migration during progenitor divisions, and more recently was shown to directly regulate spindle assembly and cell division in vitro [26], [27]. KIAA1009 is deleted and duplicated in four patients (Decipher ID: 254057, 1878, 248173 and 4070; Table 4). TAOK1 regulates microtubule dynamics and checkpoint signaling [28]. AP1S2 is involved in formation of clathrin-coated vesicles and endocytosis processes and is found both deleted and mutated in patients with mental retardation (Decipher ID: 2376, [29]). Several other genes from this list are closely related to relevant processes such as synapse formation and function (NRXN3, VCAN and SYT1), transcription (PABPN, NFAT5, TRA2B, LIMD7 and RBMX), intercellular protein trafficking (NAPG) or generally involved in brain/CNS development (RBBP4, ISLR2, BRD8, MARCH7, NFID and HDAC2). These genes represent potential candidates for the neurocognitive phenotype of the reported patients with microdeletions/duplications.
Table 4. Common down-regulated genes in the Lis1, Dcx, Ywhae mutants.
| Gene symbol | Dels/Dups | Neurocognitive Phenotype | Ex. Patient ID |
| NRXN3 | 2/0 | NF | NF |
| VCAN | 1/0 | mental retardation/autism | 596 |
| SYNE2 | NF | NF | NF |
| SYT1 | 4/0 | mental retardation | 1582 |
| MAP4K4 | 4/2 | mental retardation/speech delay | 250511 |
| PABPN1 | 0/2 | speech delay | 250430 |
| PHF16 | 0/2 | mental retardation/autism/seizures | 249240 |
| NFAT5 | NF | NF | NF |
| C7orf63 | NF | NF | NF |
| RPS6KB1 | 1/0 | mental retardation | 250379 |
| MIR1279 | NF | NF | NF |
| BRD8 | 1/1 | mental retardation/speech delay | 249097 |
| SGCE | 4/1 | mental retardation/speech delay | 856 |
| HERC2 | 3/9 | mental retardation/seizures | 1639 |
| NAPG | 5/4 | mental retardation/speech delay | 1882 |
| ARGLU1 | 3/2 | mental retardation/microcephaly | 4668 |
| KIAA1009 | 3/1 | mental retardation/macrocephaly | 1878 |
| TRA2B | NF | NF | NF |
| MARCH7 | 1/0 | NF | NF |
| LIMD1 | 0/2 | mental retardation/seizures | 2727 |
| RBMX | 3/0 | NF | NF |
| NFIB | 6/1 | mental retardation | 2385 |
| SLFN13 | 0/1 | mental retardation/cerebellar hypoplasia | 254309 |
| TAOK1 | 1/0 | NF | NF |
| RBBP4 | 1/0 | speech delay | 3456 |
| TTC14 | 0/1 | mental retardation | 251819 |
| NDEL1 | 0/2 | mental retardation | 4165 |
| ISLR2 | 4/1 | mental retardation/seizures | 2853 |
| ZNF519 | 3/0 | mental retardation/lissencephaly | 1581 |
| AP1S2 | 1/0 | mental retardation/seizures | 2376 |
| GPC6 | 1/1 | mental retardation | 221 |
| HDAC2 | 1/1 | mental retardation/microcephaly/seizures | 2498 |
| TOP1 | NF | NF | NF |
| KIAA0753 | 1/3 | mental retardation | 4165 |
| GNB2L1 | 3/1 | mental retardation/macrocephaly | 248184 |
Genes that represent potential candidates for neuronal migration and proliferation defects. In bold, genes that have already been described to play a role in neuronal migration/proliferation. Most of the genes are located in deletion/duplication regions in human patients and represent potential candidate genes for mental retardation and neurocognitive disabilities. NF = not found, indicates that no patient with neurocognitive phenotype was reported at the time of the analysis.
Cell cycle regulation is commonly altered in all mutants
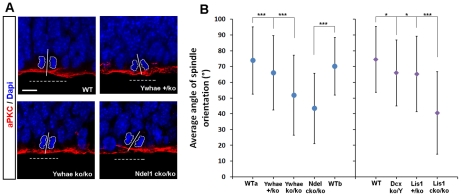
Many of the relevant findings obtained from our gene expression and bioinformatic analyses have been previously demonstrated or tested in vivo and in vitro with some of our mutants, confirming the validity of our overall approach. For example, differential expression analysis indicated enrichment of genes in WT at E14 for cell cycle processes and networks, while all mutants displayed a repression of the same processes at E14. These findings are consistent with previous publications in Lis1 [16]–[18] and Dcx mutants [14]. For example, Lis1 is essential for the regulation of spindle orientation during neural progenitor divisions [17], and loss of Dcx also leads to spindle positioning defects at the VZ during radial glial divisions [14]. To further examine this issue, we measured the spindle orientation of dividing apical progenitors from E14 Ywhae heterozygous, Ywhae homozygous and Ndel1cko/ko embryonic brains (Figure 5A) and found similar randomization of spindle orientation as we previously found in Lis1 and Dcx mutants (Figure 5B). WT embryos displayed radial glial progenitor cells with more vertical spindle orientation (73.8°±21.3°, n>150 cells WTa; 70.2°±18.3°, n>100 WTb) than radial glial progenitors from Ywhae+/ko (66.05°±23.57°, n>500 cells), Ywhaeko/ko (51.8°±25.3°, n>150 cells) and Ndel1cko/ko (43.4°±22.4°, n>100 cells) embryonic brains. The spindle orientation data across all genotypes strongly support the bio-computational finding that cell cycle is altered in these mutants. Interestingly, this data also suggested that cell cycle genes in Ndel1cko/ko were down-regulated at all phases of cell cycle. Consistently, we found a more severe spindle orientation phenotype in this mutant.
Figure 5. The average angle of spindle orientation is randomized in all mutants.
Analysis of spindle orientation and apical polarity at the ventricular surface of embryonic mouse brains. A) Spindle orientation analysis of cells in telophases and immunostaining of atypical PKC across the mutants. The localization of atypical PKC (red) is normal indicating that apical polarity is preserved. Scale bar, 10 µm. B) Quantification of the average angle of spindle positioning during radial glial mitotic divisions at E14. On the left, the average angle of spindle orientation is affected by decreasing dosage of Ywhae (100%>50%>0%) and Ndel1 (100%>35%) proteins. On the right, the average angle of spindle orientation is affected by decreasing dosage of Dcx (100%>0%) and Lis1 (100%>50%>35%) proteins as already published [14], [17]. *p-value<0.05; ***p-value<0.001.
Cell adhesion processes are commonly altered in all mutants
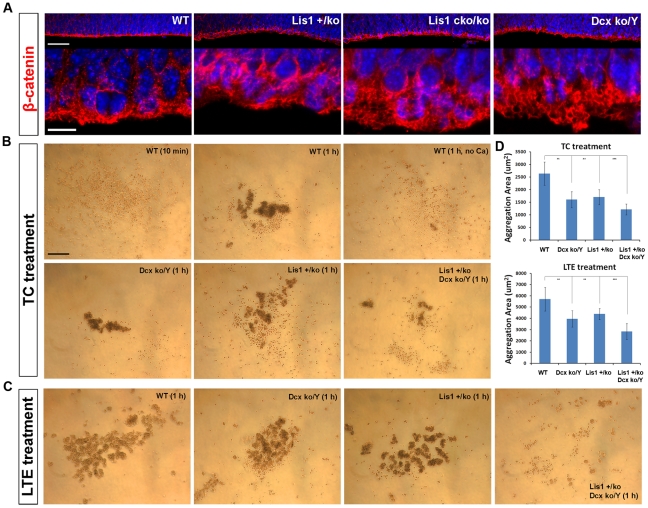
The second most represented biological process altered in all mutants at both E14 and P0 was cell adhesion. At E14 all mutants consistently displayed reduced expression of cell adhesion pathways and processes compared to WT, while at P0 the same and other cell adhesion processes were up-regulated in the mutants. Cell adhesion processes were also found enriched in several modules of the Lis1 mutants in WGCNA co-expression analysis (Figure 2E) that were not significantly represented in the WT clusters. To biologically investigate cell adhesion defects in the mutants, we stained E14 brain sections using β-catenin, an apical marker for cell adhesion. WT brains displayed a normal expression pattern of β-catenin along the VZ with most of the protein localized at the apical membrane of dividing cells (Figure 6A). Brain sections of Lis1+/ko, Lis1cko/ko and Dcxko/Y embryos displayed a broader expression pattern of β-catenin along the VZ with a less confined apical signal in dividing cells (Figure 6A). We further investigated cell adhesion in the Lis1+/ko and Dcxko/Y mutants and in the double Lis1+/ko;Dcxko/Y mutant using a functional assay to test the ability of primary cortical cells dissociated from P1 neurospheres to form aggregates in vitro (see methods). Lis1+/ko and Dcxko/Y as well as WT cells were able to form clusters in both the presence (TC) and absence (LTE) of Ca2+ (Figure 6B, 6C). The quantification of the cluster areas revealed a reduction in the clustering ability of the single mutant cells compared to WT in both the TC- and LTE- treatments (Figure 6D). The double Lis1+/ko;Dcxko/Y mutant displayed a more severe reduction of clusters that were much smaller and spread out in the wells (Figure 6B–6D). These biological data of cell adhesion support the findings we obtained with the bio-computational analysis of the mouse mutants.
Figure 6. Cell adhesion is altered in Lis1 mutants and exacerbated by Dcx mutation.
Analysis of cell adhesion by IHC and in vitro functional assay. A) β-catenin staining at the apical surface of E14 brain sections is more diffuse in the mutants compared to WT, indicating a less tightly compacted and organized surface. Scale bars, 50 µm and 100 µm. B,C) Aggregation assay (TC and LTE treatments respectively, see methods) demonstrated a reduced capacity of Lis1+/ko and Dcxko/Y neural progenitor cells to attach to each other and form aggregates. The Lis1+/ko;Dcxko/Y double mutant displayed more severe defects in cell aggregation with smaller clusters that were spread all around the wells requiring the acquisition of several images for the quantification. The other genotypes had clusters accumulating at the center of the wells requiring one image for the quantification. D) For both treatments, we quantified the areas of the aggregates and found significant differences in the mutants compared to WT (**p-value<0.01; ***p-value<0.001). The panel WT (1 h, no Ca2+) in B is the negative control for the TC treatment, in which the absence of Calcium prevents the formation of the aggregates. Scale bar, 200 µm.
Synaptogenesis is up-regulated only in Ywhae mutant at E14
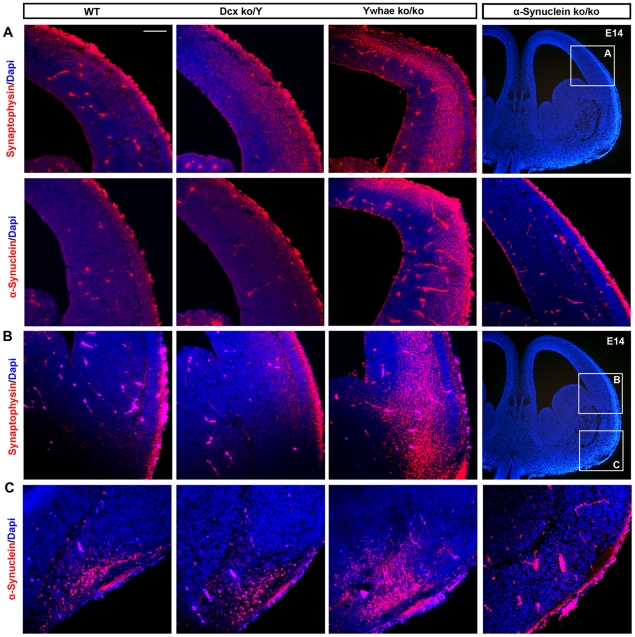
In addition to processes that are commonly altered in all mutants, there are some altered in individual mutants. For example, late developmental processes, such as synaptogenesis, synaptic contact, synaptic vesicle exocytosis and transmission of nerve impulses were found abnormally up-regulated in only the Ywhaeko/ko mutant at E14. We validated this unique finding in vivo by staining Ywhaeko/ko brain sections with the markers α-synuclein (present in the up-regulated gene list, position 32/1000) and synaptophysin. Consistent with the differential expression analysis, Ywhaeko/ko brains at E14 displayed a stronger signal for these proteins compared to WT and Dcxko/Y brains as controls (Figure 7). Increased α-synuclein staining was localized in the cortex (Figure 7A) and in the pyriform (olfactory) cortex (Figure 7C), while there was no specific staining in brains from α-synuclein knock-out mice. Increased synaptophysin was localized at the intermediate zone of the cortex and extending into the neostriatum in Ywhaeko/ko brains compared to controls (Figure 7A,7B).
Figure 7. Synaptogenesis is altered in the Ywhaeko/ko.
Immunostaining of E14 brain sections using markers specific for synaptogenesis to confirm the evidence obtained from the bio-computational analysis. A) α-Synuclein and Synaptophysin are increased in the cortex of the Ywhaeko/ko mutant. WT and Dcxko/Y are controls. B) Synaptophysin is increased in the intermediate zone of the cortex extending into the neostriatum. C) α-Synuclein is increased in the ventro-lateral area of the brain that specify for the pyriform (olfactory) cortex. Specificity of the α-synuclein antibody was confirmed on brain sections from the α-synuclein mouse knockout (right panels in A and C), while specificity of synaptophysin was tested by western-blotting (data not shown). Scale bar, 100 µm.
Discussion
In this study we sought to gain insight into normal brain development and how development was disrupted in our mouse models of neuronal migration defects. Using different bioinformatic analyses we identified networks of expressed genes that are both commonly and differentially altered across the mutants compared to WT. Some of the pathway findings are consistent with previous in vivo and in vitro findings in our mutants, whereas other novel results have been biologically confirmed in this study. We provided proof-of-principle that global gene expression and pathway analyses represent a productive and powerful approach to study gene expression alterations underlying brain development of mouse models of neuronal migration defects, even when the genes responsible for these defects code for pathways associated with cytoskeletal, and not transcriptional, processes. We also suggest that employing gene expression analysis on multiple mouse models of the neuronal migration can be used as a strategy to identify new candidate genes for human disorders of neuronal migration, and suggests that this can be used as a general strategy for other complex disease such as mental retardation, speech delay and epilepsy, for example.
Cell cycle and synaptogenesis define genotype-independent mouse brain development at E14 and P14
Analysis of WT brains at E14 and P14 using unsupervised PAM clustering revealed that cell cycle and synaptogenesis are the predominant processes and pathways differentially expressed during the development of the mouse brain at these two time points. We were unable to find processes or pathways enriched at P0, suggesting that P0 represents a transition or intermediate phase with gene expression patterns overlapping the other two developmental time points. However, we identified significant enrichment for biological processes at P0 in the differential analysis of the mutants, suggesting that genetic modification of genes important for neuronal migration and proliferation pathways lead to a delay of brain development or to a premature activation of genes normally expressed at later stages.
The finding that WT and mutant gene expression patterns were overall similar and clustered together by analysis PAM and WGCNA is consistent with the observation that WT and mutant brains display very similar overall organization at each time point. Each developmental stage is characterized by unique pathways and processes defined by specific gene expression patterns regardless of the genotype and needed for normal brain development. More focused analyses were performed to sift through these similarities to find profound genotype-dependent differences. Thus, global gene expression analysis of neuronal development using our neuronal migration mutants revealed important pathways and processes for normal development as well as the disruption of those normal patterns by these mutants.
Cell cycle and cell adhesion processes are commonly altered in all mutants
Differential expression analysis of all mutants at E14 and P0 revealed a common and significant down-regulation of cell cycle genes, consistent with our previous findings with Lis1 [17] and Dcx [14] mutants during neurogenesis and proliferation of neuronal progenitors. To support the hypothesis that LIS1, DCX, 14-3-3ε, NDEL1 proteins work in the same cell cycle pathways and genetically interact, we investigated the effects on spindle orientation of the Ndel1 and Ywhae mutations and found similar randomization of the angles in radial glial divisions at E14 as the Lis1 and Dcx mutants. These findings provide biological support that all mutants displayed down-regulation of genes involved in cell cycle processes and are consistent with our hypothesis that mutations in different genes belonging to the same pathway lead to similar phenotypes.
Similarly, differential expression analysis revealed different cell adhesion processes altered both at E14 and P0 across all mutants compared to WT. To investigate this finding biologically, we examined the localization of β-catenin in vivo and tested adhesion directly in Lis1 and Dcx mutants in vitro using a cell adhesion assay. In previous analyses of these two mutants [14], [18], we observed a broader expression pattern of aPKC with a thickened distribution at the ventricular surface, and a less organized distribution of the apical centrosomes. We hypothesized that this phenotype may be due to defects in cell adhesion molecules of neural progenitors and a disorganized apical ventricular surface. The analysis of the Lis1+/ko and Dcxko/Y single mutants in vitro suggested that the aggregation of progenitor cells was altered compared to WT cells. Moreover, double Lis1+/ko;Dcxko/Y male mutants displayed more severe defects in forming aggregates compared to single mutants, further supporting that these two genes as part of the same molecular pathway. A role of LIS1 in cell adhesion has not been previously described. DCX has been reported to interact in vivo with the ubiquitin-specific protease Drosophila fat facets-related on X chromosome (DFFRX) [30]. Additionally, β-catenin co-immunoprecipitated with both DCX and DFFRX suggesting that these three proteins can associate in the same protein complex in embryonic brain. These findings suggest that DCX may play a role in the regulation of cell adhesion during brain development and our results further support this notion. Dcx and Ndel1 mutants displayed diverse branched migration modes with multiple leading processes [14], [18], and these phenotypes may also be due to defects in cell adhesion.
Cytoskeleton rearrangement correlates with neuronal migration and proliferation phenotypes of the mutants
Co-expression analysis performed simultaneously on all datasets was used to identify possible module-correlations with quantifiable phenotypes previously published from these mouse models using both in vivo and ex vivo methodologies. Several modules were significantly correlated with particular phenotypes. Spindle orientation displayed the highest correlation score with the largest number of modules of all phenotypes examined. Cytoskeleton processes were highly enriched in all correlated modules. These findings are consistent with the known role of these processes during brain development. LIS1, DCX, NDEL1 and 14-3-3ε proteins work together in a conserved pathway that regulates cytoplasmic dynein localization and function. The interaction of these proteins promotes the recruitment, stabilization and organization of microtubules and actin cytoskeleton [10] that eventually drives neuronal migration and cell division. The prominence of cytoskeletal pathways in these analyses validates that the analytic approaches we used were sufficient to detect such known biological effects.
Genotype-specific differences and common altered genes across the mutants
In addition to finding pathways and processes commonly altered in our mouse mutants, we gained important insights into pathways and genes that were specifically altered in single mutants. For instance, IPA analysis revealed that the expression of genes regulating cell morphology was down-regulated and specific to Dcxko/Y mutants at E14 and P0. DCX is a microtubule-associated protein (MAP) localized at the growth cone of growing axons and neurites that is required for proper neuronal migration and axonal wiring. We and others have shown that Dcx mutants display morphological abnormalities of migrating neurons in the cortices of E14 brains that consist of shape changes associated with increased number of branches and a typical neurite swelling detached from the cell body [14], [31], [32]. Several other studies demonstrated that regulation of Dcx and the Dcx family at the growth cone of axon and neurites is important for their stability and extension [33]–[35]. Moreover, we surprisingly observed activation of genes involved in immune response and inflammation signaling pathways in Ndel1 mutants at P0. We have not examined these processes in Ndel1 mutants, although it is clear that these effects are genotype specific and will require further investigation.
Venn analyses revealed genes that were commonly and differentially expressed in our mutants. Many of these genes function in the development of the central nervous system. For instance, comparison of the top 20 genes at E14 revealed several genes that are responsible or associated with CNS diseases. Deficiency of Phgdh (phosphoglycerate dehydrogenase, commonly down-regulated in Ywhae and Dcx mutants) leads to mental retardation associated with microcephaly and seizures (MIM: 601815, [36]). The gene Huwe1 (down-regulated only in Ywhae mutants) is a candidate for X-linked mental retardation (MIM: 300697, [37]). The gene Cdh7 (down-regulated only in Ndel1 mutants) plays a role in the formation and function of multipotent neural crest cells by regulating cell motility (MIM: 608892, [38]). The gene Rpgrip1 (commonly up-regulated in Ndel1 and Dcx mutants) plays a role in regulating the function and organization of photoreceptors (MIM: 605446, [39]). The two genes En2 and Glo1 (upregulated in Ywhae and Ndel1 mutants, respectively) are candidates for susceptibility to autism (MIM: 131310 and 138750, [40], [41]). Lastly, the gene Fkrp (up-regulated only in Ywhae mutant) is responsible for muscle-eye-brain disease including pachigyria and hydrocephalus (LIS type II, MIM: 253280, [42]). In addition to these genes, we detected a clear enrichment for Hox genes specific for Ndel1 mutants at E14, providing an entry point for future studies. We speculate that Ndel1 might be part of multiple pathways important during development as suggested by its crucial role in the Lis1 pathway regulated by two kinases (Cdk5 and Aurora-A, [43]), as well as roles in other signaling pathways such as the Wnt-GSK3beta/beta-catenin pathway [44].
Identification of candidate genes in neuronal migration/proliferation defects and neuro-developmental disorders
The similarities of gene expression in Lis1, Dcx and Ywhae mutants at E14 revealed other genes that may be involved in neuronal migration and proliferation defects. Proof-of-principle for this was the identification of Ndel1 as a gene commonly down-regulated in these mutants. In addition, SYNE2 is important for neurogenesis and neuronal migration in mouse [25], AP1S2 is mutated in X-linked mental retardation [26], and KIAA1009 and TAOK1 regulate cell division, spindle assembly and check-point signaling [27], [28]. We extended this analysis to determine whether these similarities in gene expression could uncover candidates for neuro-developmental disorders such as mental retardation with/without brain abnormalities or associated with neurological dysfunctions such as speech delay and seizures. Most of the identified genes (25/35 see Table 4) were indeed located in deleted/duplicated regions of patients with mental retardation. Several of these genes were also associated with brain abnormalities (ARGLU1, KIAA1009, SLFN13, ZNF519, HDAC2, GNB2L1), speech delay (MAP4K4, PABPN1, BRD8, SGCE, SGCE, RBBP4) and seizures (RBBP4, HERC2, LIMD1, ISLR2, AP1S2, HDAC2). These findings suggest that our study using different mutants of the same molecular pathway (neuronal migration) can be used as a valid and novel approach for the identification of new candidate genes in neuronal migration as well as genes that may be implicated in mental retardation and related conditions such as speech delay and epilepsy.
Conclusions
In this study, we employed GO enrichment analysis to compare differentially expressed canonical pathways and biological processes in mouse models for human neuronal migration defects. Although this approach provides several novel insights, several limitations remain due to the variable quality of functional gene annotations used to define pathways, gene networks and processes; limitations in sources of annotation information and the possible discrepancies in curation. The available gene ontogeny databases are curated based on in vitro data using cancer cell lines, so there is a lack of annotation based on whole animal phenotypes. We used two hand-curated databases with the purpose of comparing and complementing results from each database, and found consistent pathway/process data with high enrichment scores. As an example, we detected one inconsistent enrichment for cell cycle processes in IPA. This pathway contained a few growth factor genes rather than primary cell cycle regulatory genes, while in Metacore no enrichment was found for the same processes. As gene ontogeny databases improve, we expect that the use of pathway analysis will improve as well. Future studies to further investigate these novel common and unique pathways and genes uncovered by this global gene expression analysis of our mouse mutants will expand our understanding of brain development, provide insight into gene function, and may identify novel candidate genes for human neuro-developmental disorders.
Materials and Methods
Mouse mutants
Mice were housed in standard cages in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities at University of California, San Francisco (UCSF). All experiments followed the guidelines of University's animal care and use committee. Mice were maintained on a 12∶12 h light/dark cycle at 22°C. All mice were in a predominantly 129S6 background, but because of difficulties in getting viable mice for Lis1, Ywhae and Dcx mutations in a completely inbred background, there was some NIH Black Swiss contribution in these crosses. Therefore, WT littermates were used to control for background effects, and were designated as inbred WT129 (129S6), WTmixA (herein as WTa for Lis1, Ywhae and Dcx) and WTmixB (herein as WTb for Ndel1). WT mice were mated with Dcx+/ko females [20] to generate Dcxko/Y mice. WT and Lis1cko/cko males [16] were mated with Lis1+/ko females [15] to generate Lis1+/ko and Lis1cko/ko mice respectively. WT and Ndel1cko/cko males were mated with Ndel1+/ko females to generate Ndel1+/ko and Ndel1cko/ko mice respectively. WT and Ywhae+/ko males [8] were mated with Ywhae+/ko females to generate Ywhaeko/ko mice.
Microarrays and expression analysis
Gene expression levels were assessed with Affymetrix 430 2.0 mouse microarray chips. This chipset represents more than 45,000 genes and SNPs from the mouse genome. Three brains were collected for each genotype at each time point (E14, P0 and P14). Ywhaeko/ko and Dcxko/Y samples were collected and analyzed only at E14 and P0, due to the neonatal lethality of these mutants; WT129S6 only at P0 and P14. All the other WTs and mutants have been collected and analyzed at the three time points. RNA samples were extracted from whole brains at E14 and from dissected hemispheres at P0 and P14 using Trizol reagent (Invitrogen). The RNA was labeled and hybridized to the array by the NIH Neuroscience Microarray Consortium (http://arrayconsortium.tgen.org/np2/home.do). Microarray data was pre-processed via the Robust Multichip Average (RMA) method [45], as implemented in the Bioconductor suite of programs (http://www.bioconductor.org/) implemented in the R statistical programming language (http://www.r-project.org/). RMA pre-processing consists of three steps: background adjustment, normalization, and summarization [45]. Analysis of expression was performed using bio-weight analysis [46]. Bio-weight is designed to produce a higher true positive rate compared to traditionally used t-test and p-value-based methods of analysis, especially for smaller sample sets. Bio-weight takes into consideration the across-replicate impact of traditional fold change analysis as well as the negative decimal log of p-values for the gene specific t-tests. This method allows the user to both retain over- and under-expressed genes that may be of biological significance and retain statistical stringency corresponding to a small p-value. Therefore, the bio-weight value can be described as the product of absolute average fold change and negative log of p-value, accounting for small p-values and large fold changes, and providing a smooth transition between the two. For a detailed explanation of bio-weight analysis, see Rosenfeld et al. [46]. Expression levels of genes obtained from the different time points in the WTa and WTb mice were examined for patterns of increasing or decreasing differential expression over the three time points. The genes that exhibited different expression patterns over time were then ranked by bio-weight analysis. A similar approach was employed for determining dosage dependent differential expression of genes for Lis1 and Ndel1 mutants.
Unsupervised clustering and WGCNA analysis
Expression values associated with individual probes were collapsed into overall gene expression values by averaging the expression values of all probes that belong to the same gene. Gene expression values from the different genotypes at the three time points (averaged across three samples) were temporally ordered, scaled and centered to control for differences in absolute levels of expression for individual genes. The gene expression ‘profiles’ over time for the different genotypes were then clustered using ‘partitioning around medoids (PAM),’ a more robust version of K-means clustering, using Euclidean distance as implemented in the function ‘pam()’ implemented in the R statistical package (http://cran.r-project.org/). The optimal number of clusters was chosen based on the visual examination of silhouette plots to obtain the highest average silhouette width with the least amount of “leaking” data points outside of the clusters, and this occurred using four clusters.
We performed weighted gene co-expression network analysis (WGCNA) using the WGCNA package [47] by constructing a signed (i.e., bidirectional) network and used a hybrid dynamic branch cutting method to assign individual genes to modules. This method combines the advantages of hierarchical clustering and partitioning around medoids. We chose a value of 0.1 as a cutoff parameter that indirectly impacts the number of modules in WGCNA (mergeCutHeight parameter), as this value yielded modules that were more highly enriched in biologically relevant pathways and processes compared to the default value of 0.25. All other parameters were kept at default values.
We also pursued phenotype correlation analysis by assessing the Pearson correlation coefficient between available phenotypes measured on a subset of strains and the relevant elements of each of modules eigengene obtained from the WGCNA analysis using routines in the R statistical package (http://cran.r-project.org/) using the rcorr() function of Hmisc package. In addition, we examined the correlation structure of the genes using Cytoscape (http://www.cytoscape.org/) and routines in R as well. Correlation matrices were transformed in the circle-shaped networks using Cytoscape (http://www.cytoscape.org/), where the thicker lines represent higher correlation matrices (Figure 3A, 3C). We color-coded the WTs (red) and the two mutants (green), overlapped the networks and displayed the relative changes in correlation by color preponderance (i.e., red is higher in WT and green is higher in mutants compared to mustard, which reflects no change or a change <10%). A change in the direction of the correlation is represented by a solid (direct correlation) or dashed (inverse correlation) line. Lis1/WTa and Ndel1/WTb Correlation matrices were also transformed into heat maps using Heatmap builder (http://ashleylab.stanford.edu/tools_scripts.html) (Figure S4A) to facilitate visual representation.
Gene enrichment analysis and visualization tools
Gene Ontology (GO) enrichment analysis was performed using Ingenuity Pathway Analysis (IPA Ingenuity Systems, www.ingenuity.com) and Metacore (www.genego.com/metacore.php). All reported pathways and biological processes are listed according to their GO enrichment score provided by the two software packages as −Log (p-values) and with a False Discovery Rate (FDR) <0.05%. Additionally, the significance of the p-values by FDR was also checked using the spreadsheet FDR-calculator available at http://www.rowett.ac.uk/~gwh/fdr.html. Heat maps were generated using the Heatmap builder (http://ashleylab.stanford.edu/tools_scripts.html).
Immuno-histochemistry and spindle orientation
Analyses were performed on experimental animals with age-matched litter-mate controls. Brains were fixed in 4% paraformaldehyde and cryoprotected in 30% sucrose for freezing or treated with 70% ethanol for paraffin embedding. At least three brains were analyzed for each experiment and matched sections were used for immuno-histochemistry (IHC) and counter-stained with DAPI (blue). Sections were roughly matched by counting the number of coronal sections starting at the rostral-most edge of the brain, and then more closely matched by anatomical landmarks. Primary antibodies [atypical PKC, α-Synuclein (mouse, Sigma) and Synaptophysin (mouse, Chemicon)] were diluted 1∶200 in blocking agent (PBS, 0.2% Triton X-100, 5% Normal Goat Serum) and incubated over-night. Secondary antibodies were diluted 1∶500 and incubated for 1 h. Embryonic day 14 (E14) brains sections were prepared at 12 µm thickness and counter-stained with DAPI to observe telophase mitotic cells at the ventricular surface. Captured images were analyzed with ImagePro v.6 to determine the angles of the spindle orientation by drawing a line parallel to the ventricular surface and a line parallel to the axis of the dividing cells (see Figure 5A) and average angle values were calculated from these measurements [also see 14], [17], [18].
Cell aggregation assay
Primary embryonic cortical cultures were established from E14 dissociated brains and cultured to generate neurospheres using a modification [18] of previously described methods [48]–[50]. Neurospheres were collected for Ca2+-dependent (TC-treatment) and Ca2+-independent (LTE-treatment) cell adhesion assays using previously described methods with slight modifications [51]. The two systems use different concentrations of trypsin with/without Ca2+ (respectively) for proteolysis protection. The Ca2+-dependent system is represented by a single family of adhesion molecules, the cadherins, while the Ca2+-independent system may include multiple classes of adhesion proteins. In brief, neurospheres were washed with HBSS and treated with TC and LTE solutions separately (for recipes, see [51]) for 30 min with shaking every 5–10 min. Cell clusters were then dispersed into single cells by pipetting with fire-polished Pasteur pipette. Single cells were centrifuged and resuspended in HBSS with 0.5% (w/v) of Soybean trypsin inhibitor solution and counted using a hematocytometer. For both assays (TC and LTE) we plated 5×106 cells/mL in each well of separate plates (12 well plates; 1 assay/brain; 3 brains/genotype). In each well of the TC- and LTE- treatment, DNase (100 mg/mL) and MgCl2 (100 mM) were added, while in the TC-wells only we added CaCl2 (1 M). Plates were placed on a gyrating shaker at ∼80 rpm and incubated for 1 h at 37°C. Aggregates areas were quantified using ImageJ (http://rsbweb.nih.gov/ij/). Of note, WT and single mutant aggregates were always found in the center of the well, requiring one single image for the quantification; while double mutant aggregates were always spread out in the well requiring the acquisition of multiple images for a precise quantification of the cell clusters.
Supporting Information
Strategy of the gene expression and bio-computational analyses used to define the biological processes and/or molecular functions during normal brain development and altered in mouse models for neuronal migration defects. Brains were isolated from E14 embryos, and cortices were isolated from P0 and P14 neonates. RNA was purified, labeled and hybridized to Affymetrix 430 2.0 mouse microarray for global analysis of RNA expression. We hypothesized that E14 brain gene expression will be characterized by biological pathways and processes important for early developmental processes such as neurogenesis and neuronal migration; while P0 and P14 brain gene expression will be characterized by biological pathways and processes important for brain maturation. In step 1, we defined the pathways and processes that are up-/down-regulated during normal brain development at each stage using differential expression (DE) analysis of WT datasets (see methods). In step 2, we investigated the potential similarities or unexpected differences in gene expression patterns of all genotypes at all time points using PAM and WGCNA analyses. In step 3, we investigated the gene expression differences between Lis1 and Ndel1 mutants using WGCNA analysis, in the Lis1 mutant alone and across all mutants using differential expression analysis. All lists of genes from the DE analysis, expression patterns and modules were analyzed by Gene Ontology (GO) enrichment to define the biological processes or molecular functions affected. Part of the bio-computational findings has been tested in-vivo or in-vitro to provide biological confirmation.
(0.23 MB TIF)
Cell cycle processes define early brain development in mouse. Differential expression analysis followed by pathway enrichment via Metacore (A, B) and Ingenuity (C) software defines the canonical maps (A,B) or molecular functions (C) that are up-/down-regulated at each developmental stage versus the others. A) Up-regulated (blue) and down-regulated canonical maps (yellow) sorted by intensity at E14. B) Up-regulated and down-regulated canonical maps sorted by intensity at P14. No gene expression signature is characteristic of P0. C) Up-regulated and down-regulated canonical maps sorted by pathway category. Intensity values refer to enrichment values provided by Metacore (A,B) or Ingenuity (C) and calculated as −Log(p-value) (FDR<0.05). Cell cycle regulatory genes in WT from IPA analysis: ANLN, BRCA1, BUB3, CCNB1, CCND1, CCNF, CDC123, CDC14A, CDC25C, CENPF, CHEK1, CHEK2, CHUK, CSNK1D, DDX11, DLG7, E2F2, E2F5, HIPK2, HK2, INCENP, JOSD3, JUB, KIF22, KIFC1, MCM10, MTCP1, NEK2, NEUROG1, NPAT, NR2E1, NUSAP1, ORC2L, PA2G4, PAWR, PCAF, PCM1, PLAGL1, PSMG2, PTPN2, PTTG1, RABGAP1, RASSF1, RPA1, SUGT1, TAF1, TCEB3, TERF1, TGIF1, TTK.
(2.01 MB TIF)
Four clusters best fit the unsupervised PAM clustering analysis. We determined the best fit for the number of clusters before running the PAM analysis. A) Silhouette analysis indicated that 4 was the optimal number of clusters, since we obtained the highest average silhouette width with the least amount of leaking data points (bars pointing leftwards). This optimal clustering was confirmed by projecting the gene expression profiles as linear functions of the variables (genotypes and time-points) in two dimensional space. B) Visual representation of the clustering of the expression ‘profiles’, where each point (character) represents the profile of one gene summarized by two discriminant coordinates (principal components), and colored according to which cluster the gene has been assigned to. This projection of the data points is mapped onto two dimensional space to demonstrate the quality of the clustering. Data points are distributed close to each other in contiguous clusters with very little overlap between each cluster. C) Co-expression network analysis by WGCNA clustered gene expression data into 25 modules. Fourteen modules displayed significant enrichment (Table 1), and these modules contained genes, pathways and processes consistent with the PAM analysis results. The leaves in the dendrogram represent individual genes and the closer the leaves are to each other, the more highly correlated their expression is, measured at the three time-points across the various genotypes. The assignment of genes into modules is shown in the band plot at the bottom of the figure, where each gene along the x-axis is represented by a vertical line colored according to the module to which the gene has been assigned by WGCNA. D) Overlap analysis of the top 18 modules (containing at least 100 genes from the WGCNA) with the four PAM clusters. The four colored bars shown for each module represent the proportion of the genes in each of four clusters identified by PAM analysis that compose each module. All the modules were included in the PAM clusters and displayed consistent gene enrichment.
(2.32 MB TIF)
Gene expression in the Lis1cko/ko mutant is more disrupted than Ndel1cko/ko mutants compared to WT. A) Analysis of the correlation changes in the pair-wise comparisons revealed more severe alteration in the Lis1 mutant than Ndel1. B) The results of the correlation matrices were transformed in heat maps to easily identify correlation differences between the paired genotypes (Lis1cko/ko versus WTa; Ndel1cko/ko versus WTb). As shown in the quantification graph (Figure 2B, 2D), for instance Translation in mitochondria displays several changes in the WTa/Lis1 comparison, while not as much in the WTb/Ndel1 comparison.
(1.42 MB TIF)
LIS1 protein reduction causes gene expression alterations at early and later developmental time-points. Ingenuity pathways analysis (IPA) of the differential gene expression analysis with decreasing dosage levels of LIS1 (100%>50>35%). Up-regulated (blue) and down-regulated (yellow) processes are sorted by category and by intensity according to the time points (top left, E14; to right P0; bottom left, P14). IPA analysis confirmed most of the Metacore pathways and biological processes altered in the Lis1 mutants and displayed additional differences. For instance, down-regulation of genes involved in migration of neurons and movement of neurons were evident in the Lis1 mutant and not identified with the Metacore software. Intensity values refer to enrichment values provided by IPA and calculated as −Log(p-value) (FDR<0.05).
(0.95 MB TIF)
Biological processes are commonly and specifically altered in the mutants. IPA analysis confirmed most of the Metacore pathways and biological processes commonly altered in the mutants and displayed additional differences. For instance, cell morphology was found down-regulated exclusively in the Dcx mutant and not identified using the Metacore software. Up-regulated (blue) and down-regulated (yellow) processes are sorted by category and by intensity according to the time-points (E14; P0). A) IPA analysis across the mutants at E14. B) IPA analysis across the mutants at P0. Intensity values refer to enrichment values provided by IPA and calculated as −Log(p-value) (FDR<0.05).
(1.29 MB TIF)
Acknowledgments
We thank Lana Bogdanova and Donna Holland for animal handling, Jeehee Hong and Patricia LaPorte for technical assistance, all members of the Wynshaw-Boris' laboratory for the useful comments during the manuscript revision, Peter Langfelder for his repeated useful advice regarding the use of Weighted Gene Co-expression Network analysis tool (WGCNA), Feng He for assisting in the statistical expression analysis, Traver Hart for the use of Cytoscape, and Robert Nussbaum and Yien-Ming Kuo for providing the α-synuclein knockout mouse.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by the NIH (NS041310 and a supplement to fund the array studies to AW-B). NJS and OL are funded in part by UL1 RR025774-03. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 2.Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, et al. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 3.Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 4.des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Dobyns WB. Lissencephaly and the molecular basis of neuronal migration. Hum Mol Genet. 2003;12(Spec No 1):R89–R96. doi: 10.1093/hmg/ddg086. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, et al. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 7.Niethammer M, Smith DS, Ayala R, Peng J, Ko J, et al. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 8.Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, et al. 14–3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 9.Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: a genetic look at neocortical development. Nat Rev Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- 10.Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 11.Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- 12.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, et al. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–21. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pramparo T, Youn YH, Yingling J, Hirotsune S, Wynshaw-Boris A. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J Neurosci. 2010;30:3002–12. doi: 10.1523/JNEUROSCI.4851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 16.Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, et al. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youn YH, Pramparo T, Hirotsune S, Wynshaw-Boris A. Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice. J Neurosci. 2009;29:15520–15530. doi: 10.1523/JNEUROSCI.4630-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–27. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbo JC, Deuel TA, Long JM, LaPorte P, Tsai E, et al. Doublecortin is required in mice for lamination of the hippocampus but not the neocortex. J Neurosci. 2002;22:7548–7557. doi: 10.1523/JNEUROSCI.22-17-07548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, et al. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- 22.Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–84. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Y, Yu W, Li Y, Yang Z, Yan X, et al. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–66. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16:1535–48. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–87. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leon A, Omri B, Gely A, Klein C, Crisanti P. QN1/KIAA1009: a new essential protein for chromosome segregation and mitotic spindle assembly. Oncogene. 2006;25:1887–95. doi: 10.1038/sj.onc.1209215. [DOI] [PubMed] [Google Scholar]
- 28.Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–64. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 29.Tarpey PS, Stevens C, Teague J, Edkins S, O'Meara S, et al. Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet. 2006;Dec 79:1119–24. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friocourt G, Kappeler C, Saillour Y, Fauchereau F, Rodriguez MS, et al. Doublecortin interacts with the ubiquitin protease DFFRX, which associates with microtubules in neuronal processes. Mol Cell Neurosci. 2005;28:153–64. doi: 10.1016/j.mcn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Kappeler C, Saillour Y, Baudoin JP, Tuy FP, Alvarez C, et al. Branching and nucleokinesis defects in migrating interneurons derived from doublecortin knockout mice. Hum Mol Genet. 2006;15:1387–1400. doi: 10.1093/hmg/ddl062. [DOI] [PubMed] [Google Scholar]
- 32.Koizumi H, Higginbotham H, Poon T, Tanaka T, Brinkman BC, et al. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- 33.Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, et al. DCX, a new mediator of the JNK pathway. EMBO J. 2004;23:823–32. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deuel TA, Liu JS, Corbo JC, Yoo SY, Rorke-Adams LB, et al. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 2006;49:41–53. doi: 10.1016/j.neuron.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 35.Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, et al. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129:579–91. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaeken J, Detheux M, Van Maldergem L, Foulon M, Carchon H, et al. 3-Phosphoglycerate dehydrogenase deficiency: an inborn error of serine biosynthesis. Arch Dis Child. 1996;74:542–545. doi: 10.1136/adc.74.6.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Molec Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 40.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junaid MA, Kowal D, Barua M, Pullarkat PS, Sklower Brooks S, et al. Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor. Am J Med Genet. 2004;131A:11–17. doi: 10.1002/ajmg.a.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beltran-Valero de Bernabe D, Voit T, Longman C, Steinbrecher A, Straub V, et al. Mutations in the FKRP gene can cause muscle-eye-brain disease and Walker-Warburg syndrome. J Med Genet. 2004;41:e61. doi: 10.1136/jmg.2003.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wynshaw-Boris A, Pramparo T, Youn YH, Hirotsune S. Lissencephaly: mechanistic insights from animal models and potential therapeutic strategies. Semin Cell Dev Biol. 2010;21:823–30. doi: 10.1016/j.semcdb.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh KK, Ge X, Mao Y, Drane L, Meletis K, et al. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 46.Rosenfeld S, Wang T, Kim Y, Milner J. Numerical deconvolution of cDNA microarray signal: simulation study. Ann N Y Acad Sci. 2004;1020:110–23. doi: 10.1196/annals.1310.012. [DOI] [PubMed] [Google Scholar]
- 47.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rietze RL, Reynolds BA. Neural stem cell isolation and characterization. Methods Enzymol. 2006;419:3–23. doi: 10.1016/S0076-6879(06)19001-1. [DOI] [PubMed] [Google Scholar]
- 50.Ishii Y, Matsumoto Y, Watanabe R, Elmi M, Fujimori T, et al. Characterization of neuroprogenitor cells expressing the PDGF beta-receptor within the subventricular zone of postnatal mice. Mol Cell Neurosci. 2008;37:507–518. doi: 10.1016/j.mcn.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Takeichi M, Nakagawa S. Cadherin-dependent cell-to-cell adhesion. 1998. Current Protocols in Cell Biology 9.3.1–9.3.15 by John Whiley & Sons, Inc. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strategy of the gene expression and bio-computational analyses used to define the biological processes and/or molecular functions during normal brain development and altered in mouse models for neuronal migration defects. Brains were isolated from E14 embryos, and cortices were isolated from P0 and P14 neonates. RNA was purified, labeled and hybridized to Affymetrix 430 2.0 mouse microarray for global analysis of RNA expression. We hypothesized that E14 brain gene expression will be characterized by biological pathways and processes important for early developmental processes such as neurogenesis and neuronal migration; while P0 and P14 brain gene expression will be characterized by biological pathways and processes important for brain maturation. In step 1, we defined the pathways and processes that are up-/down-regulated during normal brain development at each stage using differential expression (DE) analysis of WT datasets (see methods). In step 2, we investigated the potential similarities or unexpected differences in gene expression patterns of all genotypes at all time points using PAM and WGCNA analyses. In step 3, we investigated the gene expression differences between Lis1 and Ndel1 mutants using WGCNA analysis, in the Lis1 mutant alone and across all mutants using differential expression analysis. All lists of genes from the DE analysis, expression patterns and modules were analyzed by Gene Ontology (GO) enrichment to define the biological processes or molecular functions affected. Part of the bio-computational findings has been tested in-vivo or in-vitro to provide biological confirmation.
(0.23 MB TIF)
Cell cycle processes define early brain development in mouse. Differential expression analysis followed by pathway enrichment via Metacore (A, B) and Ingenuity (C) software defines the canonical maps (A,B) or molecular functions (C) that are up-/down-regulated at each developmental stage versus the others. A) Up-regulated (blue) and down-regulated canonical maps (yellow) sorted by intensity at E14. B) Up-regulated and down-regulated canonical maps sorted by intensity at P14. No gene expression signature is characteristic of P0. C) Up-regulated and down-regulated canonical maps sorted by pathway category. Intensity values refer to enrichment values provided by Metacore (A,B) or Ingenuity (C) and calculated as −Log(p-value) (FDR<0.05). Cell cycle regulatory genes in WT from IPA analysis: ANLN, BRCA1, BUB3, CCNB1, CCND1, CCNF, CDC123, CDC14A, CDC25C, CENPF, CHEK1, CHEK2, CHUK, CSNK1D, DDX11, DLG7, E2F2, E2F5, HIPK2, HK2, INCENP, JOSD3, JUB, KIF22, KIFC1, MCM10, MTCP1, NEK2, NEUROG1, NPAT, NR2E1, NUSAP1, ORC2L, PA2G4, PAWR, PCAF, PCM1, PLAGL1, PSMG2, PTPN2, PTTG1, RABGAP1, RASSF1, RPA1, SUGT1, TAF1, TCEB3, TERF1, TGIF1, TTK.
(2.01 MB TIF)
Four clusters best fit the unsupervised PAM clustering analysis. We determined the best fit for the number of clusters before running the PAM analysis. A) Silhouette analysis indicated that 4 was the optimal number of clusters, since we obtained the highest average silhouette width with the least amount of leaking data points (bars pointing leftwards). This optimal clustering was confirmed by projecting the gene expression profiles as linear functions of the variables (genotypes and time-points) in two dimensional space. B) Visual representation of the clustering of the expression ‘profiles’, where each point (character) represents the profile of one gene summarized by two discriminant coordinates (principal components), and colored according to which cluster the gene has been assigned to. This projection of the data points is mapped onto two dimensional space to demonstrate the quality of the clustering. Data points are distributed close to each other in contiguous clusters with very little overlap between each cluster. C) Co-expression network analysis by WGCNA clustered gene expression data into 25 modules. Fourteen modules displayed significant enrichment (Table 1), and these modules contained genes, pathways and processes consistent with the PAM analysis results. The leaves in the dendrogram represent individual genes and the closer the leaves are to each other, the more highly correlated their expression is, measured at the three time-points across the various genotypes. The assignment of genes into modules is shown in the band plot at the bottom of the figure, where each gene along the x-axis is represented by a vertical line colored according to the module to which the gene has been assigned by WGCNA. D) Overlap analysis of the top 18 modules (containing at least 100 genes from the WGCNA) with the four PAM clusters. The four colored bars shown for each module represent the proportion of the genes in each of four clusters identified by PAM analysis that compose each module. All the modules were included in the PAM clusters and displayed consistent gene enrichment.
(2.32 MB TIF)
Gene expression in the Lis1cko/ko mutant is more disrupted than Ndel1cko/ko mutants compared to WT. A) Analysis of the correlation changes in the pair-wise comparisons revealed more severe alteration in the Lis1 mutant than Ndel1. B) The results of the correlation matrices were transformed in heat maps to easily identify correlation differences between the paired genotypes (Lis1cko/ko versus WTa; Ndel1cko/ko versus WTb). As shown in the quantification graph (Figure 2B, 2D), for instance Translation in mitochondria displays several changes in the WTa/Lis1 comparison, while not as much in the WTb/Ndel1 comparison.
(1.42 MB TIF)
LIS1 protein reduction causes gene expression alterations at early and later developmental time-points. Ingenuity pathways analysis (IPA) of the differential gene expression analysis with decreasing dosage levels of LIS1 (100%>50>35%). Up-regulated (blue) and down-regulated (yellow) processes are sorted by category and by intensity according to the time points (top left, E14; to right P0; bottom left, P14). IPA analysis confirmed most of the Metacore pathways and biological processes altered in the Lis1 mutants and displayed additional differences. For instance, down-regulation of genes involved in migration of neurons and movement of neurons were evident in the Lis1 mutant and not identified with the Metacore software. Intensity values refer to enrichment values provided by IPA and calculated as −Log(p-value) (FDR<0.05).
(0.95 MB TIF)
Biological processes are commonly and specifically altered in the mutants. IPA analysis confirmed most of the Metacore pathways and biological processes commonly altered in the mutants and displayed additional differences. For instance, cell morphology was found down-regulated exclusively in the Dcx mutant and not identified using the Metacore software. Up-regulated (blue) and down-regulated (yellow) processes are sorted by category and by intensity according to the time-points (E14; P0). A) IPA analysis across the mutants at E14. B) IPA analysis across the mutants at P0. Intensity values refer to enrichment values provided by IPA and calculated as −Log(p-value) (FDR<0.05).
(1.29 MB TIF)