Abstract
Cytoplasmic male sterility (CMS) has been identified in numerous plant species. Brassica napus CMS plants, such as Polima (pol), MI, and Shaan 2A, have been identified independently by different researchers with different materials in conventional breeding processes. How this kind of CMS emerges is unclear. Here, we report the mitochondrial genome sequence of the prevalent mitotype in the most widely used pol-CMS line, which has a length of 223,412 bp and encodes 34 proteins, 3 ribosomal RNAs, and 18 tRNAs, including two near identical copies of trnH. Of these 55 genes, 48 were found to be identical to their equivalents in the “nap” cytoplasm. The nap mitotype carries only one copy of trnH, and the sequences of five of the six remaining genes are highly similar to their equivalents in the pol mitotype. Forty-four open reading frames (ORFs) with unknown function were detected, including two unique to the pol mitotype (orf122 and orf132). At least five rearrangement events are required to account for the structural differences between the pol and nap sequences. The CMS-related orf224 neighboring region (∼5 kb) rearranged twice. PCR profiling based on mitotype-specific primer pairs showed that both mitotypes are present in B. napus cultivars. Quantitative PCR showed that the pol cytoplasm consists mainly of the pol mitotype, and the nap mitotype is the main genome of nap cytoplasm. Large variation in the copy number ratio of mitotypes was found, even among cultivars sharing the same cytoplasm. The coexistence of mitochondrial mitotypes and substoichiometric shifting can explain the emergence of CMS in B. napus.
Introduction
The major contribution of the mitochondrion to plant growth and development is the production of energy [1]. In some higher plants, the economic significance of mitochondria is boosted by its role in the determination of cytoplasmic male sterility (CMS). To outline characteristics of the plant mitochondrial genome and identify CMS-related genes, the mitochondrial genomes of more than 10 fertile or sterile plant species have been sequenced, including Arabidopsis thaliana [2], Beta vulgaris [3], [4], Oryza sativa [5], [6], [7], Brassica napus [8], Zea mays [9], [10], Nicotiana tabacum [11], Triticum aestivum [12], Vitis vinifera [13], Citrullus lanatus, and Cucurbita pepo [14]. Research revealed that unique deleterious genes inserted into the mitochondrial genome confer CMS traits on natural plant germplasm [15], [16].
In higher plants, natural CMS has been identified in more than 150 species over the past century [17]. CMS emergence undoubtedly stems from alteration of the mitochondrial genome [15], [17], but the question is whether this kind of genomic alteration is the result of toxic genes being inserted during the past century or from the substoichiometric shifting of mitochondrial DNA molecules. Research in a few crops has demonstrated that substoichiometric shifting results in the inherent CMS-triggering mitotype dominating mitochondrial genome constituents [18]–[20], which results in the occurrence of CMS. In the common bean, a progenitor mitochondrial form containing a three-molecule CMS-associated configuration is universally present in the germplasm [18], and substoichiometric shifting can explain the emergence of CMS. In cybrids between CMS radish and B. napus, mitochondrial genomic components of the two species coexist in stable rapeseed progenies, and the phenomena of substoichiometric shifting has been observed between male fertile and sterile plants [21].
In B. napus, at least five natural occurrences of CMS have been independently reported [22]–[27]. Genetic experiments and restriction mapping have shown that the CMS lines can be classified as two cytoplasmic groups, one including T-CMS and S-CMS [22]–[24], which is commonly referred to as the “nap” cytoplasm present in normal fertile accessions, and the other group as “pol” cytoplasm including pol-CMS, MI CMS, and shaan 2A CMS [25]–[27], which has been the most widely applied line in the world. During the past few decades, the fact that the grouped CMS accessions from multiple independent findings have the same genetic regulation has been a perplexing issue for researchers [28]. In order to identify the mechanism for natural CMS in B. napus, the main mitochondrial genome of pol-CMS was sequenced and compared to the sequence of Westar (nap cytoplasm), uncovering structural and evolutionary differences. Based on the mitotype-specific sequences of the mitochondrial genome in B. napus, the constituents of the mitochondrial genome were detected by PCR amplification.
Results
The pol mitochondrial genome
A single circular mitochondrial genome 223,412 bp (EMBL accession number: FR715249) in size was obtained by shotgun sequencing. Because of heteroplasmy in B. napus, we refer to the mtDNA sequence of the fertile rapeseed variety Westar as the nap mitotype, in contrast to the polima-derived pol mitotype. The pol mitotype sequence is 1,559 bp longer than that of the nap mitotype, and the G+C content of the two mitotypes is 45.22% and 45.19%, respectively. Sequence alignment showed that 3.63% of nap and 4.53% of pol is mitotype-specific.
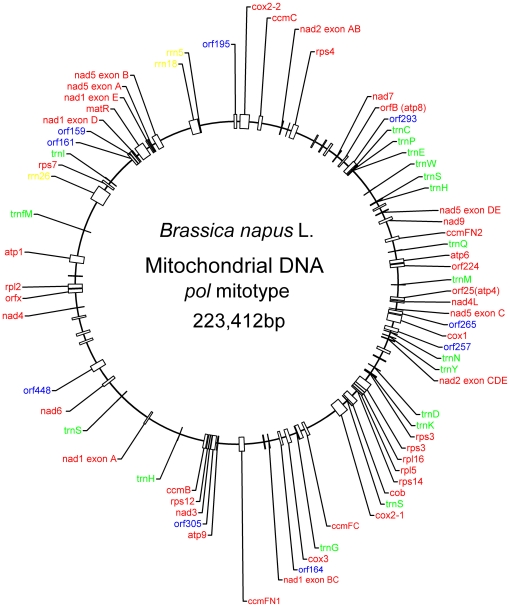
The pol sequence encodes 34 proteins, three ribosomal RNAs, and 18 tRNAs, constituting 17.34% of the genome (Figure 1). The sequence includes two near identical copies of trnH that differ by only one base pair, whereas only a single copy is present in the nap mitotype. Of the 55 genes present in pol, 48 have an identical copy in nap, implying that the known mitochondrial genes are well conserved in B. napus. Five of the six remaining genes in nap differ only marginally from their counterparts in pol (the exception was orf224). The atp1 sequences differ by one synonymous single nucleotide polymorphism (SNP), as do the trnC and trnE sequences. One non-synonymous SNP in cox1 produces a proline to leucine switch, but this has no effect on gene function. Similarly, a single SNP produces a proline to serine switch in cox2. The alignment of the pol and nap versions of orf224, which is generally accepted as the determinant of CMS in pol, revealed 84% nucleotide similarity and 77% peptide similarity.
Figure 1. Organization of the pol mitotype in B. napus.
A total of 55 genes with known function were identified, including 34 protein-coding genes (red), 3 ribosomal RNA genes (yellow), and 18 tRNA genes (green). Nine (out of 44) putative ORFs encoding at least 150 amino acids are marked in blue.
We found that the pol mitotype comprises 44 putative open reading frames (ORFs) encoding at least 100 amino acid residues. Of these ORFs, 35 are identical to their equivalents in nap, but the other ORFs are different (Table S1). Orf122 and orf132 are both present in pol but not in nap, whereas orf117b is unique in nap. None of these three sequences aligned with any known sequence in GenBank. Thus, the sequences were inferred to be the result of large insert sequences in the mitochondrial genome. Orf261 in nap and orf265 in pol partially aligned and were inferred to be generated by the rearrangement of syntenic regions in the mitochondrial genome.
Reorganization of the mitochondrial genome
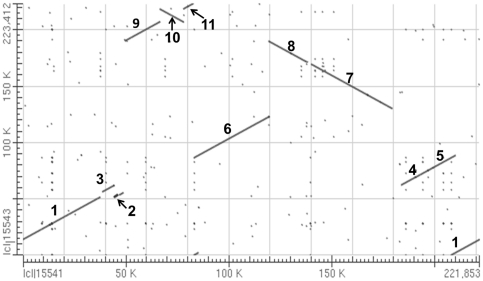
Analysis of the syntenic regions of pol and nap using the bl2seq algorithm suggested that the sequence as a whole consists of 11 syntenic regions (similarity >99%, length >2 kb). The orientation of eight of the regions is identical, but in the other three it is reversed (Figure 2). The syntenic regions are largely discrepant in the two genomic positions, though the genetic sequences are well conserved (Figure 2). A minimum of five recombination events is required to account for the structural differences between the two mitotypes.
Figure 2. Alignment of the nap and pol mitotype genomes.
The numbers refer to the syntenic regions with the pol mitotype sequence as a reference. Comparisons between nap (horizontal axis) and pol (vertical axis) indicated that the nucleotide sequences of the syntenic region are highly conserved, but the syntenic orders and directions were evolutionally rearranged.
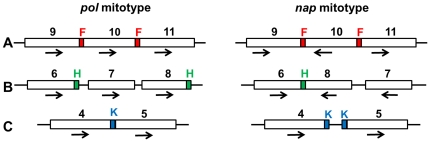
Repeated sequences in the pol mitotype were analyzed. A pair of large repeats 2427 bp in size identified in pol are also found in nap [8] and related to the formation of the multipartite structure of the Brassica mitochondrial genome, including one master circle and two smaller subgenomic circles through homologous recombination [29], [30]. The pol sequence also contains multiple copies of various 30–500 bp repeats; applying a 90% sequence similarity criterion to define a repeat, these sequences account for 4.75% of the genome and are arranged in both direct and inverse orientation. The repeats probably provide the opportunity for mtDNA sequence evolution as illustrated in Figure 3. The pair of inverted 146 bp repeats (F) on either side of syntenic region 10 may account for the changed orientation of this region in the two mitotypes (Figure 3A). A pair of inverted 233 bp repeats (H) is present at the end of syntenic regions 8 and 6 in pol, whereas the whole of regions 8 and 7 is inverted in nap following rearrangement, accompanied by the deletion of one short repeat (Figure 3B). Short repeats seem to be associated with inserted or deleted sequences of mtDNA because the 50 bp sequence (K) between syntenic regions 4 and 5 in pol is present as two copies in the nap mitotype, along with the insertion of an anonymous 44 bp sequence (Figure 3C). Short repeats providing cues to the genome reorganization may correspond to recombination hotspots in plant mitochondrial genomes [31].
Figure 3. Syntenic reordering and structural genomic changes caused by short repetitive sequences.
The numbers refer to the syntenic regions as in Figure 2. The orientation of the sequence is shown by an arrow. (A) A pair of inverted repeats (in red, F) may have induced the re-orientation of region 10. (B) In pol, a pair of inverted repeats (in green, H) is located at the end of linked regions 8 and 7, but in nap, this region is re-orientated and contains a small deletion of a sequence of unknown origin and fewer repeats due to an overlap of H at the terminus of region 6. (C) A 50 bp sequence (in blue, K) is located between syntenic regions 4 and 5 in pol, whereas two copies of K are present at this location in nap, accompanied by a 44 nucleotide insertion of unknown origin.
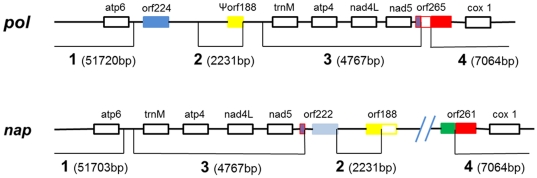
A more detailed analysis of the orf224 region, which is reported to be the CMS-associated region for pol [32], was carried out. The pol and nap mitotypes differed by at least two recombination events (Figure 4). The first recombination event, within orf265 of pol mitotype, broke the region into three parts. In the nap mitotype, the frontal 78 bp and terminal 467 bp of the orf265 sequence were separated, together with regions 3 and 4, respectively, whereas a 253 bp segment in the middle of the ORF was lost in the course of the rearrangement. The 467 bp orf265 sequence was then attached to a neighboring sequence, generating the nap form of orf261. The second recombination event occurred at the end of region 2. Another putative gene, orf188, in the nap mitotype appeared because of the relocation of region 2. Only 90 bp of the nap orf188 sequence remained in pol, probably due to evolutionary events that resulted in a sequence loss. Summarily, this region is more frequently rearranged than other regions in genomic sequences.
Figure 4. Structural polymorphism in the region surrounding orf224.
The numbers refer to the syntenic regions as in Figure 2. At least two recombination events are inferred in this region. One of these, located within orf265 in pol, splits this sequence into three segments, and in nap the terminus of orf265 generates orf261 by fusing to a neighboring sequence; the second event occurs between regions 2 and 3 resulting in the formation of orf188, a combination of the terminal 90 bp of region 2 and a new neighboring sequence in the nap mitotype.
Substoichiometrically different pol and nap mitotypes coexist in B. napus
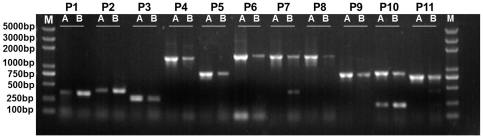
In order to demonstrate our inference about the coexistence of pol and nap mitotypes in B. napus cytoplasm, we designed 11 pairs of mitotype-specific PCR primers according to the two entire mitotype sequences. Using mtDNA extracted from pol-CMS line NH3A and its nap cytoplasm maintainer line NH3B as templates, PCR amplification showed that all primers amplified fragments regardless of the line (Figure 5). The profile generated by the multiple-primer P10 consisted of two bands of contrasting intensity(Figure 5). The results indicated that the mitochondrial population in both cytotypes was mixed. In addition, 14 materials with different origins were used in the PCR reactions. PCR with the P1 (nap-specific) and P5 (pol-specific) primers indicated that both primers were effective at amplifying all of the templates (Figure S1 and S2), demonstrating the coexistence of the two mitotypes. Sequencing of the various amplicons generated from the 11 primer pairs confirmed that all PCRs amplified their target sequences. The PCR profiles successfully established the coexistence of the two mitotypes within one B. napus plant. However, these results do not mean that the main genomes in the cytoplasm of different genetic lines are the same. Under identical PCR conditions with the same controlled mtDNA template concentration, the amplification was greater for the pol-CMS line than its maintainer when pol mitotype-specific primers were used (Figure 5). On the other hand, when nap mitotype-specific primers were used, the amplification was greater in the maintainer line compared to the CMS line. These results indicated that differences exist at the substoichiometric level of mtDNA molecules between the pol-CMS line and its maintainer. The main genome in the pol-CMS plant is the pol mitotype, whereas the main genome in the nap cytoplasm is nap. In other words, the cytoplasmic difference may be explained by the substoichiometric complexity of the rapeseed mitochondrial genome, such as that observed in the common bean [18], [19]. These interesting and surprising truths upset the conceptual framework of the rapeseed mitochondrial genome in regards to the pol and nap cytotypes.
Figure 5. PCR amplification of the pol-CMS line and its maintainer.
The pol and nap mitotypes are both present in pol-CMS line NH3A (A) and its maintainer NH3B (B), but are substoichiometrically differentiated in these cytotypes. Primer pairs P1 and P2 targeted the nap mitotype-specific sequences orf117b and orf222, respectively, whereas P3-9 and P11 targeted pol mitotype-specific sequences. P10 amplified two distinct fragments 841 bp (pol-specific) and 226 bp (nap-specific) in size. All PCR reactions used the 10 ng of mtDNA template. All primer pairs were able to amplify target fragments in both lines, but amplification differed in the two lines. The PCR assay indicated that the nap and pol mitotypes coexisted in rapeseed, but the content was substoichiometrically different in the two cytoplasm types. M: DNA ladder.
Orf222 to orf224 copy number ratio in B. napus
The ratio of orf224 and orf222 copy numbers, which are popularly accepted as CMS-related genes, was estimated using a TaqMan qPCR platform. The results indicated that the ratio was cytoplasm-dependent. In pol-CMS lines, the orf224 copy number was greater than that of orf222, but the predominant gene was orf222 in maintainer lines and other male fertile cultivars carrying nap cytoplasm (Table 1). These quantifying results are consistent with the PCR assays mentioned above. Obviously, some substoichiometric shifting of orf224/orf222 occurred between pol-CMS lines and their maintainers with nap cytoplasm. Substoichiometric shifting from orf222 to orf224 (or from orf224 to orf222) in different materials represents a cytoplasmic difference, and the substoichiometric shifting appears to be the determinant of the male fertility/sterility phenotype in B. napus.
Table 1. Copy number ratio of orf222 and orf224 in different accessions of B. napus.
| pol cytoplasm | orf224/orf222 | nap cytoplasm | orf222/orf224 |
| NH3A | 15.00 | NH3B | 1097.71 |
| NH12A | 1929.68 | NH12B | 7479.48 |
| NH15A | 287.25 | NH15B | 15771.91 |
| NH18A | 88.48 | NH18B | 1630.26 |
| NH21A | 15652.38 | NH21B | 4401.30 |
| NH923A | 18.39 | NH923B | 445.26 |
| Westar | 11337.91 | ||
| Tapidor | 36.17 | ||
| Huaiyin 16 | 155209.38 | ||
| Zheshuang 72 | 135117.61 | ||
| Westar in 2010 | 164059.11* | ||
| Tapidor in 2010 | 746.43* |
* repeated determination in 2010. The rest were determined in 2009.
However, we also noted variation in the copy number ratio among cultivars sharing the same cytotype. In cultivars carrying pol cytoplasm, the orf224:orf222 ratio ranged from 15 to >15,000; in cultivars with nap cytoplasm, the orf222:orf224 ratio ranged from 36 to >150,000 (Table 1). The abundance of sublimons (i.e. alternative genomes) was roughly two to six orders of magnitude less than that of the prevalent mitotype. This dramatic variation in the substoichiometric ratio among accessions sharing the same cytoplasm may be under nuclear control. The orf222:orf224 copy number ratio in the Westar cultivar was consistently higher than that of the Tapidor cultivar, though the ratio was not stable from year to year (Table 1), presumably as a result of an interaction with the environment.
Sequence evolution in B. napus
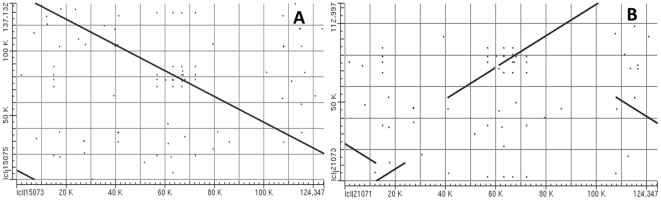
A sequence comparison of the pol and nap mitotypes identified 197 SNPs (102 transitions, 95 transversions), equivalent to a polymorphism rate of 8.9 bp per 10,000 bases. As many as 104 of the SNPs were located within the orf224/orf222 genes. Only six SNPs were identified in other genes of known function, indicating a much higher level of sequence conservation. Although reconstructing the evolution of the pol and nap mitotypes is not possible, the former is likely more primitive. The evidence for this conclusion is that the structure of the pol mitotype is closely related to that of a 124 kb Brassica rapa mitochondrial BAC clone (GenBank accession number AC172860.1) that is the likely ancestor of B. napus (Figure 6). Another reason is that restorer in the natural germplasm population is sparse for the pol-CMS line, but popular for the nap-CMS line, implying that nap cytoplasm is more accommodated evolutionarily than pol cytoplasm.
Figure 6. Alignment of the B. rapa mitochondrial segment with the two B. napus mitotype sequences.
The 124 kb B. rapa mitochondrial segment was plotted on the horizontal axis against the pol subgenome (A) or nap subgenome (B) on the vertical axis.
Discussion
The coexistence of mitotypes has been discovered by only a few gene detection experiments in wheat [33], maize [34], pearl millet [20], and cybrids [21], and is probably common in higher plants. In the common bean, coexisting mitochondrial genome molecules have been systematically demonstrated through restriction mapping. We showed here for the first time using a combination of genome sequencing and PCR assays that the pol and nap mitotypes coexist within one B. napus plant.
The mitochondrial genome in higher plants is very stable, maintaining plant survival in natural environments to achieve its important functions in living activities, so it is not probable that so many cases of CMS emerge only by evolution of the mitochondrial genome over one century. Substoichiometric shifting of coexisting mitotypes in plants may be an explanation of the phenomena related to CMS findings. The mitotype diversification cannot be explained by evolutionary events occurring over a short time, and the mitotypes are probably evolutionary products of a long time-span and coexisting in the mitochondrial population. The presence of mitochondria carrying alternative, rearranged genomes (sometimes referred to as “sublimons”) has been associated with phenotypic switching, tissue differentiation, cytotype evolution, and various nuclear-cytoplasmic interactions in higher plants [35]. The means by which sublimons are maintained, propagated, and transmitted largely remain unexplored. However, the sublimons do appear to be stable and, thus, able to affect pollen fertility via substoichiometric shifting.
We provided evidence that the B. napus mitochondrial genome includes at least two distinct mitotypes that are present in variable ratios in both CMS and male fertile cultivars. B. napus cultivars sharing the same cytotype can vary considerably with respect to the copy number ratio; therefore, the value of the copy number ratio is controlled, at least to some extent, by the nuclear genotype. In the common bean, a dominant allele in a nuclear gene can act to limit the copy number of mtDNA containing the CMS-associated pvs-orf239 sequence. Environmental factors also exert an influence on this ratio [18]. Both the nap and pol CMS trait was selected during the course of conventional breeding, apparently because in a particular confluence of nuclear genotype and environment, the copy number of the critical sublimon amplified sufficiently to exert a phenotypic effect (Figure S3). Our results, as well as previous conclusions in plant species [19], show that the conditions for the substoichiometric shifting of mitotypes are natural and common. Generally, when the alternative mitotype with malicious genes is accompanied by particular nuclear genotypes, the plants may manifest CMS phenotypes [18]. The model for the natural emergence of pol-CMS is shown in Figure S3 and probably applies to other plants.
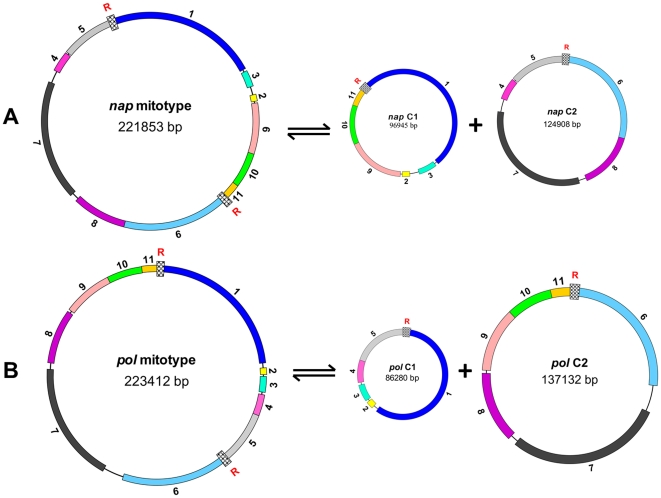
In higher plants, mtDNA molecules exist in many forms [35], but a multiple partite structure may still not be denied [36]. Based on the two 2.4 kb sequence repeats in the mitotypes, the pol mitotype is inferred as containing one master circle accompanied by two smaller circles 86.2 kb and 137.1 kb in size (Figure 7B), which is different from the nap mitotype (Figure 7A). Assuming that the two mitotypes are present in an equimolar amount in an oilseed rape cell, the mitochondrial genome as a whole can be expected to form a six-circle structure comprising two master and four smaller circles.
Figure 7. The tripartite mitochondrial genomic structure of the mitotypes.
The numbers refer to the syntenic regions as in Figure 2. Highly or completely homologous regions are indicated by color. (A) nap; (B) pol. R: 2.4 kb repetitive sequence.
A major difference between nap and pol mitotypes is localized sequence rearrangement. The short repeated sequences in higher plant mitochondria are usually inactive and play a central role in irreversible recombination to produce a new stable mitochondrial genome structure [36]. The pol mitotype also contains numerous short repeated sequences, including direct repeats and inverted repeats; those located at the edge of rearranged syntenic regions may represent the vestiges of past recombination events that restructured the genome. Nap and pol mitotypes also have significant differences at the nucleotide level. In the pol-CMS of B. napus, expression analysis has shown that the co-transcription of orf224 and atp6 alters the expression of atp6, which is associated with CMS [32], [37]. In addition to the sequence polymorphism within orf224, both orf118 and orf265 differ between pol and nap, whereas orf122 and orf132 are pol-specific. Whether the pol-specific ORFs play any determining role in CMS is still unknown.
Materials and Methods
Plant material
Six pol-CMS lines, their corresponding maintainers, and four commercial cultivars were analyzed for mitotype constitution and copy number: NH3A/B (CMS line/maintainer), NH12A/B, NH15A/B, NH21A/B, NH18A/B, and NH923A/B (bred by the present authors). The four commercial cultivars were Westar (from Canada), Tapidor (from France), and Huaiyin 16 and Zheshuang 72 (from China). The source of mitochondrial DNA for genome sequencing was pol-CMS line NH12A. These plants were planted in Jiangpu experimental station of Nanjing Agricultural University in the Autumn of 2008, harvested in 2009, and mitotype copy number determined in the Summer of 2009 with their abundant seeds. The determinations for Westar and Tapidor were repeated in 2010.
Isolation of mtDNA
The mtDNA was purified from 7-day-old etiolated seedlings germinated on an artificial medium in the dark at 25°C. The DNA isolation method was a modification of the method used in tobacco [38]. All of the extraction steps were performed at 4°C unless stated otherwise. A 250 g sample of seedling material was homogenized in 1000 ml buffer (0.5 M sucrose, 50 mM Tris-HCl, 10 mM EDTA, 1% bovine serum albumin, 5 mmol/L β-mercaptoethanol, 1.5% polyvinyl-pyrrolidone, pH 7.5). The homogenate was filtered through a layer of cheesecloth and two layers of miracloth, and centrifuged at 3000 g for 15 min. The resulting supernatant was then centrifuged at 17,000 g for 15 min and the pellet re-suspended in buffer A (0.5 M sucrose, 50 mM Tris-HCl, 10 mM MgCl2, and 25 µg/ml DNase (Roche 104159), pH 7.5) at a 5∶1 (g/ml) ratio and held at room temperature for 1 h. The suspension was then centrifuged at 18,000 g for 20 min and the pellet re-suspended in 30 ml buffer B (0.6 M sucrose, 10 mM Tris-HCl, 20 mM EDTA, pH 7.5). The suspension was centrifuged once more at 18,000 g for 20 min. Finally, the pellet was re-suspended in the same buffer and layered onto a step gradient consisting of 1.45 M and 1.2 M sucrose solution containing 10 mM Tris-HCl and 20 mM EDTA (pH 7.5). The gradient was centrifuged at 72,000 g for 90 min. Purified mitochondria were removed from the 1.45 M-1.2 M interphase, diluted with two volumes of buffer C (10 mM Tris-HCl, 20 mM EDTA, pH 7.5), and centrifuged at 18,000 g for 20 min. The final pellet was gently re-suspended in 10 ml lysis buffer (50 mM Tris–HCl, 10 mM EDTA, 1% SDS, and 200 µg/ml proteinase K(Sigma)) and incubated at 37°C for 3 h. Ammonium acetate was added to reach a concentration of 0.8 M, and the preparation was extracted twice in phenol/chloroform. The mtDNA was precipitated overnight in 2.5 volumes of 100% ethanol and centrifuged at 18,000 g for 20 min. The pellet was washed twice with 70% ethanol, and then air-dried and re-suspended in sterile distilled water.
Sequencing strategy
The mtDNA was sonicated randomly and size-fractionated by agarose gel electrophoresis. The 1.6–4 kb fraction was cloned into the pUC19 vector. The inserts were subjected to cycle sequencing (BigDye terminator v3.1 Cycle Sequencing kit, Applied Biosystems, USA). A set of 2,592 reads was obtained, which is equivalent to 11x coverage of the B. napus mitochondrial genome. The individual reads were assembled into five contigs using Phred-Phrap software (http://www.phrap.org/). Gaps between the contigs were filled by PCR.
Detection of coexisting mitotypes
A set of 11 PCR primer pairs (Table S2) was based on the pol and nap mitotype sequences, of which eight (P3-9, P11) are pol-specific. P1 targeted orf117b and P2 orf222, both of which are nap-specific. P10 was a multiple-primer designed to amplify both pol and nap mitotype templates, specifically generating an 841 bp pol-specific and 226 bp nap-specific amplicon.
The mtDNA samples were amplified using the primers in 25 µl PCR reactions containing 50 µM dNTPs, 0.2 µM of each primer, 1 µl template, 2.5 µl Taq polymerase buffer, and 1 U Taq polymerase (TIANGEN, China). The PCR was programmed as follows: an initial step of 94°C/5 min, then 30 cycles of 94°C/30 s, 53°C–60°C/30 s, and 72°C/90 s, followed by a final amplification step of 72°C/10 min. All PCR products were sequenced directly.
Determination of copy number ratios
A quantitative PCR platform (TaqMan) was used to estimate the copy number of specific mitotypes. The sequences of the relevant primers and TaqMan probe were designed from the orf224 and orf222 sequences, which were specific for the pol and nap mitotypes, respectively (Table S3). The probe was 5′-labeled with FAM and 3′-labeled with TAMRA. All primers and probes were obtained from Invitrogen Biotechnology (China). Each 20 µl PCR contained 10 µl premix EX Taq (2x) (TaKaRa), 0.3 µM of each primer, 0.2 µM TaqMan probe, 0.4 µl Rox reference dye II, and 5 µl template. The two step PCR was carried out on an ABI PRISM 7500 Fast sequence detection system (Applied Biosystems) using the following cycling program: 95°C/50 s, then 40 cycles of 95°C/3 s, 58.5°C/30 s. The analysis of each mtDNA sample was performed in triplicate. In order to check the amplification efficiency (E) of each primer pair/probe, validation experiments were performed using a 10-fold serial dilution of mtDNA. The E of each primer pair/probe was 99.7% and 100.1%, respectively. The copy number ratio of orf224 and orf222 was determined from the Ct values and E.
Sequence analysis
The NCBI database was searched using the blast network service and bl2seq (http://www.ncbi.nlm.nih.gov/). A tRNA gene search was carried out using the tRNA scan-SE service (http://www.genetics.wustl.edu/eddy/tRNAscan-SE/). Small dispersed repeats were defined as sequences of identical length (30–500 bp) that were found more than once in the genome and at least 90% identical. SNPs were identified by applying default settings in Mauve software [39]. Peptide and nucleotide sequences were aligned by ClustalX 1.81 using default settings [40].
Supporting Information
PCR analysis of a panel of male fertile and pol-CMS cultivars using P1. Analysis was based on the nap-specific primer pair P1, which targeted orf117b, but amplification occurred in both nap- and pol-CMS lines. Huaiy16: Huaiyin16; Zhesh72: Zheshuang72; M: DNA ladder.
(TIF)
PCR analysis of a panel of male fertile and pol-CMS cultivars using P5. Analysis was based on the pol-specific primer pair P5. Amplification of the target fragment occurred in both nap- and pol-CMS lines. Huaiy16: Huaiyin16; Zhesh72: Zheshuang72; M: DNA ladder.
(TIF)
The coexistence and stoichiometric shifting of the nap and pol mitotypes in B. napus. Nap and pol mitotypes coexisted in B. napus, but the relative content of the two mitotypes was different in the two cytoplasm types. In the nap cytotype, nap is the prevalent mitotype and pol is present as a sublimon. In the pol cytotype, pol is the prevalent mitotype and nap is present as a sublimon. Under certain conditions, including nuclear genotypes and environmental stress, sublimons may be amplified and accumulate to take over the role of the main genome.
(TIF)
Different ORFs between nap and pol mitotypes.
(DOC)
PCR primer sequences.
(DOC)
Primers and probes for TaqMan qPCR.
(DOC)
Acknowledgments
The authors wish to thank Shanghai Majorbio Bio-pharm Biotechnology Company (China) for their help with the shotgun sequencing.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Basic Research Program of China (973 Program) (2011CB109300); National Natural Science Foundation of China (30970289); National Key Technology R&D Program (2010BAD01B02) in China; and the Ministry of Agriculture of China for Transgenic Research (2009ZX08009-040B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 3.Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, et al. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, et al. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics. 2004;272:247–256. doi: 10.1007/s00438-004-1058-9. [DOI] [PubMed] [Google Scholar]
- 5.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 6.Tian X, Zheng J, Hu S, Yu J. The rice mitochondrial genomes and their variations. Plant Physiol. 2006;140:401–410. doi: 10.1104/pp.105.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics. 2010;11:209. doi: 10.1186/1471-2164-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. . Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen JO, Fauron CM, Minx P, Roark L, Oddiraju S, et al. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics. 2007;177:1173–1192. doi: 10.1534/genetics.107.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 12.Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 14.Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linke B, Borner T. Mitochondrial effects on flower and pollen development. Mitochondrion. 2005;5:389–402. doi: 10.1016/j.mito.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 16.He SC, Abad AR, Gelvin SB, Mackenzie SA. A cytoplasmic male sterility-associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proc Natl Acad Sci USA. 1996;93:1176–11768. doi: 10.1073/pnas.93.21.11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlsson J, Leino M, Sohlberg J, Sundstrom JF, Glimelius K. Mitochondrial regulation of flower development. Mitochondrion. 2008;8:74–86. doi: 10.1016/j.mito.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell. 1998;10:1163–1180. doi: 10.1105/tpc.10.7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, et al. Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng X, Kaur AP, Mackenzie SA, Dweikat IM. Substoichiometric shifting in the fertility reversion of cytoplasmic male sterile pearl millet. Theor Appl Genet. 2009;118:1361–1370. doi: 10.1007/s00122-009-0986-5. [DOI] [PubMed] [Google Scholar]
- 21.Bellaoui M, Martin-Canadell A, Pelletier G, Budar F. Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol Gen Genet. 1998;257:177–185. doi: 10.1007/s004380050637. [DOI] [PubMed] [Google Scholar]
- 22.Thompson KF. Cytoplasmic male sterility in oil-seed rape. Heredity. 1972;29:253–257. [Google Scholar]
- 23.Shiga T, Bata S. Cytoplasmic male sterility in rape plants oilseed (Brasscica napus). Japanese Journal of Breeding. 1971;21:16–17. [Google Scholar]
- 24.Shiga T, Bata S. Cytoplasmic male sterility in rape (Brasscica napus), and its utilization to breeding. Japanese Journal of Breeding. 1973;23:187–197. [Google Scholar]
- 25.Fu T. Production of and research in rapeseed in China. Eucarpia Cruciferae Newsletter. 1981;6:6–7. [Google Scholar]
- 26.Li D. Preliminary report on breeding of male sterile, maintainer and restorer lines in Brassica napus L. Shaanxi Journal of Agricultural Sciences. 1980;1:26–29. (in Chinese with English abstract) [Google Scholar]
- 27.Fu S, Qi C, Tang J. Breeding of Cytoplasmic male sterile line MICMS in Brassica napus L. Acta Agronomica Sinica. 1989;15:305–309. (in Chinese with English abstract) [Google Scholar]
- 28.Yang G, Fu T, Brown G. The genetic classification of cytoplasmic male sterility systems in Brassica napus L. Scientia Agricultura Sinica. 1998;31:27–31. (in Chinese with English abstract) [Google Scholar]
- 29.Palmer JD, Shields CR. Tripartite structure of the Brassica campestris mitochondrial genome. Nature. 1984;307:437–440. [Google Scholar]
- 30.Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 31.Scotti N, Marechal-Drouard L, Cardi T. The rpl5-rps14 mitochondrial region: a hot spot for DNA rearrangements in Solanum spp. somatic hybrids. Curr Genet. 2004;45:378–382. doi: 10.1007/s00294-004-0496-6. [DOI] [PubMed] [Google Scholar]
- 32.Singh M, Brown GG. Suppression of cytoplasmic male sterility by nuclear genes alters expression of a novel mitochondrial gene region. Plant Cell. 1991;3:1349–1362. doi: 10.1105/tpc.3.12.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hattori N, Kitagawa K, Takumi S, Nakamura C. Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics. 2002;160:1619–1630. doi: 10.1093/genetics/160.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small ID, Suffolk R, Leaver CJ. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 35.Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes-though this be madness, yet there's method in't. J Exp Bot. 2010;61:657–671. doi: 10.1093/jxb/erp361. [DOI] [PubMed] [Google Scholar]
- 36.Andre C, Levy A, Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- 37.L'Homme Y, Brown GG. Organizational differences between cytoplasmic male sterile and male fertile Brassica mitochondrial genomes are confined to a single transposed locus. Nucleic Acids Res. 1993;21:1903–1909. doi: 10.1093/nar/21.8.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bland MM, Matzinger DF, Levings CS. Comparison of the mitochondrial genome of Nicotiana tabacum with its progenitor species. Theor Appl Genet. 1985;69:535–541. doi: 10.1007/BF00251100. [DOI] [PubMed] [Google Scholar]
- 39.Darling AE, Mau B, Perna NT. progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR analysis of a panel of male fertile and pol-CMS cultivars using P1. Analysis was based on the nap-specific primer pair P1, which targeted orf117b, but amplification occurred in both nap- and pol-CMS lines. Huaiy16: Huaiyin16; Zhesh72: Zheshuang72; M: DNA ladder.
(TIF)
PCR analysis of a panel of male fertile and pol-CMS cultivars using P5. Analysis was based on the pol-specific primer pair P5. Amplification of the target fragment occurred in both nap- and pol-CMS lines. Huaiy16: Huaiyin16; Zhesh72: Zheshuang72; M: DNA ladder.
(TIF)
The coexistence and stoichiometric shifting of the nap and pol mitotypes in B. napus. Nap and pol mitotypes coexisted in B. napus, but the relative content of the two mitotypes was different in the two cytoplasm types. In the nap cytotype, nap is the prevalent mitotype and pol is present as a sublimon. In the pol cytotype, pol is the prevalent mitotype and nap is present as a sublimon. Under certain conditions, including nuclear genotypes and environmental stress, sublimons may be amplified and accumulate to take over the role of the main genome.
(TIF)
Different ORFs between nap and pol mitotypes.
(DOC)
PCR primer sequences.
(DOC)
Primers and probes for TaqMan qPCR.
(DOC)