Abstract
Ornithine lipids (OLs) are widespread among gram-negative bacteria. Their basic structure consists of a 3-hydroxy fatty acyl group attached in amide linkage to the α-amino group of ornithine and a second fatty acyl group ester-linked to the 3-hydroxy position of the first fatty acid. OLs can be hydroxylated within the secondary fatty acyl moiety and this modification has been related to increased stress tolerance. Rhizobium tropici, a nodule-forming α-proteobacterium known for its stress tolerance, forms four different OLs. Studies of the function of these OLs have been hampered due to lack of knowledge about their biosynthesis. Here we describe that OL biosynthesis increases under acid stress and that OLs are enriched in the outer membrane. Using a functional expression screen, the OL hydroxylase OlsE was identified, which in combination with the OL hydroxylase OlsC is responsible for the synthesis of modified OLs in R. tropici. Unlike described OL hydroxylations, the OlsE-catalyzed hydroxylation occurs within the ornithine moiety. Mutants deficient in OlsE or OlsC and double mutants deficient in OlsC/OlsE were characterized. R. tropici mutants deficient in OlsC-mediated OL hydroxylation are more susceptible to acid and temperature stress. All three mutants lacking OL hydroxylases are affected during symbiosis.
INTRODUCTION
Membranes of the gram-negative model organism Escherichia coli only contain three major phospholipids, that is phosphatidylethanolamine, phosphatidylglycerol and cardiolipin (Heath et al., 2002). Some other bacteria also form the membrane lipids phosphatidylinositol or phosphatidylcholine (Jackson et al., 2000, Sohlenkamp et al., 2003). In addition to phospholipids, many bacteria also present phosphorus-free membrane lipids such as ornithine lipids (OLs), diacylglyceryl-N, N, N-trimethylhomoserine (DGTS) or sulpholipids (SLs) in their membranes (López-Lara et al., 2003, Geiger et al., 2010). In some cases, like for example Rhodobacter sphaeroides or Sinorhizobium meliloti, the formation of these phosphorus-free membrane lipids is induced by phosphate-limiting growth conditions (Benning et al., 1995, Geiger et al., 1999). Some bacteria such as Brucella abortus (Comerci et al., 2006, Bukata et al., 2008) or Rhizobium tropici (Rojas-Jiménez et al., 2005, Sohlenkamp et al., 2007) also form significant amounts of OLs during growth in standard laboratory media such as LB which contain phosphate in concentrations that are not growth-limiting.
OLs are widespread among gram-negative bacteria and have also been reported in some gram-positive bacteria, like Mycobacterium and Streptomyces species, but seem to be absent from Archaea and Eukarya (López-Lara et al., 2003, Geiger et al., 2010). OLs contain a 3-hydroxy fatty acyl group that is attached in amide linkage to the α-amino group of ornithine. A second fatty acyl group is ester-linked to the 3-hydroxy position of the first fatty acid. It has been reported that in some bacteria the ester-linked fatty acid is hydroxylated at the 2 or 3 position (Asselineau, 1991). The genes olsB and olsA encoding the two enzymes essential for OL biosynthesis from ornithine and acyl-ACPs have been first described in S. meliloti (Weissenmayer et al., 2002, Gao et al., 2004). Although OLs are probably found in both membranes of gram-negative bacteria, they seem to be enriched in the outer membrane as was shown in the acid-resistant species Thiobacillus thiooxidans (Dees & Shively, 1982). Therefore, Dees and Shively speculated about a role of OLs in acid resistance (Dees & Shively, 1982).
R. tropici CIAT899 is highly tolerant to many environmental stresses such as acidity or high temperatures. It can grow on acidified media down to pH 4.0, and it is a good competitor for nodule occupancy in Phaseolus vulgaris (common bean) and other hosts under acidic conditions (Martínez-Romero et al., 1991). A gene responsible for the hydroxylation of OL has been isolated in R. tropici using a transposon mutagenesis approach looking for mutants affected in their capacity to grow at pH 4.5 (Rojas-Jiménez et al., 2005, Vinuesa et al., 2003). Rojas-Jiménez et al. described the presence of four different species of OL in R. tropici membranes which were called S1, S2, P1 and P2. They showed that the putative hydroxylase OlsC is responsible for the formation of P1 and P2, presumably from OLs S1 and S2 functioning as substrates (Fig. 1), but they did not investigate on the position of the OlsC-dependent hydroxylation. No acid growth phenotype was observed for the olsC-deficient mutant, but constitutive expression of olsC was associated with the inability of the strain to grow at pH 4.5. Upon inoculation of the olsC mutant onto bean plants only poorly developed nodules were observed (Ndv−) 21 days after inoculation of the plants (Rojas-Jiménez et al., 2005). In an earlier study, Taylor et al. (1998) had observed an increased formation of hydroxylated OLs at an elevated temperature in Burkholderia cepacia. These two previous results indicated a role of modified OLs in stress tolerance, and prompted us to investigate the synthesis of modified OLs and their role in stress tolerance in R. tropici in more detail. In this study we describe the isolation of the OL hydroxylase OlsE and the construction of R. tropici mutants deficient in the hydroxylation of OLs. We show that OlsC is introducing a hydroxyl group in the 2 position of the secondary fatty acid of OLs and that OlsE introduces a hydroxylation in the ornithine moiety of OLs. The characterization of these mutants shows that hydroxylated OLs are important for adaptation to stress conditions in R. tropici.
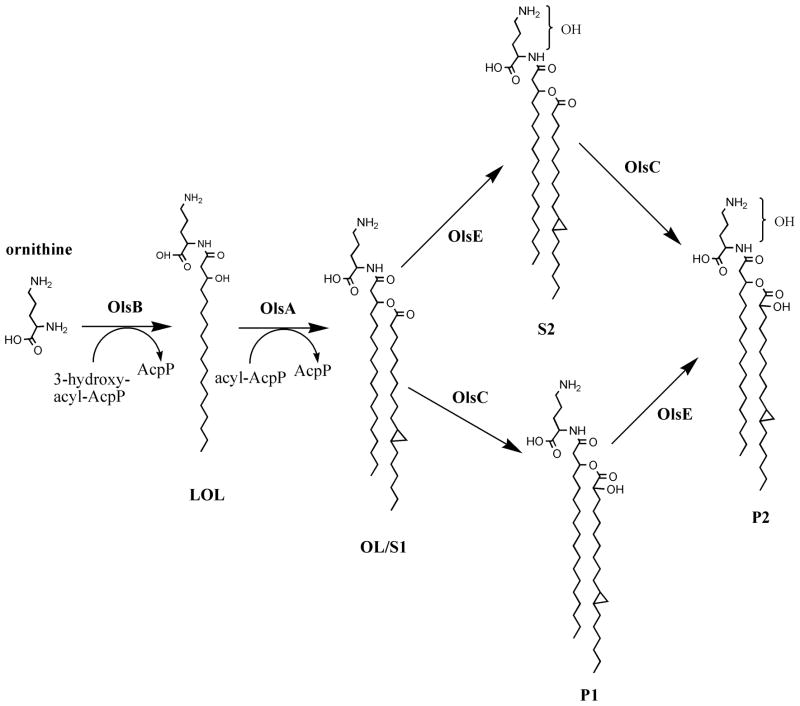
Figure 1.
Biosynthesis of ornithine lipids in Rhizobium tropici CIAT899. The genes coding for OlsB and OlsA have been first identified in Sinorhizobium meliloti, whereas the gene encoding the OL hydroxylase OlsC has been described first in Rhizobium tropici. Here we describe that the hydroxylation introduced by OlsC is in the 2 position of the secondary fatty acid. We also describe the identification of the gene encoding the OL hydroxylase OlsE introducing a hydroxyl group in the ornithine moiety of OL. Lyso-ornithine lipid (LOL), ornithine lipid (OL).
RESULTS
Stress conditions alter the amount of modified ornithine lipids in R. tropici indicating a role of OLs in stress adaptation
R. tropici CIAT899 is a nodule-forming rhizobium well-known for its ability to resist stress conditions such as acidic pH or high temperatures (Martínez-Romero et al., 1991). In an earlier study Rojas-Jiménez et al. (2005) had observed that R. tropici forms four different ornithine lipids (OLs). In addition to the unmodified OL which was named S1 (for substrate 1) three additional modified OLs probably derived from S1 are present. Taylor et al. (1998) had observed an increase in the relative amounts of hydroxylated OL when Burkholderia cepacia was grown at increased temperatures. To find out if the modification of OL also occurs as a stress response in R. tropici and if these modifications might have a role in stress adaptation, R. tropici CIAT899 was grown at 30 °C, 37 °C and 42 °C and its lipid composition was analyzed. (Fig. 2A, B, C, table 1). At the standard growth temperature of 30 °C, all four OLs can be detected, with P1 being the most abundant OL. An increase in growth temperature to 37 °C causes a decrease in the OLs S2 and P2 and a simultaneous increase in S1. When grown at 42 °C the amounts of S1 and P1 decrease slightly. The OLs S2 and P2 cannot be detected in cells grown at 42 °C. An unknown lipid which migrates similarly as the sulpholipid sulphoquinovosyl diacylglycerol is apparently formed at 42 °C but not at lower growth temperatures. The decrease in OLs is accompanied by changes in the phospholipid composition: phosphatidylethanolamine (PE) decreases whereas phosphatidylcholine (PC), phosphatidylglycerol (PG) and cardiolipin (CL) increase. R. tropici CIAT899 was also grown in complex TY medium adjusted to different pH values (compare Fig. 2A, D, E, table 2). In R. tropici cells grown at pH 4.5 the OLs S1 and S2 are not detectable, whereas P2 is increased and no changes are detected for P1. When grown at pH 4.0 again OLs S1 and S2 cannot be detected, but P1 increases drastically and becomes the major membrane lipid (Fig. 2E).
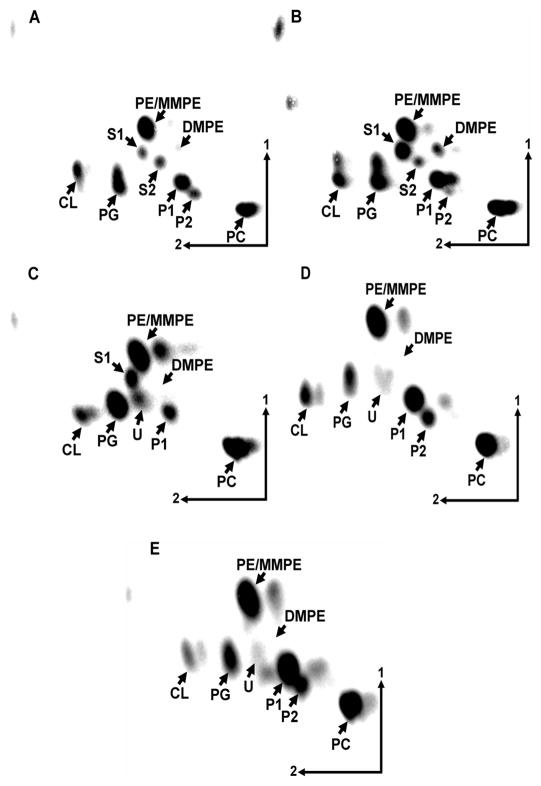
Figure 2.
Separation of [14C]acetate-labeled lipids from Rhizobium tropici CIAT899 grown in complex TY medium at 30 °C (A), at 37 °C (B), at 42 °C (C), at 30 °C at pH 4.5 (D) or at 30 °C at pH 4.0 (E) by two-dimensional thin layer chromatography. The phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL), monomethyl PE (MMPE), dimethyl PE (DMPE) and the ornithine lipids (OLs) S1, S2, P1 and P2 are indicated. U (unknown lipid).
Table 1.
Membrane lipid composition of Rhizobium tropici wild type CIAT 899, olsE-deficient mutant MAV04, olsC-deficient mutant 899-olsCΔ1, and olsC/olsE-deficient double mutant MAV05 after growth on complex TY medium at 30 °C, 37 °C or 42 °C. The values shown are mean values ± standard deviation derived from at least three independent experiments.
| Lipid | Composition (% of total 14C) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 °C | 37 °C | 42 °C | ||||||||||
| CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | |
| PC | 22.6±0.6 | 24.8±2.9 | 20.7±0.7 | 24.8±0.1 | 28.9±4.0 | 27.2±4.8 | 23.5±4.3 | 24.6±1.2 | 26.8±1.2 | 28.5±1.6 | 27.0±2.0 | 27.5±2.1 |
| PE | 25.0±0.1 | 24.6±1.4 | 24.9±2.1 | 25.6±0.6 | 23.5±5.0 | 23.2±3.7 | 19.1±1.0 | 20.4±4.8 | 16.2±1.3 | 17.9±1.9 | 11.9±1.3 | 11.8±1.1 |
| DMPE | 1.1±0.1 | 1.7±0.2 | 1.6±0.6 | 1.6±0.4 | 1.4±0.6 | 1.1±0.2 | 1.4±0.4 | 1.3±0.2 | 0.4±0.0 | 0.4±0.1 | 0.6±0.2 | 0.7±0.1 |
| PG | 15.8±0.8 | 13.9±2.8 | 17.4±1.5 | 16.0±0.6 | 12.5±0.9 | 11.8±1.2 | 18.8±1.7 | 16.9±1.6 | 23.8±0.3 | 22.1±0.8 | 33.9±1.0 | 29.2±1.1 |
| CL | 4.0±0.1 | 4.8±0.1 | 3.0±0.1 | 5.6±0.3 | 5.3±1.3 | 4.8±1.0 | 8.0±2.0 | 6.7±2.4 | 7.5±0.5 | 8.2±0.3 | 11.1±2.5 | 10.7±1.3 |
| S1 | 2.9±0.2 | 6.8±1.0 | 26.5±1.7 | 26.4±1.8 | 7.3±1.0 | 9.0±1.4 | 27.7±0.9 | 30.1±5.6 | 5.6±0.2 | 4.7±0.9 | 10.8±0.8 | 16.0±1.5 |
| S2 | 3.5±0.5 | n.d. | 5.9±0.5 | n.d. | 1.6±0.3 | n.d. | 1.5±0.2 | n.d. | n.d. | n.d. | n.d. | n.d. |
| P1 | 20.6±0.3 | 23.4±1.8 | n.d. | n.d. | 18.2±2.8 | 22.9±0.1 | n.d. | n.d. | 15.2±0.5 | 15.2±0.7 | n.d. | n.d. |
| P2 | 4.5±0.3 | n.d. | n.d. | n.d. | 1.3±0.4 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| U | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 4.5±0.5 | 3.0±0.7 | 4.7±0.7 | 4.1±1.0 |
PC (phosphatidylcholine), PE (phosphatidylethanolamine), MMPE (monomethyl phosphatidylethanolamine), DMPE (dimethyl phosphatidylethanolamine) PG (phosphatidylglycerol), CL (cardiolipin), S1 (substrate 1, unmodified ornithine lipid); S2, P1, P2 (hydroxylated ornithine lipids), U (unidentified lipid). n.d.- not detected.
Table 2.
Membrane lipid composition of Rhizobium tropici wild type CIAT899, olsE-deficient mutant MAV04, olsC-deficient mutant 899-olsCΔ1, and olsC/olsE-deficient double mutant MAV05 after growth on complex TY medium adjusted to pH 7.0, pH 4.5 or pH 4.0. The values shown are mean values ± standard deviation derived from at least three independent experiments. For abbreviations see Table 1.
| Lipid | Composition (% of total 14C) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 7.0 | pH 4.5 | pH 4.0 | ||||||||||
| CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | CIAT899 | MAV04 | 899-olsCΔ1 | MAV05 | |
| PC | 27.6±0.7 | 25.0±0.2 | 21.3±0.3 | 24.6±0.4 | 27.7±0.2 | 28.1±1.7 | 23.3±2.2 | 23.8±3.8 | 26.2±0.3 | 26.6±0.4 | 19.5±0.5 | 18.1±2.0 |
| PE | 26.7±0.3 | 26.5±0.1 | 21.7±0.1 | 24.7±0.5 | 27.4±0.3 | 27.3±2.1 | 12.8±1.5 | 10.9±0.4 | 22.3±0.4 | 25.6±0.3 | 14.9±0.4 | 12.3±2.0 |
| DMPE | 0.4±0.1 | 0.6±0.1 | 0.6±0.1 | 0.9±0.0 | 0.8±0.1 | 0.5±0.1 | 1.0±0.2 | 1.0±0.1 | 0.8±0.2 | 0.2±0.0 | 0.2±0.1 | 0.6±0.2 |
| PG | 12.2±0.4 | 10.0±0.1 | 13.1±0.2 | 11.5±0.1 | 8.2±0.2 | 4.7±0.4 | 10.1±0.9 | 9.2±0.9 | 6.8±0.1 | 7.4±1.3 | 12.4±1.2 | 12.9±1.1 |
| CL | 4.4±0.2 | 5.7±0.4 | 5.5±0.3 | 3.7±0.3 | 6.9±0.3 | 2.4±0.2 | 4.2±0.7 | 4.3±0.1 | 3.7±0.5 | 4.2±1.0 | 5.7±0.4 | 4.7±0.7 |
| S1 | 2.9±1.7 | 8.9±0.2 | 32.5±0.2 | 34.6±0.2 | n.d. | 2.0±0.5 | 40.6±2.4 | 46.0±2.9 | n.d. | 1.6±0.3 | 40.7±1.1 | 44.6±3.6 |
| S2 | 3.3±0.3 | n.d. | 5.3±0.5 | n.d. | n.d. | n.d. | 2.5±0.1 | n.d. | n.d. | n.d. | n.d. | n.d. |
| P1 | 18.0±0.4 | 23.3±0.1 | n.d. | n.d. | 19.4±0.3 | 32.0±1.0 | n.d. | n.d. | 30.2±0.1 | 31.9±2.5 | n.d. | n.d. |
| P2 | 4.5±0.4 | n.d. | n.d. | n.d. | 8.3±0.2 | n.d. | n.d. | n.d. | 8.1±0.0 | n.d. | n.d. | n.d. |
| U | n.d. | n.d. | n.d. | n.d. | 1.3±0.1 | 3.0±0.3 | 5.5±1.0 | 4.8±0.5 | 1.9±0.1 | 2.5±0.4 | 6.6±0.6 | 6.8±0.5 |
OLs are enriched in the outer membrane of R. tropici CIAT899
Dees and Shively (1982) had shown that in the acid-resistant species Thiobacillus thiooxidans OL is present mainly in the outer membrane (OM) and they had therefore speculated that it might play a role in conferring acid resistance to these bacteria. If such a hypothesis were true one would expect an accumulation of OLs also in the OM of the acid-tolerant bacterium R. tropici. Inner membrane (IM) and OM from R. tropici were separated and the lipids of both membranes were extracted and separated using two 2D-TLC (Fig. 3). The protein content of the fractions was estimated using absorption measurements at 280 nm. The protein-enriched fractions formed two peaks corresponding to the IM and OM (Fig. 3A). KDO (2-keto-3-deoxyoctanoate)-content and NADH oxidase activity that were used as markers for the OM and IM, respectively, indicated that the IM was contaminated to some extent by the OM, but that the OM was almost free of contamination by the IM (Fig. 3B). The TLC analysis of the Bligh-Dyer extracts showed that phospholipids are the major membrane lipids of the IM but are present in much smaller relative amounts in the OM (Fig. 3C, D, table 3). A quantification of the lipids showed that phospholipids form more than 70 % of the membrane lipids of the IM but only about 40 % of the membrane lipids of the OM, excluding lipopolysaccharide (LPS). OLs form less than 30 % of the membrane lipids of the IM but about 60 % of the membrane lipids of the OM (again excluding LPS). Taking the contamination of the IM fractions with OM material into account the result over-estimates the real concentration of OLs in the IM. Assuming that the outer leaflet of the OM is composed mainly of the lipid A moiety of LPS, this result indicates that the major proportion of the inner leaflet of the OM is composed of OLs.
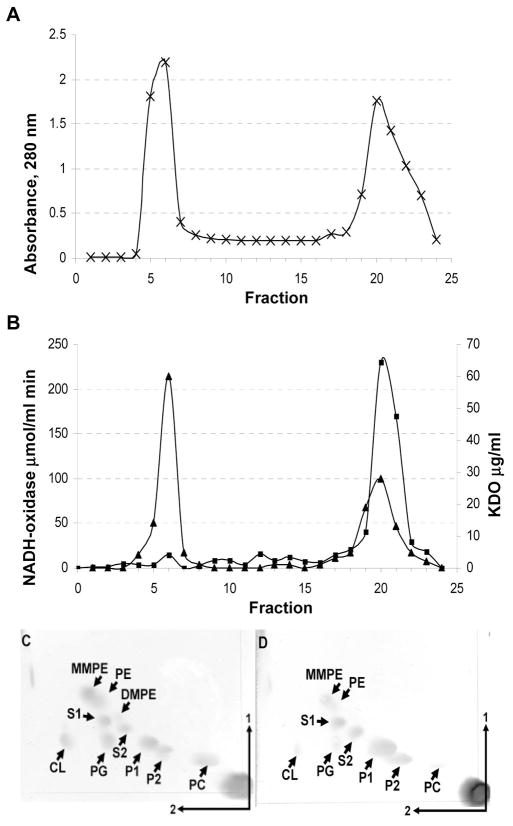
Figure 3.
Localization of OLs in membranes of wild type Rhizobium tropici CIAT899. (A and B) Results of a sucrose density gradient centrifugation of cell membranes of R. tropici CIAT899. (A) A280 readings of the gradient fractions. (B) 2-Keto-3-oxyoctonate content (closed triangles) and NADH oxidase (closed squares) activity of the fractions. Separation of membrane lipids extracted from the inner (C) and outer membrane (D). Fractions corresponding to the inner and outer membranes were pooled, lipids were extracted with 1-butanol and subsequently analyzed using 2D-TLC. Lipids were visualized by spraying with ceric sulphate in sulphuric acid. The phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL), monomethyl PE (MMPE), dimethyl PE (DMPE) and the ornithine lipids (OLs) S1, S2, P1 and P2 are indicated. A quantification of the lipids is shown in table 3.
Table 3.
Membrane lipid composition of the inner and outer membrane of R. tropici CIAT899. The data were obtained from a the TLC plates shown in figures 3C and D using the program ImageQuant. Numbers present percent of total lipids present in the TLC. For abbreviations see table 1.
| Lipid | Inner membrane | Outer membrane |
|---|---|---|
| PC | 23.9 | 9.1 |
| PE | 8.2 | 6.6 |
| MMPE | 5.4 | 5.9 |
| DMPE | 4.8 | 5.0 |
| PG | 17.2 | 7.7 |
| CL | 12.2 | 6.7 |
| S1 | 6.2 | 11.4 |
| S2 | 5.0 | 11.0 |
| P1 | 11.1 | 25.3 |
| P2 | 6.0 | 11.3 |
Expression cloning of the OL-modifying enzyme OlsE from R. tropici
The experiments described earlier indicated a possible role for the different OLs in the R. tropici stress response. In S. meliloti only one type of OL is present. In contrast, four different types of OLs called S1, S2, P1 and P2 are present in R. tropici CIAT899 (Fig. 1). The gene olsC encoding the enzyme OlsC responsible for the synthesis of OLs P1 and P2 from the substrates S1 and S2 has been described earlier (Rojas-Jiménez et al., 2005). It was not known however, which gene encodes the hypothetical enzyme OlsE responsible for the synthesis of S2 and possibly also for the synthesis of P2 (Fig. 1). We suspected that S1, corresponding to the OL present in S. meliloti, was a substrate for the OlsE-catalyzed reaction. The S. meliloti strain CS111.pNG25 lacking the ninhydrin-positive lipid PE and producing increased amounts of the OL S1 was constructed and transconjugants of CS111.pNG25 harboring cosmids containing R. tropici CIAT899 genomic DNA were assayed for the presence of a second ninhydrin-positive lipid in addition to S1. In the transconjugant referred to as CS111.pNG25.pCos94, two ninhydrin-positive lipids with the expected Rf values for S1 and S2 were detected. A restriction analysis of pCos94 showed that it contains about 18–20 kb of inserted DNA. Restriction fragments of the pCos94 insert were subcloned into a broad host range vector and again conjugated into CS111.pNG25. The resulting transconjugants were analyzed as described above for the cosmid bank (data not shown). A plasmid conferring the formation of the OL S2 was identified and its insert was sequenced. In addition to three predicted complete ORFs it contained two incomplete ORFs (GenBank accession number HM010770). BLAST searches using the NCBI database with the amino acid sequences of the three complete ORFs as query were made (Altschul et al., 1997). The first ORF was annotated as a putative acetyltransferase, the second ORF as a putative aminoglycoside N(6′) acetyltransferase, and the third ORF as a putative hydroxylase. The three candidate ORFs were cloned into a broad host range plasmid and the resulting plasmids were conjugated into CS111.pNG25. Labelling of the lipids of the three transconjugants with [14C]acetate showed that ORF3 codes for the putative hydroxylase OlsE which is responsible for the formation of S2 (Fig. 4).
Figure 4.
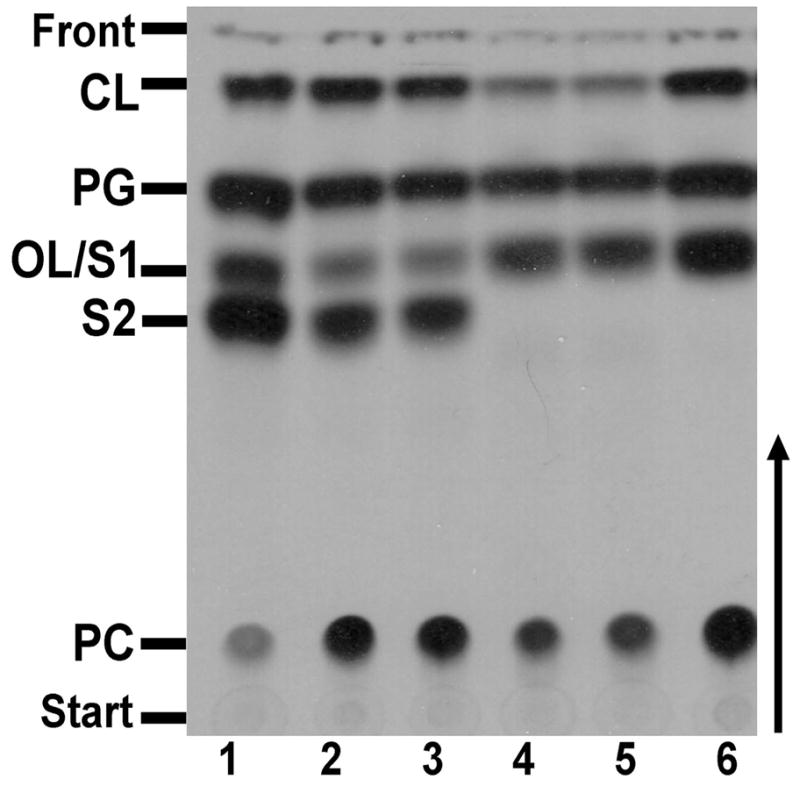
Expression cloning of olsE from R. tropici. Lipids of Sinorhizobium meliloti CS111.pNG25 containing different plasmids or cosmids were radiolabeled with [14C]acetate and separated by one-dimensional TLC. The following strains were analyzed: CS111.pNG25.pCos94 (cosmid, lane 1), CS111.pNG25.pERMAV04 (ORF1 to 3/3.5 kb insert, lane 2), CS111.pNG25.pERMAV13 (ORF3, lane 3), CS111.pNG25.pERMAV12 (ORF2, lane 4), CS111.pNG25.pERMAV11 (ORF1, lane 5), and CS111.pNG25.pERMAV06 (negative control, lane 6). The phospholipids phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL) and the ornithine lipids (OLs) S1 and S2 are indicated.
OlsE belongs to the fatty acyl hydroxylase superfamily and is responsible for the hydroxylation of OL within the ornithine moiety
The gene olsE encodes a very hydrophobic protein of 330 amino acids predicted to form between four and six transmembrane helices. An analysis of the amino acid sequence shows that OlsE belongs to the fatty acyl hydroxylase superfamily (cl01132) which is characterized by the presence of two copies of the HXHH motif. This superfamily includes fatty acid and carotene hydroxylases, sterol desaturases (Mitchell & Martin, 1997), C-5 sterol desaturase (Arthington et al., 1991) and C-4 sterol methyl oxidase (Bard et al., 1996, Kennedy et al., 2000). A similar motif (HX3–4H, HX2–3HH, HX2–3H) can be found in membrane-bound fatty acid desaturases such as OLE1 from Saccharomyces cerevisiae and is also present in bacterial alkane hydroxylase (Kok et al., 1989) and x ylene monooxygenase (Suzuki et al., 1991). In these proteins the conserved histidine residues act to coordinate an oxo-bridged diiron cluster (Fe-O-Fe) that functions as part of the reaction center (Fox et al., 1993, Shanklin et al., 1994).
The annotation of OlsE as a fatty acyl hydroxylase indicates that OlsE introduces a hydroxyl group into OL at an unknown position. To localize the hydroxyl group on the OL S2, lipids were extracted according to Bligh and Dyer (Bligh & Dyer, 1959) from a one liter culture of the olsC-deficient R. tropici mutant 899-olsCΔ1. OLs S1 and S2 were purified from the total lipid extract and analysed by normal phase LC coupled electrospray ionization (ESI) mass spectrometry (MS) in the negative ion mode. Prior to fragmentation ions with m/z 691 and 707 corresponding to OLs S1 and S2 were detected. The molecular ion was shifted in case of S2 to an m/z 16 amu higher in comparison to S1 indicating the presence of an additional oxygen suggesting the presence of an additional hydroxyl group. Comparing the fragmentation patterns of S1 and S2 it was observed that the modification present in S2 is located within the ornithine moiety and not in the fatty acyl chains (Fig. 5). When assaying the two-dimensional TLC plates with R. tropici lipids with ninhydrin it was noticed that S2 and P2 react with delay in comparison to S1 and P1, and that the developed colour is different. While S1 and P1 upon reaction with ninhydrin develop a red to purple colour, the reaction of S2 and P2 causes the formation of an orange colour.
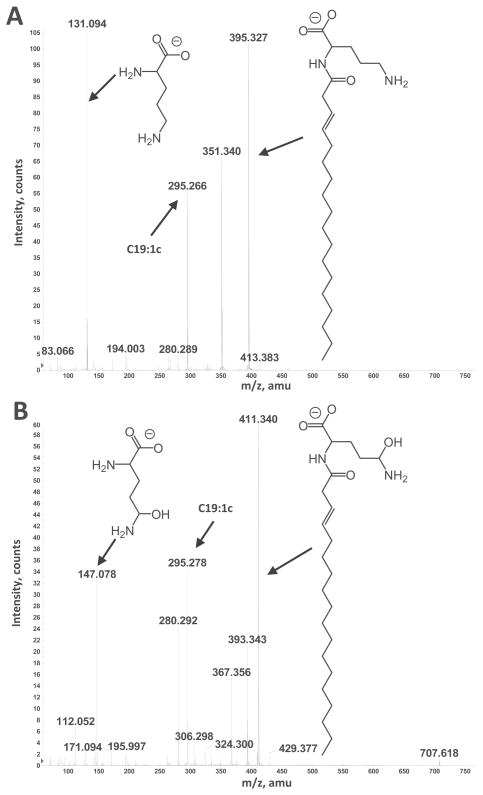
Figure 5.
Collision-induced dissociation mass spectra of ornithine lipids S1 and S2 detected in lipid extract of R. tropici mutant 899-olsCΔ1. Negative ion collision induced dissociation mass spectra of [M-H]− ions at m/z 671 (A) obtained from OL S1 and m/z 707 (B) obtained from OL S2. The structures of major fragment ions are indicated. The position of the hydroxyl group introduced in the ornithine moiety is assigned tentatively. Complete structures of the OLs are shown in Fig. 1.
OlsC introduces a hydroxyl group at the 2 position of the secondary fatty acid of ornithine lipid
OlsC is a homolog of the hydroxylase LpxO from Salmonella typhimurium that is responsible for the addition of a 2-hydroxy group to the myristate residue present at the 3′ position of lipid A. Rojas-Jimenez et al. (2005) had discovered the gene olsC and had shown that OlsC is a putative hydroxylase responsible for the formation of the ornithine lipids P1 and P2 from the ornithine lipids S1 and S2 in R. tropici (Fig. 1). However, it was not known in what part of the OL structure the OlsC-dependent hydroxylation occurs. To localize the hydroxyl group on the OL P1, OLs S1 and P1 were purified from the total lipid extracts and analyzed by normal phase LC coupled electrospray ionization (ESI) mass spectrometry (MS) in the negative ion mode. Prior to fragmentation ions with m/z 691 and 707 corresponding to OLs S1 and P1 were detected. The molecular ion of P1 was shifted to an m/z 16 amu higher in comparison to S1 indicating the presence of an additional oxygen suggesting the presence of an additional hydroxyl group. Comparing the fragmentation patterns of S1 and P1 it was observed that the modification present in P1 is located within the secondary fatty acyl chain (data not shown) which in case of S1 is mainly lactobacillic acid and in case of P1 hydroxy lactobacillic acid. In order to determine the position of the OlsC-dependent hydroxylation in P1 its fatty acids were transmethylated before the hydroxyl groups were derivatized to TMS ethers similar to as described by Gibbons et al. (2008). Alpha- and beta-hydroxy fatty acid standards of 16 and 18 carbons were processed in parallel with the samples (Fig. S1A–D). GC/MS analysis of the derivatized fatty acids shows the presence of three peaks present in the samples derived from P1 that are not present in the samples derived from S1 (Fig. S1E–F). Their fragmentation pattern indicates that the OlsC-dependent hydroxylation occurs in the 2 position (Fig. S1G).
Lipid composition analysis of olsE and olsE/olsC mutants
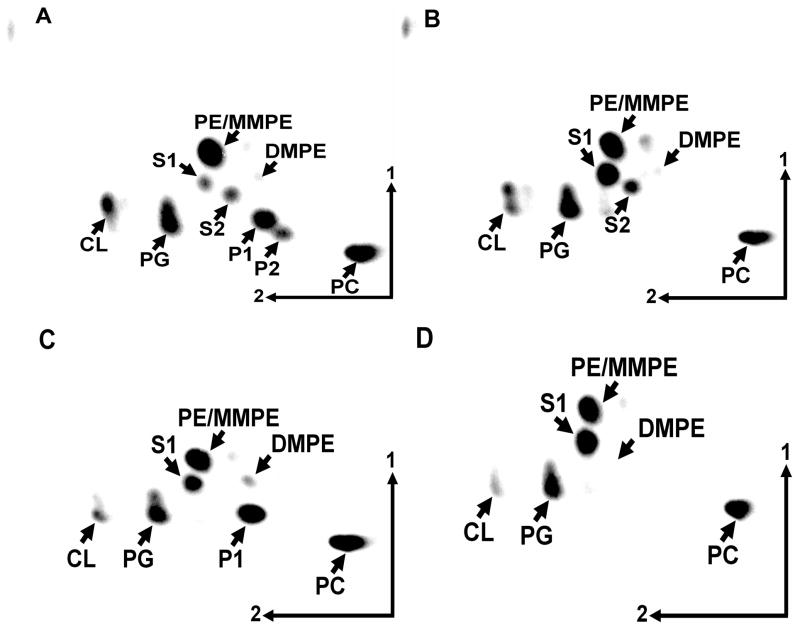
To study the role of OLs in R. tropici in more detail, mutants deficient in olsE and double mutants deficient in olsC and olsE were constructed. Their lipid compositions were compared to the wild type strain CIAT899 and the OlsC-deficient mutant 899-olsCΔ1 (Fig. 6, Table 1). As expected the olsE-deficient mutant MAV04 lacked the OLs S2 and P2, the olsC-deficient mutant 899-olsCΔ1 lacked P1 and P2 and in the double mutant MAV05 (ΔolsCΔolsE) no S2, P1 or P2 were detectable. Apparently, the amount of OLs, being the sum of S1, S2, P1 and P2, is more or less stable between 20 and 35 % when R. tropici is grown in complex TY medium at 30 °C. No significant differences in the relative amounts of the phospholipids PE, PC, PG and CL were observed between the different strains. To show that the observed phenotypes were caused by the absence of the deleted genes, mutants MAV04 (ΔolsE) and MAV05 (ΔolsCΔolsE) were also complemented. When olsE was present in trans in MAV04 again formation of S2 and P2 was detected and when mutant MAV05 was complemented with olsE the OLs S2 and P2 could be detected, whereas S1 and P1 did not accumulate (data not shown). Constitutive expression of olsC and olsE together in MAV05 caused the accumulation of P2 while only trace amounts of the other OLs were observed (data not shown). Such an over-complementation leading to the accumulation of the reaction product(s) while almost completely consuming the substrate(s) had also been observed earlier for the complementation of the olsC-deficient mutant 899-olsCΔ1 (Rojas-Jiménez et al., 2005).
Figure 6.
Analysis of membrane lipid composition of R. tropici wild type CIAT899 (A), olsC-deficient mutant 899-olsCΔ1 (B), olsE-deficient mutant MAV04 (C), and olsC/olsE-deficient double mutant MAV05 (D). Lipids were labelled with [14C]acetate during growth in complex TY medium at 30 °C and separated using two-dimensional TLC. The phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL), monomethyl PE (MMPE), dimethyl PE (DMPE) and the ornithine lipids (OLs) S1, S2, P1 and P2 are indicated.
Growth and characterization of the lipid composition of mutants 899-olsCΔ1, MAV04 and MAV05: OlsC is important in conferring stress resistance
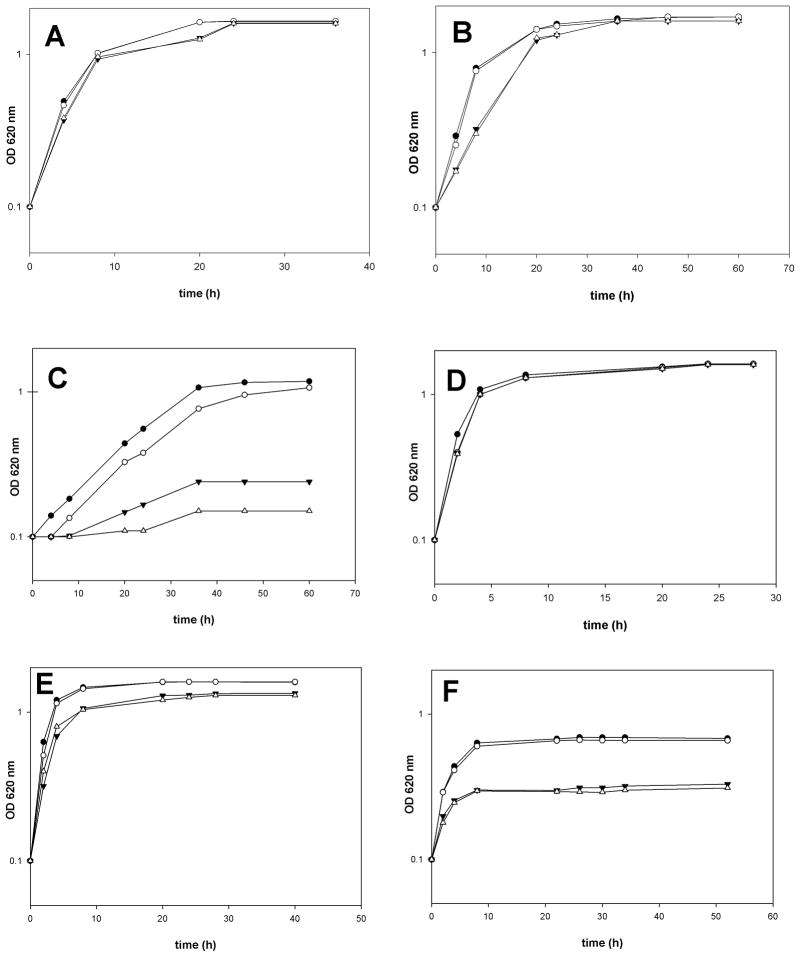
An earlier study in Burkholderia cepacia (Taylor et al., 1998) had shown an increase in formation of hydroxylated OL when the bacteria were grown at higher temperature. In this study we have shown that the relative amounts of the different OLs shift in response to a change in growth temperature or pH of the medium. Rojas-Jimenez et al. (2005) had observed that a R. tropici strain constitutively expressing olsC was not able to grow at pH 4.5 anymore. Therefore, it was expected that the R. tropici mutants deficient in OL hydroxylation would show a phenotype under conditions of acid or temperature stress. The wild type R. tropici CIAT899 and the three mutants 899-olsCΔ1, MAV04 and MAV05 were cultivated in complex TY medium at pH 4.0, 4.5 and 7.0. At pH 7.0 all four strains divide at a similar rate (Fig. 7A). At pH 4.5 the wild type CIAT899 and the mutant MAV04 (ΔolsE) grow at a similar rate compared to pH 7.0 whereas the other two mutants seem to present a longer generation time (Fig. 7B). At pH 4.0 the wild type CIAT899 and the mutant MAV04 (ΔolsE) grow significantly slower than at pH 4.5 but still both cultures reach a final optical density larger than 1.0, whereas the mutants 899-olsCΔ1 and MAV05 (ΔolsCΔolsE) at most undergo one single division (Fig. 7C). To determine if the observed differences are related to changes in lipid composition, wild type and mutant cells were grown and labelled in the corresponding media and analyzed by TLC in two dimensions (Table 2). At pH 7.0 all four strains show similar concentrations of phospholipids and the distinct patterns of the different OLs typical for each mutant described above. At pH 4.5 both OlsC-deficient mutants (899-olsCΔ1 and MAV05) show a drastic reduction in PE content and a strong increase in S1 to up to more than 40%. At pH 4.0 again, both OlsC-deficient mutants show a very similar lipid composition with S1 being the major membrane lipid and PE being drastically reduced. The wild type CIAT899 apparently forms more P1 under these conditions. It seems that low pH conditions cause the accumulation of OLs in all strains: in the wild type and the mutant MAV04 (ΔolsE) the major lipid accumulated is P1, whereas in the mutants 899-olsCΔ1 and MAV05 (ΔolsCΔolsE) the major lipid is S1. When the wild type CIAT899 and the three mutants deficient in OL hydroxylation were cultivated in TY medium at 30 °C, no differences in generation time can be observed between them (Fig. 7D). At 37 °C, both strains lacking olsC (899-olsCΔ1 and MAV05) seem to grow slightly slower than the other two strains (Fig. 7E). At 42 °C wild type CIAT899 and mutant MAV04 (ΔolsE) grow slower than at the lower temperature and reach a final OD620 of only 0.65 to 0.68 (Fig. 7F). The mutants 899-olsCΔ1 and MAV05 (ΔolsCΔolsE) divide distinctly slower at 42 °C than the two former strains and reach a final OD620 of only 0.3.
Figure 7.
Growth of R. tropici mutants lacking olsC is affected under stress conditions. R. tropici wild type CIAT899 and mutants were grown in complex TY medium adjusted to pH 7.0 (A), pH 4.5 (B) or pH 4.0 (C) at 30 °C or in complex TY medium at 30 °C (D), 37 °C (E) or 42 °C (F). The result of a typical experiment is shown. CIAT 899- closed circles, MAV04- open circles, MAV05-open triangles, 899-olsCΔ1- closed triangles.
The lipid composition of the three mutants deficient in OL hydroxylation and the wild type CIAT899 was also analyzed at the different temperatures (Table 1). For each of the four strains the lipid compositions are very similar at 30 °C and 37 °C. At 42 °C the amount of PG is increased by about 10 to 15% and also CL seems to be a bit more abundant at the higher temperature. The total of the four OLs is decreasing in all four strains. Whereas at 30 °C and 37 °C the sum of S1, S2, P1 and P2 is about 30%, at 42 °C the strains contain only between 10 and 20% OLs.
R. tropici mutants deficient in OlsC cause an increase in nodule number that is reverted by the deletion of olsE
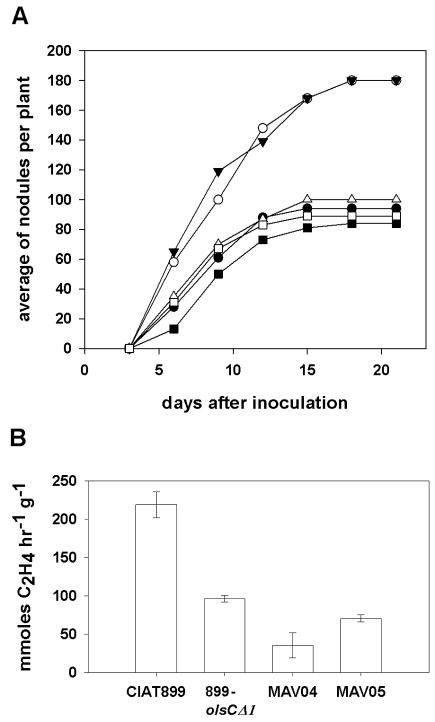
The R. tropici mutant deficient in OlsC (899-olsCΔ1) formed nodules on bean plants that were poorly developed 21 days after inoculation with the bacteria, lacked lenticels, and presented a twofold reduction in nitrogen fixation (Rojas-Jiménez et al., 2005). These results suggested that the R. tropici mutants MAV04 deficient in olsE and double mutants MAV05 deficient in olsC and olsE might also show nodulation phenotypes. Nodulation assays were performed in an agar-based medium in order to be able to observe the kinetics of nodule formation over time. While wild type R. tropici CIAT899 and MAV04 (ΔolsE) produced reproducibly between 80 and 100 nodules per plant, the mutant 899-olsCΔ1 caused the formation of more than 160 nodules per plant. When the olsC-deficient mutant was complemented with the olsC gene, it again formed nodules in numbers similar to the wild type (Fig. 8A). Surprisingly, the double mutant MAV05 (ΔolsCΔolsE) formed a similar number of nodules as the wild type. The nodules were sectioned and while the wild type caused almost exclusively the formation of nodules that were red inside indicating the formation of leghemoglobin, while the mutants caused formation of many small nodules that were whitish on the inside indicating the absence of leghemoglobin (Figure S2). Roots from plants infected with the olsC-deficient mutant 899-olsCΔ1 presented few red nodules and many whitish nodules and roots from plants infected with the olsE-deficient mutant MAV04 presented even less red and more white nodules. On roots infected with the olsC/olsE-deficient double mutant MAV05 almost no red nodules were formed (Fig. S2). Nitrogen fixation per hour and nodule fresh weight was affected in all three mutants in comparison to the wild type (Fig. 8B). These results indicate that the absence of hydroxylated OLs strongly interferes with the development of functional nodules during R. tropici-bean symbiosis.
Figure 8.
Symbiotic phenotypes of R. tropici wild type CIAT899 and strains deficient in OL modification on bean plants. (A) Nodulation assay. Nodules were counted every second or third day. Plants were harvested 21 dpi, nodules were assayed for nitrogen fixation activity. The experiment was repeated three times with five plants for each strain. The result of a typical experiment is shown. CIAT899 (closed circles), 899-olsCΔ1 (open circles), MAV04 (closed squares), MAV05 (open squares), 899-olsCΔ1.pERMAV05 (closed triangles), 899-olsCΔ1.pERMAV15 (open triangles). Uninoculated plants did not develop nodules. (B) Mean acetylene reduction of nodulated bean roots inoculated with wild type R. tropici CIAT899 and mutants 899-olsCΔ1, MAV04 and MAV05. Values are the mean ± SD of three repetitions.
DISCUSSION
Although OLs are widespread in eubacteria (López-Lara et al., 2003, Geiger et al., 2010) the genes olsB and olsA responsible for OL biosynthesis were only recently described in S. meliloti (Weissenmayer et al., 2002, Gao et al., 2004). In addition to the unmodified OL consisting of a 3-hydroxy fatty acid linked in an amide bond to the α-amino group of ornithine and a second fatty acid bound in an ester linkage to the first, several hydroxylated forms of OL have been described in organisms diverse as Burkholderia cepacia (Taylor et al., 1998), Rhizobium tropici (Rojas-Jiménez et al., 2005), Flavobacterium (Kawai et al., 1988), Thiobacillus (Knoche & Shively, 1972), Streptomyces (Asselineau, 1991) and some Ralstonia (Galbraith et al., 1999) species. The fact that the genes coding for the enzymes responsible for the hydroxylation of OLs have not been identified except for the case of R. tropici where the gene olsC was described (Rojas-Jiménez et al., 2005), has made it difficult to study the function of these hydroxylated forms of OL.
Apparently OL and especially their hydroxylated forms play a role in stress response as has been observed by Rojas-Jiménez et al. (2005) and Taylor et al. (1998). R. tropici mutants deficient in the formation of the hydroxylated OL P1 (899-olsCΔ1 and MAV05) are affected in growth at low pH and at high temperature in comparison to the wild type. It has to be mentioned that in an earlier study the mutant 899-olsCΔ1 grew as well as the wild type (Rojas-Jiménez et al., 2005). The explanation for this difference is unknown, but possibly slight differences in the pH of the medium cause drastic differences in the growth behaviour of the mutant. At pH 4.0 and 4.5 a drastic increase in the formation of OLs was observed when compared to growth at neutral pH. In the wild type CIAT899 and in the mutant MAV04 (ΔolsE) especially P1 is increased, whereas in the olsC-deficient mutants unable to form P1 the substrate S1 is accumulating. This probably means that under acid growth conditions OL biosynthesis via OlsB and OlsA is induced. It is less clear what happens at the elevated growth temperature. Although the concentration of OLs is decreased during growth at 42 °C in comparison to 30 °C, again the presence of P1 seems to be of importance as olsC-deficient mutants show a growth phenotype under this condition. The elevated temperature also seems to interfere with OlsE activity as S2 and P2 cannot be detected.
Dees and Shively (1982) made the observation that in the extreme acid tolerant bacterium Thiobacillus oxidans OLs are accumulated in the outer membrane and therefore speculated about a role for OL in acid resistance in this organism (Dees & Shively, 1982). From the growth phenotype of the mutants unable to form P1 it is apparent that the hydroxylation at the 2 position of the secondary fatty acid is of importance under acid growth conditions. Our localization study confirms that although OLs seem to be present in both membranes, they show a higher relative abundance in the outer membrane. Both studies therefore agree that OLs play a role in acid resistance, but it is not clear by which mechanism this effect of OLs is exerted. The hydroxyl group introduced by OlsC in the 2 position of the secondary fatty acid may increase hydrogen bonding between neighbouring OL molecules similarly as has been suggested for LpxO-hydroxylated lipid A in Salmonella and hydroxylated sphingolipids (Nikaido, 2003, Murata et al., 2007). These additional hydrogen bonds should result in bilayer stabilization and a decrease in membrane permeability which could explain the decrease in acid and temperature resistance of OlsC-deficient mutants.
In this study we identified the OL hydroxylase OlsE using a functional expression screening. OlsE belongs to the fatty acyl hydroxylase superfamily, unlike the other OL hydroxylase OlsC from R. tropici which belongs to the aspartyl-/asparaginyl β-hydroxylase protein family to which also the lipid A-myristate β-hydroxylase LpxO from Salmonella typhimurium belongs (Gibbons et al., 2000, Gibbons et al., 2008). The closest homologs to OlsC from R. tropici are present in the α-proteobacteria Agrobacterium radiobacter, Agrobacterium vitis, Ochrobactrum anthropi, Brucella species, and in several cyanobacteria. Unlike other hydroxylations described in OL, the hydroxylation introduced by OlsE seems to be unique because it occurs in the ornithine moiety, but not in the fatty acid moieties as has been described for example in T. thiooxidans, Burkholderia cepacia or R. tropici (this study). Unrelated ornithine hydroxylases like for example PvdA from Pseudomonas aeruginosa have been described and studied in some detail (Meneely et al., 2009, Visca et al., 1994). PvdA is involved in pyoverdin biosynthesis and introduces a hydroxyl group in the δ-amino group of ornithine but is unrelated on sequence level to OlsE. It is not clear yet in which position the OlsE-catalyzed hydroxylation occurs, but apparently the newly introduced hydroxyl group is close enough to the δ-amino group to change its reactivity with ninhydrin. As other members of the fatty acyl hydroxylase superfamily introduce hydroxyl groups at carbon atoms but not at nitrogen atoms OlsE possibly introduces a hydroxyl group at the δ-carbon. It is not clear how the OlsE-dependent hydroxylation might affect membrane characteristics. Possibly the OlsE-dependent hydroxylation enables the OLs S2 and P2 to form a lactone ring within the ornithine headgroup, the presence of which should change its biophysical properties drastically.
The closest OlsE homologs are present in some α-proteobacteria and more distant homologs are present in several actinobacteria, a few γ-proteobacteria and a few other α-proteobacteria. Possibly several of the closer homologs also function as OL hydroxylases. For the OlsE homolog Atu0318 from A. tumefaciens we could show that is responsible for the formation of the OL S2 (data not shown). Distant OlsE-homologs such as the one in Bradyrhizobium japonicum may use distinct substrates. One example for bacterial lipids that are frequently hydroxylated are the hopanoids. In B. japonicum, an α-proteobacteria that forms hopanoids but no OL (Perzl et al., 1998, López-Lara et al., 2003) the OlsE homolog might be responsible for the hydroxylation of hopanoids.
The R. tropici mutants deficient in OL hydroxylation showed nodulation phenotypes, indicating that an adequate concentration of the correct OLs is required for the establishment of a successful symbiosis. It is possible that the nodulation phenotype is partly a consequence of the acid sensitivity phenotype, as during establishment of the symbiosis between rhizobia and legumes the bacteria are exposed to low pH conditions in the rhizosphere and later again inside symbiosomes (Udvardi & Day, 1997). Other aspects, however, seem to be important as well as the OlsE-deficient mutant grows like the wild type in media at pH 4.0, but still presents a severe nodulation phenotype. Modification of OL might be also of importance for the animal pathogen Brucella that has to survive acid pH conditions in the range of 4.0 to 4.5 inside phagosomes (Kohler et al., 2002). Brucella species form OL in a constitutive manner (Comerci et al., 2006, Bukata et al., 2008) and additionally have a close homolog to OlsC from R. tropici which makes it probable that they can form the hydroxylated OL P1. If hydroxylated OLs really play a role in conferring acid resistance then Brucella mutants deficient in their OlsC homolog might be affected in their survival inside phagosomes.
The exact function of OL S1 and its hydroxylated forms is still not known, although our data argue for an important role in stress resistance. The knowledge of the complete scheme of OL biosynthesis in R. tropici should facilitate future functional studies on the role of OLs. In addition, the phenomenon of over-complementation described above allows the construction of R. tropici strains principally accumulating one specific class of OL. Characterization of these strains should make it possible to assign roles to the different forms of OL.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in the present work and their relevant characteristics are shown in Table 4. Rhizobium tropici strains were grown in complex TY medium that contained 10 mM CaCl2 (Beringer, 1974) at 30 °C, 37 °C or 42 °C. Acidic media at pH 4.0 and 4.5 were buffered with 25 mM Homopipes (Research Organics, Cleveland, OH, U.S.A.) adjusted to the respective pH with NaOH, and media at pH 7.0 were buffered with 25 mM Hepes (Sigma). Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37 °C (Sambrook & Russell, 2001). When needed, antibiotics were added at the following final concentrations (μg/mL): kanamycin (Km) 50; carbenicillin (Cb) 100; tetracycline (Tc) 10; nalidixic acid (Nal) 20; and chloramphenicol (Cm) 60.
Table 4.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Rhizobium tropici strains | ||
| CIAT899 | wild type; Acid tolerant, Nalr | (Martínez-Romero et al., 1991) |
| 899-olsCΔ1 | CIAT899 carrying a 211 bp nonpolar deletion in olsC | (Rojas-Jiménez et al., 2005) |
| MAV04 | CIAT899 carrying a deletion in olsE | This work |
| MAV05 | CIAT899 carrying deletions in olsC and olsE | This work |
| Sinorhizobium meliloti strains | ||
| CS111 | pssA-deficient mutant of wild type 1021 | (Sohlenkamp et al., 2004) |
| Burkholderia cenocepacia strains | ||
| J2315 | wild type | (Holden et al., 2009) |
| Escherichia coli strains | ||
| DH5α | recA1, Φ80 lacZΔM15; cloning strain | (Hanahan, 1983) |
| S17-1 | thi pro recA hsdR− hsdM+ RP4 integrated in the chromosome, 2-Tc::Mu, Km::Tn7(Tpr/Smr) | (Simon et al., 1983) |
| Plasmids | ||
| pET17b | Expression vector, Cbr | (Studier, 1991) |
| pET9a | Expression vector, Kanr | (Studier, 1991) |
| pRK404 | Broad-host-range vector, tetracycline resistant | (Ditta et al., 1985) |
| pBBR1MCS | Broad-host-range plasmid, chloramphenicol-resistant | (Kovach et al., 1994) |
| pUC18 | Cloning vector, ampicillin-resistant | (Yanisch-Perron et al., 1985) |
| pRK2013 | Helper plasmid; Kmr | (Ditta et al., 1985) |
| pVK102 | cosmid vector | (Vargas et al., 1990) |
| pK18mobsacB | conjugative suicide vector, kanamycin resistant | (Schäfer et al., 1994) |
| pNG23 | olsB of Burkholderia cenocepacia cloned in pET17b | This work |
| pNG25 | olsB of B. cenocepacia subcloned as a BglII/HindIII fragment from pNG23 into BamHI/HindIII-digested pBBR1MCS | This work |
| pCCS98 | olsC of R. tropici in pET9a | This work |
| pCos94 | pVK102 derivative containing the olsE gene | This work |
| pEMAV01 | 1 kb fragment upstream of olsE, cloned as SmaI/BamHI fragment in pUC18 | This work |
| pEMAV02 | 1 kb fragment downstream of olsE, cloned as BamHI/HindIII fragment in pUC18 | This work |
| pEMAV03 | 1 kb upstream and 1kb downstream sequences flanking olsE, cloned into pUC18 | This work |
| pPMAV04 | Suicide vector for construction of mutant MAV04 and MAV05 | This work |
| pURMAV03 | olsE-containing 3.5 kb fragment of pCos94 cloned as PstI/PstI fragment in pUC18 | This work |
| pERMAV04 | olsE-containing 3.5 kb fragment of pCos94 cloned as PstI/PstI fragment in pRK404 | This work |
| pERMAV06 | pET9a cloned as a BamHI fragment into pRK404 | This work |
| pEMAV07 | ORF1 in pET9a | This work |
| pEMAV08 | ORF2 in pET9a | This work |
| pEMAV09 | olsE in pET9a | This work |
| pERMAV11 | pEMAV07 cloned as a BamHI fragment into pRK404 | This work |
| pERMAV12 | pEMAV08 cloned as a BamHI-fragment into pRK404 | This work |
| pERMAV13 | pEMAV09 cloned as a BamHI-fragment into pRK404 | This work |
| pERMAV15 | pCCS98 cloned as a BamHI fragment into pRK404 | This work |
| pEMAV16 | olsC cloned as a BamHI/BglII fragment into BamHI-digested pEMAV09 | This work |
| pERMAV17 | pEMAV16 cloned as a BamHI fragment into pRK404 | This work |
DNA manipulations
Recombinant DNA techniques were performed according to standard protocols (Sambrook & Russell, 2001). The cosmid subclone containing olsE and PCR products were sequenced at Eurofins Medigenomix by the chain termination method. The DNA region containing olsE was analyzed using the NCBI (National Center for Biotechnology Information) BLAST network server (Altschul et al., 1997). Oligonucleotide sequences are listed in table S1.
Expression cloning of the Rhizobium tropici ornithine lipid hydroxylase gene olsE
A cosmid library of R. tropici CIAT899 made in pVK102 using partially digested HindIII genomic DNA fragments (Vargas et al., 1990) was mobilized into Sinorhizobium meliloti CS111.pNG25 by triparental mating using pRK2013 as the helper plasmid (Figurski & Helinski, 1979). CS111.pNG25 was used to facilitate the screening: CS111 is a phosphatidylserine synthase deficient mutant (Sohlenkamp et al., 2004) derived from the wild type 1021 which is constitutively expressing the gene olsB from Burkholderia cenocepacia. CS111.pNG25 will form increased amounts of S1 which is one of the suspected substrates of OlsE while lacking the ninhydrin-positive membrane lipid phosphatidylethanolamine. Plasmid pNG25 was constructed as follows: The oligonucleotide primers oLOP111 and oLOP112, introducing NdeI and HindIII sites, respectively, were used in the PCR to amplify the gene olsB from B. cenocepacia J2315 using genomic DNA as template. After digestion of the PCR product the obtained fragment was cloned into the plasmid pET17b previously digested with the same enzymes to yield the plasmid pNG23. To obtain plasmid pNG25, the BglII/HindIII fragment containing olsB of B. cenocepacia together with the T7 promoter of pET17b was subcloned from pNG23 and cloned into BamHI/HindIII-digested pBBR1-MCS. Via diparental mating using E. coli S17-1 as a donor strain, pNG25 was introduced into S. meliloti CS111 to obtain CS111.pNG25 which was used as a receptor strain for the cosmid bank. Cosmid transconjugants were selected on TY containing the following antibiotics: tetracycline 10 μg/mL; nalidixic acid 20 μg/mL; chloramphenicol 60 μg/mL. Four hundred individual S. meliloti transconjugants harbouring random fragments of the library were picked and streaked for subsequent lipid analysis in small patches (1 cm by 1 cm) on fresh plates. After growth for three days, cells from each patch were collected with a toothpick and swirled in 60 μL chloroform-methanol (1:1, v/v) as described previously (Benning & Somerville, 1992). After the addition of 20 μL of 1 M KCl-0.2 N H3PO4, the tubes were vortexed and centrifuged to separate the organic and aqueous phases. A 10 μL aliquot from the lipid-containing lower phase was spotted on a HPTLC silica gel 60 plate (Merck). The TLC was developed in one dimension using the solvent system chloroform-methanol-glacial acetic acid (130:50:20, v/v). Under these conditions unmodified OL was readily separated from the modified OL we were looking for and from other polar lipids such as PC, PG and CL. Lipids were detected first with iodine and subsequently primary amine containing lipids were visualized by spraying the plates with a solution of 0.2% ninhydrin in acetone and heating the plates at 120 °C. A transconjugant containing a gene modifying S1 should have two ninhydrin-positive lipids being either S2 and S1 similar to the lipid profile of the R. tropici mutant 899-olsCΔ1 or S1 and P1. Once S. meliloti CS111.pNG25.pCos94 had been identified, cosmid pCos94 was isolated and re-introduced by conjugation into CS111.pNG25 to confirm that the lipid phenotype was caused by the presence of the cosmid and not by an independent mutation leading to the activation of an endogenous S. meliloti gene. In this independent transconjugant again the presence of S2 was observed. Next, the insert of pCos94 was digested with PstI. The resulting PstI/PstI-fragments were subcloned into the broad host vector pRK404 and again mobilized into CS111.pNG25 repeating the lipid analysis described above. A pRK404-derived plasmid containing an approximately 3.5 kb insert was identified (pERMAV04) and its insert sequenced after subcloning into pUC18.
Expression of the three candidate ORFs from R. tropici CIAT899
The three candidate ORFs from plasmid pERMAV04 were separately amplified using genomic DNA from R. tropici CIAT899 as a template and XL-PCR polymerase (Applied Biosystems). Specific oligonucleotide primers incorporating NdeI and BamHI sites into the final PCR products were used (oORF1_01 and oORF1_02 for ORF1; oORF2_01 and oORF2_02 for ORF2; oORF3_01 and oORF3_02 for ORF3). After digestion with the respective enzymes, the PCR products were cloned as NdeI/BamHI fragments into pET9a to yield the plasmids pEMAV07, pEMAV08 and pEMAV09, respectively. These three plasmids and pET9a were linearized with BamHI and were cloned into the BamHI site of pRK404, similarly to an earlier description (Gao et al., 2004) yielding the plasmids pERMAV11, pERMAV12, pERMAV13 and pERMAV06, respectively. Plasmids were mobilized into S. meliloti CS111.pNG25 and the lipids of the transconjugants were assayed as described above.
Deletion of the olsE gene from R. tropici CIAT899
Oligonucleotide primers oOlsX899ar1 and oOlsX899ar2 were used in a PCR (XL-PCR kit; Applied Biosystems) to amplify about 1.0 kb of genomic DNA upstream of the putative olsE gene from R. tropici CIAT899, introducing SmaI and BamHI sites into the PCR product. Similarly, primers oOlsX899ab1 and oOlsX899ab2 were used to amplify about 1.0 kb of genomic DNA downstream of the putative olsE gene from R. tropici CIAT899, introducing BamHI and HindIII sites into the PCR product. After digestion with the respective enzymes, PCR products were cloned as SmaI/BamHI or BamHI/HindIII fragments into pUC18 to yield the plasmids pUMAV01 and pUMAV02, respectively. Then, the BamHI/HindIII fragment from pUMAV02 was subcloned into pUMAV01 to yield pUMAV03. Plasmid pUMAV03 was digested with SmaI and HindIII to subclone the regions usually flanking the rhizobial olsE gene into the suicide vector pK18mobsacB (Schäfer et al., 1994) to yield pPMAV04. Via diparental mating using E. coli S17-1 (Simon et al., 1983) as a mobilizing strain, pPMAV04 was introduced into the wild type strain R. tropici CIAT899. Transconjugants were selected on TY medium containing neomycin to select for single recombinants in a first step. The plasmid pK18mobsacB contains the sacB gene (Selbitschka et al., 1993), which confers sucrose sensitivity to many bacteria. Growth of the single recombinants on high sucrose will therefore select for double recombinants and the loss of the vector backbone of pK18mobsacB from the bacterial genome. Single recombinants were grown under nonselective conditions in complex medium for 1 day before being plated on TY medium containing 12% (w/v) sucrose. Several large and small colonies grew after 5 days, and the membrane lipids of eight candidates were analyzed by in vivo labelling during growth on complex medium with [14C]acetate and subsequent TLC (data not shown). Four clones lacking S2 and P2 were identified. Southern blot analysis confirmed that the S2- and P2-deficient strains were indeed double recombinants in which the gene olsE was deleted (data not shown).
Construction of a double mutant deficient in olsE and olsC
To construct a R. tropici double mutant deficient in olsE and olsC, the suicide plasmid pPMAV04 was conjugated into the olsC-deficient mutant 899-olsCΔ1 (Rojas-Jiménez et al., 2005). The selection for double recombinants was performed in two steps as described above. Ten isolated colonies were chosen and their lipids were labelled with [14C]acetate (see below). We used R. tropici CIAT899 and the mutant 899-olsCΔ1 as control strains. The lipids were analyzed by TLC. One strain presented the expected phenotype which is the absence of the OLs S2, P1 and P2. Therefore this colony was called MAV05. Southern blot analysis confirmed that MAV05 was indeed a double recombinant in which the genes olsC and olsE were deleted (data not shown).
Complementation of the R. tropici mutants MAV04, MAV05 and 899-olsCΔ1
To show that the observed mutant phenotypes were caused by the introduced deletion and not by a secondary independent mutation, the mutants were complemented. The olsE-deficient mutant MAV04 was complemented with the plasmid pERMAV13. In this construct olsE is expressed under control of the T7 promoter. In earlier work we had observed constitutive expression from this promoter in different Rhizobiaceae. In the study published by Rojas-Jiménez et al. (2005) the mutant 899-olsCΔ1 was complemented by olsC under its endogenous promoter, but in order to be able to compare the results from the complementation of the olsC-deficient mutant to the complementations of the mutants MAV04 and MAV05 a new plasmid was constructed.
The gene olsC was amplified using genomic DNA from R. tropici CIAT899 as a template and XL-PCR polymerase (Applied Biosystems). Specific oligonucleotide primers incorporating NdeI and BamHI sites into the final PCR product were used (o5B_olsC and o3_olsC). The digested PCR product was cloned into pET9a to yield the plasmid pCCS98. Plasmid pCCS98 was linearized with BamHI and cloned into BamHI-digested pRK404 to yield pERMAV15. To complement the double mutant MAV05 a plasmid containing both olsC and olsE under the control of the T7 promoter was constructed. A DNA fragment containing olsC under the control of the T7 promoter was subcloned from pCCS98 as BamHI/BglII-fragment into the BamHI-digested pEMAV09 yielding plasmid pEMAV16. Plasmid pEMAV16 therefore contains the genes olsC and olsE, both under the control of separate T7 promoters. Subsequently, pEMAV16 was linearized with BamHI and cloned into BamHI-linearized pRK404 to yield pERMAV17.
In vivo labeling of S. meliloti and R. tropici with [14C]acetate and quantitative analysis of lipid extracts
The lipid compositions of bacterial strains were determined following labelling with [1-14C]acetate (Amersham Biosciences). Cultures (1 mL) of wild-type and mutant strains were inoculated from precultures grown in the same medium. After addition of 0.5 μCi [14C]acetate (60 mCi/mmol) to each culture, the cultures were incubated for 4 h. The cells were harvested by centrifugation, washed with 500 μL water and re-suspended in 100 μL water, and lipid extracts were obtained according to Bligh and Dyer (1959). Aliquots of the lipid extracts were spotted on high-performance TLC silica gel 60 (Merck, Poole, U.K.) plates and were separated in two dimensions using chloroform/methanol/water (140:60:10, v/v) as a mobile phase for the first dimension and chloroform/methanol/glacial acetic acid (130:50:20, v/v) for the second dimension. Primary amine-containing lipids were visualized by spraying the plates with a solution of 0.2 % ninhydrin in acetone and subsequent treatment at 120°C for 10 min. To visualize the membrane lipids, developed 2D-TLC plates were exposed to autoradiography film (Kodak) or to a PhosphorImager screen (Amersham Biosciences). The individual lipids were quantified using ImageQuant software (Amersham Biosciences).
Separation of inner and outer membrane and determination of their respective lipid compositions
Membrane separation was performed as described previously (de Maagd & Lugtenberg, 1986, Klüsener et al., 2009), with minor modifications. A 400 mL culture R. tropici CIAT899 was grown in TY medium at 30°C overnight to an OD600 of 0.5 to 0.6. Cells were harvested by centrifugation at 10,000 × g, 4°C, for 10 min. The cells were re-suspended in 24 mL lysis buffer (50 mM Tris-HCl, pH 7.5, 20% (w/v) sucrose, 0.2 M KCl, 0.2 mM dithiothreitol (DTT), 0.2 mg/mL DNase I, 0.2 mg/mL RNase A) and disrupted by two passages through a pre-chilled French pressure cell at 16,000 lb/in2. The lysate was treated with 0.5 mg/mL lysozyme for 1 h on ice and centrifuged at 10,000 × g for 20 min, 4°C, to remove the unbroken cells. The supernatant was centrifuged at 150,000 × g (SW40Ti), 4°C, for 1 h to collect the membranes. The resulting membrane pellet was carefully re-suspended in 2 mL of 20% (w/v) sucrose containing 5 mM EDTA, pH 7.5, and 0.2 mM DTT. Material that was not completely suspended was removed by centrifugation for 5 min at 16,000 × g. The gradient was prepared by layering 7.5 mL 53% (w/v) sucrose over a cushion of 2.5 mL 70% (w/v) sucrose. Both sucrose solutions contained 5 mM EDTA, pH 7.5. The membrane suspension was layered on the top of the gradient, and sucrose density gradient ultracentrifugation was carried out at 100,000 × g (SW40Ti), 4°C, for 16 h. After ultracentrifugation, the separated membranes were fractionated in 500 μL aliquots. For each fraction the protein concentration was estimated, and the density, the NADH activity, and the 2-keto-3-deoxyoctonate (KDO) content were determined. The protein distribution was estimated using absorption measurements at 280 nm (Scopes, 1987). The NADH oxidase activity was determined by the method of Osborn et al. (Osborn et al., 1972) and the KDO content was determined as described earlier after the fractions had been precipitated twice with 10 % (w/v) TCA (Karkhanis et al., 1978). NADH oxidase activity and KDO content were used as marker for the inner and outer membrane, respectively. Fractions corresponding to the inner and the outer membranes were pooled and the lipids were extracted with 1-butanol (Bremer, 1963). Lipids were analyzed using two-dimensional TLC as described above and the lipids were detected by oxidative charring using ceric sulphate in sulphuric acid (Villaescusa & Pettit, 1972). The lipid spots were quantified using the program ImageQuant (Applied Biosystems).
ESI-MS/MS analysis of lipids S1 and S2
In order to identify in which part of the OL S2 the modification is encountered, a 1 L culture of the mutant 899-olsCΔ1 (Rojas-Jiménez et al., 2005) was grown to an optical density at 620 nm of 1.0 in TY medium, and lipids were extracted according to a modified Bligh-and-Dyer procedure (Bligh & Dyer, 1959). Lipids were fractionated using a silica column and chloroform/methanol/water (140:60:8, v/v) as a mobile phase. Fractions were analyzed by one-dimensional TLC using chloroform/methanol/water (140:60:8, v/v) as a mobile phase. Fractions containing OLs were identified by iodine and ninhydrin staining as described above. OL-containing fractions were dried under N2 stream and re-dissolved in methanol/chloroform (1:1, v/v). LC-ESI/MS of lipids was performed using an Agilent 1200 Quaternary LC system coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). An Ascentis® Si HPLC column (5 μm, 25 cm × 2.1 mm) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v). Mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min, and finally returned to 100% A over 0.5 min and held at 100% A for 5 min. The total LC flow rate was 300 μL/min. The post-column splitter diverted ~10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer, with MS settings as follows: IS = −4500 V, CUR = 20 psi, GS1 = 20 psi, DP = −55 V, and FP = −150 V. Nitrogen was used as the collision gas. Data acquisition and analysis were performed using Analyst QS software version 1.1.
Determination of the position of the hydroxyl group introduced by OlsC into ornithine lipids
Large cultures (4 L) of the R. tropici mutant MAV05 and the strain MAV05.pERMAV15 were grown to an OD620 of 0.9in TY medium. MAV05 only forms S1 and MAV05.pERMAV15 forms preferentially P1. Cells were harvested and lipids were extracted from the cell pellets according to a modified Bligh and Dyer method. OLs S1 and P1 were purified using preparative TLC using Si500F plates (Baker) in two steps. First chloroform/methanol/water (140:60:10, v/v) was used as a mobile phase and the OLs were purified from the silica. Enriched OLs were further purified by a second preparative TLC using chloroform/methanol/glacial acetic acid (130:50:20, v/v) as mobile phase. ESI-MS/MS analysis of S1 and P1 was performed as described above.
The derivatization of the lipids was performed essentially as described by Gibbons et al. (2008). Purified OLs S1 and P1 were hydrolyzed in acidic methanol, and then converted to trimethylsilyl (TMS) ethers. Hydroxy fatty acid standards (α- and β-hydroxy palmitic acid, α- and β-hydroxy stearic acid) were processed and analyzed in parallel with the samples. Typically, about 1 mg of sample was dried in a Reacti-vial and samples were hydrolyzed by adding 300 μL of 1 M HCl in methanol and heated at 80 °C for 16 h. The reactions were cooled and solvents were evaporated under a stream of nitrogen. Next, 200 μL Tri-Sil HTP reagent (Thermo) was added to the dried samples. After incubation for one hour at 25 °C a 20 μL aliquot was diluted 1:6 in hexane and transferred to a new vial for GC/MS analysis. GC/MS was performed using a Clarus 600T MS instrument coupled to a Clarus 600 gas chromatography system (Perkin Elmer). The column was a Elite-5 MS (0.32 mm internal diameter and 0.25 μm phase thickness) from Perkin Elmer. The temperature program of the GC was as follows: the column oven temperature was initially held at 140 °C for 6 min, increased to 250 °C at a rate of 4 °C/min and finally held at 250 °C for 5 min. The total run time was 38.5 min. The injector was operated in the split mode, and the temperature of the injector was kept at 250 °C. Helium was the carrier gas at a constant pressure of 7 psi. The instrument was operated in the electron impact (EI) mode with the electron energy set at 70 eV.
Plant tests
P. vulgaris seeds were surface-sterilized with 1.2% sodium hypochlorite and were germinated on 1% agar-water plates as described (Vinuesa et al., 1999). Seedlings were transferred to 250-ml flasks filled 220 ml nitrogen-free nutrient solution (Fahraeus, 1957) containing agar at 0.7 % and were inoculated with about 50000 CFU/ml per plant. Plants were grown in a controlled growth chamber at 28 °C with a 15 h day/9 h night cycle and harvested 21 d after inoculation. Nitrogenase activity of nodulated roots was determined by the acetylene reduction assay as described previously (Martínez et al., 1985). Nitrogen fixation activity per plant was normalized with respect to the nodule fresh weight per plant.
Supplementary Material
Acknowledgments
This research was supported by grants from CONACyT-Mexico (46020-N) and DGAPA/UNAM (IN217907 and IN201310). Z.G. and the mass spectrometry facility in the Department of Biochemistry, Duke University Medical Center, were supported by a LIPID MAPS glue grant (GM-069338) from the National Institutes of Health. M. Á. V.-G. is a PhD student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México and is a recipient of a scholarship from the Consejo Nacional de Ciencia y Tecnología, Mexico. We are grateful to Alfonso Leija Salas for his support in nodule analysis.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthington BA, Bennett LG, Skatrud PL, Guynn CJ, Barbuch RJ, Ulbright CE, Bard M. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- Asselineau J. Bacterial lipids containing amino acids or peptides linked by amide bonds. Fortschr Chem Org Naturst. 1991;56:1–85. doi: 10.1007/978-3-7091-9084-5_1. [DOI] [PubMed] [Google Scholar]
- Bard M, Bruner DA, Pierson CA, Lees ND, Biermann B, Frye L, Koegel C, Barbuch R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci U S A. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Huang ZH, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bremer J. Carnitine in Intermediary Metabolism. The Biosynthesis of Palmitylcarnitine by Cell Subfractions. J Biol Chem. 1963;238:2774–2779. [PubMed] [Google Scholar]
- Bukata L, Altabe S, de Mendoza D, Ugalde RA, Comerci DJ. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J Bacteriol. 2008;190:8197–8203. doi: 10.1128/JB.01069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerci DJ, Altabe S, de Mendoza D, Ugalde RA. Brucella abortus synthesizes phosphatidylcholine from choline provided by the host. J Bacteriol. 2006;188:1929–1934. doi: 10.1128/JB.188.5.1929-1934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maagd RA, Lugtenberg B. Fractionation of Rhizobium leguminosarum cells into outer membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J Bacteriol. 1986;167:1083–1085. doi: 10.1128/jb.167.3.1083-1085.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees C, Shively JM. Localization of quantitation of the ornithine lipid of Thiobacillus thiooxidans. J Bacteriol. 1982;149:798–799. doi: 10.1128/jb.149.2.798-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang XW, Finlay DR, Guiney D, Helinski DR. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Fahraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BG, Shanklin J, Somerville C, Munck E. Stearoyl-acyl carrier protein delta 9 desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci U S A. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith L, Jonsson MH, Rudhe LC, Wilkinson SG. Lipids and fatty acids of Burkholderia and Ralstonia species. FEMS Microbiol Lett. 1999;173:359–364. [Google Scholar]
- Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, López-Lara IM, Geiger O. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol. 2004;53:1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49:46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Geiger O, Röhrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Lin S, Cotter RJ, Raetz CR. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275:32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Reynolds CM, Guan Z, Raetz CR. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A. Biochemistry. 2008;47:2814–2825. doi: 10.1021/bi702457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Jackowski S, Rock CO. Fatty acid and phospholipid metabolism in prokaryotes. In: Vance DE, Vance JE, editors. Biochemistry of lipids, lipoproteins and membranes. Elsevier; 2002. pp. 55–92. [Google Scholar]
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- Karkhanis YD, Zeltner JY, Jackson JJ, Carlo DJ. A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of gram-negative bacteria. Analytical Biochemistry. 1978;85:595–601. doi: 10.1016/0003-2697(78)90260-9. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Yano I, Kaneda K. Various kinds of lipoamino acids including a novel serine-containing lipid in an opportunistic pathogen Flavobacterium. Their structures and biological activities on erythrocytes. Eur J Biochem. 1988;171:73–80. doi: 10.1111/j.1432-1033.1988.tb13760.x. [DOI] [PubMed] [Google Scholar]
- Kennedy MA, Johnson TA, Lees ND, Barbuch R, Eckstein JA, Bard M. Cloning and sequencing of the Candida albicans C-4 sterol methyl oxidase gene (ERG25) and expression of an ERG25 conditional lethal mutation in Saccharomyces cerevisiae. Lipids. 2000;35:257–262. doi: 10.1007/s11745-000-0521-2. [DOI] [PubMed] [Google Scholar]
- Klüsener S, Aktas M, Thormann KM, Wessel M, Narberhaus F. Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes. J Bacteriol. 2009;191:365–374. doi: 10.1128/JB.01183-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoche HW, Shively JM. The structure of an ornithine-containing lipid from Thiobacillus thiooxidans. J Biol Chem. 1972;247:170–178. [PubMed] [Google Scholar]
- Kohler S, Porte F, Jubier-Maurin V, Ouahrani-Bettache S, Teyssier J, Liautard JP. The intramacrophagic environment of Brucella suis and bacterial response. Vet Microbiol. 2002;90:299–309. doi: 10.1016/s0378-1135(02)00215-8. [DOI] [PubMed] [Google Scholar]
- Kok M, Oldenhuis R, van der Linden MP, Raatjes P, Kingma J, van Lelyveld PH, Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J Biol Chem. 1989;264:5435–5441. [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- López-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol Plant Microbe Interact. 2003;16:567–579. doi: 10.1094/MPMI.2003.16.7.567. [DOI] [PubMed] [Google Scholar]
- Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol. 1991;41:417–426. doi: 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- Martínez E, Pardo MA, Palacios R, Cevallos MA. Reiteration of nitrogen fixation gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J Gen Microbiol. 1985;131:1779–1786. [Google Scholar]
- Meneely KM, Barr EW, Bollinger JM, Jr, Lamb AL. Kinetic mechanism of ornithine hydroxylase (PvdA) from Pseudomonas aeruginosa: substrate triggering of O2 addition but not flavin reduction. Biochemistry. 2009;48:4371–4376. doi: 10.1021/bi900442z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AG, Martin CE. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J. Mechinism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Perzl M, I, Reipen G, Schmitz S, Poralla K, Sahm H, Sprenger GA, Kannenberg EL. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim Biophys Acta. 1998;1393:108–118. doi: 10.1016/s0005-2760(98)00064-2. [DOI] [PubMed] [Google Scholar]
- Rojas-Jiménez K, Sohlenkamp C, Geiger O, Martínez-Romero E, Werner D, Vinuesa P. A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol Plant Microbe Interact. 2005;18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Scopes RK. Measurement of enzyme activity. In: Cantor CR, editor. Protein Purification-Principles and Practice. New York: Springer; 1987. pp. 253–283. [Google Scholar]
- Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Selbitschka W, Niemann S, Pühler A. Construction of gene replacement vectors for gram-negative bacteria using a genetically modified sacRN as a positive selection marker. Appl Microbiol Biotechnol. 1993;38:615–618. [Google Scholar]
- Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A braod host range mobilization system fir in vivo genetic engeneering: Transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Sohlenkamp C, de Rudder KE, Geiger O. Phosphatidylethanolamine is not essential for growth of Sinorhizobium meliloti on complex culture media. J Bacteriol. 2004;186:1667–1677. doi: 10.1128/JB.186.6.1667-1677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Galindo-Lagunas KA, Guan Z, Vinuesa P, Robinson S, Thomas-Oates J, Raetz CR, Geiger O. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol Plant Microbe Interact. 2007;20:1421–1430. doi: 10.1094/MPMI-20-11-1421. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, I, López-Lara M, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog Lipid Res. 2003;42:115–162. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Hayakawa T, Shaw JP, Rekik M, Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CJ, Anderson AJ, Wilkinson SG. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology. 1998;144(Pt 7):1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA. Metabolite Transport across Symbiotic Membranes of Legume Nodules. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:493–523. doi: 10.1146/annurev.arplant.48.1.493. [DOI] [PubMed] [Google Scholar]
- Vargas C, Martinez LJ, Megias M, Quinto C. Identification and cloning of nodulation genes and host specificity determinants of the broad host-range Rhizobium leguminosarum biovar phaseoli strain CIAT899. Mol Microbiol. 1990;4:1899–1910. doi: 10.1111/j.1365-2958.1990.tb02039.x. [DOI] [PubMed] [Google Scholar]
- Villaescusa FW, Pettit GR. Steroids and related natural products. 69. Synthesis of 20(22)-Dihydro-23-deoxodigitoxigenin. J Org Chem. 1972;37:569–572. doi: 10.1021/jo00969a010. [DOI] [PubMed] [Google Scholar]
- Vinuesa P, Neumann-Silkow F, Pacios-Bras C, Spaink HP, Martínez-Romero E, Werner D. Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol Plant Microbe Interact. 2003;16:159–168. doi: 10.1094/MPMI.2003.16.2.159. [DOI] [PubMed] [Google Scholar]
- Vinuesa P, Reuhs BL, Breton C, Werner D. Identification of a plasmid-borne locus in Rhizobium etli KIM5s involved in lipopolysaccharide O-chain biosynthesis and nodulation of Phaseolus vulgaris. J Bacteriol. 1999;181:5606–5614. doi: 10.1128/jb.181.18.5606-5614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme L-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenmayer B, Gao JL, López-Lara IM, Geiger O. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol Microbiol. 2002;45:721–733. doi: 10.1046/j.1365-2958.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.