Abstract
CRE-loxP-mediated inactivation and activation of genes in mouse mammary epithelium have been widely used to study genetic pathways in normal development and neoplastic transformation in vivo. In 1997 we generated three distinct mouse lines carrying an identical MMTV-Cre transgene (lines A, D and F). Since the presence of CRE recombinase can adversely affect the physiology of non-mammary cells, we have explored whether transgenic females display lactational defects. While dams from line D nurse their pups and display overtly normal mammary development, line A shows some impairment in lactation and females from line F completely fail to nurse their litters. The ability to nurse a litter correlates with the extent of alveolar development and differentiation. This study demonstrates the importance of including appropriate “Cre-only” controls and provides guidelines to avoid problems in data interpretation.
Keywords: mouse mammary tumor virus, long terminal repeat, CRE recombinase, mammary gland, lactation, transgenic mice
Introduction
Loss-of-gene studies in mice have been instrumental in deciphering protein function in development, physiology and cancer. In particular, CRE recombinase-mediated deletion of genes targeted with loxP sites has provided detailed information on cell-specific roles of genes. Over the past decade several distinct transgenic mouse strains expressing CRE recombinase in mammary epithelium have been established. These strains are based on three regulatory elements; promoter sequences from the mouse whey acidic protein (WAP) [1; 2; 3; 4] and ovine -lactoglobulin [5] genes as well as the mouse mammary virus (MMTV) long terminal repeat (LTR) [1; 2; 6; 7; 8; 9]. It is a given that distinct lines of transgenic mice carrying a particular transgene display unique expression patterns due to the specific chromosomal integration site. Therefore, the suitability of a particular strain for a specific study has to be assessed prior to the experiment. Because of reports that CRE expression might interfere with the physiology of target cells [10; 11; 12], it is essential that “Cre-only” controls be included in these experiments. This is of special importance in cases where only minor defects on lactation are ascribed to the deletion of a gene.
MMTV-Cre mice generated in our laboratory [1; 2] have been used by over 40 research groups to inactivate genes in mammary epithelium, which in many cases resulted in impaired mammary development and lactation failure. However, there has been no comprehensive study to determine the effect of CRE expression in these lines on the development of mammary tissue during pregnancy and the ability to nurse litters. In this report we evaluate the suitability of the three strains for deletion studies in mammary epithelium of primiparous animals by analyzing the ability to support a litter and discuss advantages and disadvantages of each transgenic line.
Results and Discussion
The following MMTV-Cre transgenic lines [1] were used for a comparison by histological examination and lactational performance: From Jackson Laboratory, Line A {Tg(MMTV-cre)1MamJ; stock number 003551} and Line D {Tg(MMTV-cre)4MamJ; stock number 003553} and Line F from the mouse repository at NCI-Frederick, MD (B6.Cg-Tg(MMTV-Cre)FMam; strain 01XA9). All the strains are currently on a mixed background and have not been backcrossed to an inbred strain. Lines A and D have by far been the most frequently used (at least 7 and 20 research groups, respectively, based on the database at www.informatics.jax.org). A list of all identifiable publications in which these lines were used can be found in the supplementary references.
Pup growth
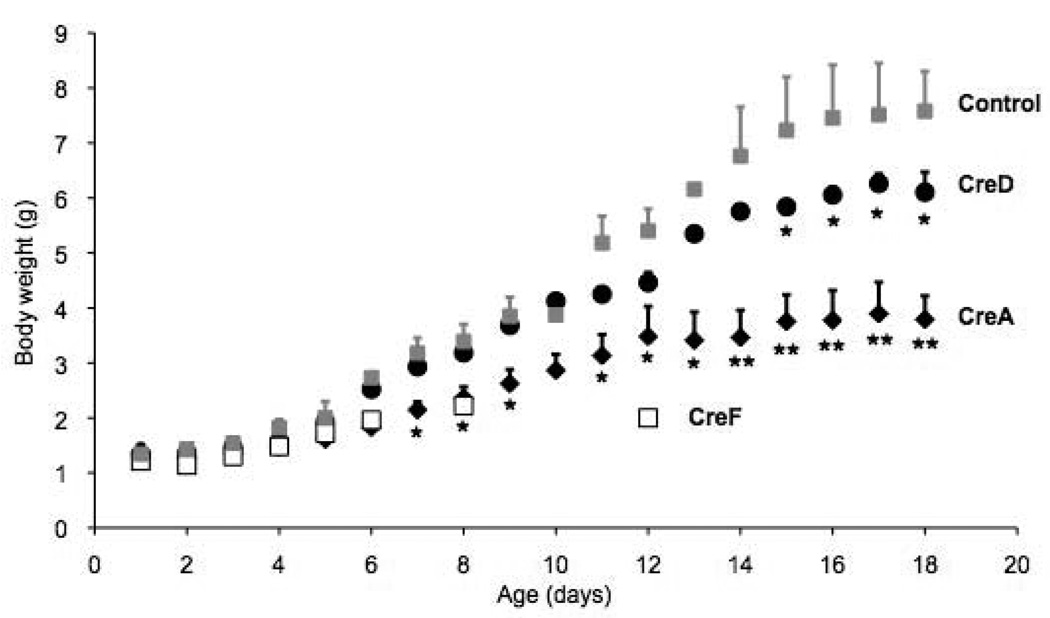
In order to detect a possible toxicity of the CRE protein on mammary epithelium we mated females of all three lines and monitored pup weights and survival of the first litter (Figure 1). Pups born to non-transgenic littermates from each of the lines served as controls and each line was compared to the growth of these pups. Lowest toxicity occurred in CreD dams with 81% of the born pups surviving. Pups from CreD dams were smaller than those nursed by control dams throughout the nursing phase but the differences were not statistically significant (Figure 1). They weighed 15% less at weaning (post partum day 21) (Table 1). Pups born to and nursed by dams from the CreA line survived to 38%. Survivors were runts, had lower weights throughout the nursing period (p value <0.05 for post natal days 7 to 18) and weighed only 40% of controls at weaning (Figure 1). None of 56 pups born to 7 dams from the CreF line survived to weaning, most died between 2 and 3 days after birth and only one pup survived for 12 days with minimal weight gain (Figure 1).
Figure 1. Weight gain of pups nursed by MMTV-Cre transgenic and control mothers.
Pup weight is shown in g/pup ± standard error of the mean.
Table 1.
Pup growth at pp day 21
| Cre line | CreA (n=9) | CreD (n=10) | CreF (n=7) | Control (n=7) |
|---|---|---|---|---|
| Weight (g/pup) | 3.3 ± 0.3 | 7.1 ± 0.2 | n.a. | 8.3 ± 1.5 |
| Survival | 25/66 (38%) | 74/91 (81%) | n.a | 31/41 (76%) |
The number of pups delivered by primiparous females was recorded at birth and pups were weight every other day. At least 7 litters were weighed per line (parentheses in first row). Weight ± standard deviation (middle row) and number of pups at weaning at 21 days post partum (pp) per number of pups born (lower row) are given. The percent of surviving pups is indicated in parentheses (lower row).
Mammary development
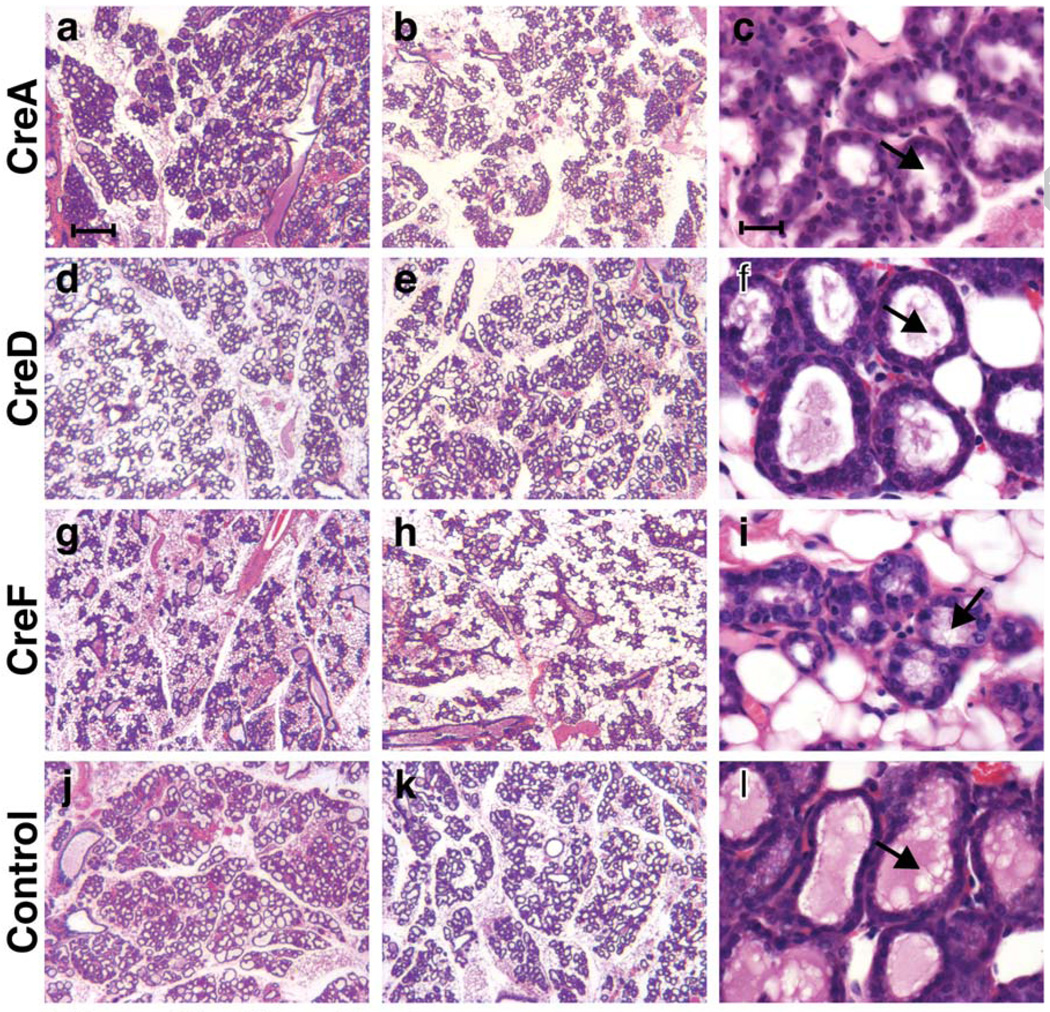
Mammary gland development during pregnancy was monitored the morning after parturition by harvesting one gland and preparing wholemounts and histological sections. Similar to samples from inbred strains some variability of mammary tissue with respect to ratio of stroma to epithelium and expansion of alveoli was observed. Thus, we failed to consistently assign the sections (Figure 2) or wholemounts (data not shown) unequivocally to a specific line in a blinded experiment conducted independently by both authors. However, the overall appearance of the tissue in the three lines correlated with their ability to nurse. Low magnification images of two mice with the most and least developed glands from each line are presented to illustrate very heterogeneous and incomplete filling of the fat pads, in particular those from line F (Figure 2g and h) and to a lesser extent line A (Figure 2a and b). The alveoli were expanded in all the samples but the presence of lipid droplets and milk, the hallmarks of differentiation, differed among the samples (Figure 2c, f, i and l).
Figure 2. Alveolar development in MMTV-Cre strains.
Representative areas of on H&E stained sections of mammary tissues collected at term from CreA (a – c), CreD (d – f), CreF (g – i) and wild type (j – l) mice are shown. Low magnification images of two mice from each line (least and most developed) are shown to illustrate mouse-to-mouse variation and regional heterogeneity (left 2 panels). Mammary epithelial cells appear morphologically differentiated at higher magnification in lines CreA (c), CreD (f), CreF (i) and controls (l) (right panel row). The lumina are filled to varying extent with proteinaceous material and milk fat globules (arrows). Scale bar = 150 min a, b, d, e, g, h, j and k; 10 min c, f, i and l.
It should be noted that the mice were not backcrossed onto a specific genetic background and this could contribute to some extent to the heterogeneity. It is known that different genetic strains of mice exhibit different lactation abilities but this has not been explored systematically. However, the inability to rear litters as seen here would be detrimental to the maintenance of a mouse strain and genetic heterogeneity alone cannot explain our observations.
Mosaicism
There are additional distinctive characteristics of the three lines that are particularly relevant for investigations of mammary gland development. In lines A and F CRE activity was detected already in glands from newborn females, as monitored by a ROSA26 LacZ reporter line [13; 14]. CRE activity in line D is initiated at about three weeks of age when pups enter puberty [2]. Despite initial mosaic deletion complete deletion was achieved in older mice probably due to activation of the MMTV-LTR in each estrous cycle (data not shown; Lu et al., 2008). Depending on the nature of the targeted gene mosaic expression can result in a selection of cells containing undeleted genes. This is illustrated in a series of experiments where CreA and CreD were used to excise the Stat5a/b locus. STAT5 is the crucial transcription factor for mammary epithelial cell development and its absence affects cell proliferation, maintenance and differentiation [17]. While Stat5fl/fl;CreD mice displayed attenuated alveolar development during pregnancy and were unable to maintain litters in the first lactation they attained the ability to sustain litters in subsequent pregnancies as a consequence of outgrowth of cells that had failed to delete the floxed Stat5 locus [15]. Absence of alveolar development was observed in Stat5fl/fl;CreA mice even after four to five pregnancies indicating uniform CRE activity in all epithelial cells [18]. In another study we observed a strong phenotype upon the deletion of Rbpsuh with CreA but not with WAP-Cre [14].
On the other hand, the mosaic nature of gene deletion allows a direct comparison of the effect on cell behavior within a tissue as employed by Lu and colleagues (2008). It also suggests that deletion should be demonstrated on the cellular level ideally by a cell based detection method such as immunostaining or crossing with a reporter strain (Yalcin-Ozuysal et al., 2010). Stochastic expression poses less of a problem when deletion of loxP-flanked sequences is used to activate oncogenes or remove tumor suppressor genes. This scenario might actually represent a more realistic condition compared to uniform overexpression of the oncogene.
It is also important to note, that in the A and F lines CRE is expressed in the female germline and leads to a deletion of floxed sequences when inherited from a MMTV-Cre transgenic mother.
The three transgenic lines described here have been generated with the same transgenic construct and the differences in expression patterns most likely are caused by the different chromosomal sites of transgene integration. It is possible that one the integration of the transgene could affect expression of a gene that plays a critical role in mammary gland development, a scenario that could be explored further.
Strength and weaknesses of each line
In addition to the three MMTV-Cre lines described, six other Cre lines have been used to disrupt or activate genes selectively in mammary epithelium (Table 2). One MMTV-Cre line was generated by Li et al. [7], two each by the laboratories of William Muller [6; 19] and Eva Lee [8] and one by Boussadia et al. [9]. The lines reported by Li et al. (2002), Lin et al. (2004), Boussadia et al. (2002) and Andrechek et al. (2000) resemble the ones we have generated in that the MMTV-LTR is controlling the Cre gene. In contrast, the line generated by Ursini-Siegel and colleagues (2008) carries both the ErbB2 oncogene and the Cre gene under the control of the MMTV-LTR, which ensures expression of CRE and ERBB2 in the same cell (Ursini-Siegel et al., 2008). Four lines use regulatory elements from whey protein genes, the beta-lactoglobulin [5] and whey acidic protein (WAP) genes [1; 3; 4]. In these lines CRE activity is tightly linked to pregnancy and is restricted to mammary alveolar cells. Especially in the lines from Ludwig et al. (2001) and Wintermantel et al. (2002) where the transgene was inserted into the WAP gene by homologous recombination.
Table 2.
Characterization of mammary specific CRE transgenic lines
| CRE line | alveoli | ducts | mosaicism | oocytes | skin | immune |
|---|---|---|---|---|---|---|
| MMTV-Cre (A) | Y | Y | N | Y | Y | Y |
| MMTV-Cre (D) | Y | Y | Y | N | Y | Y |
| MMTV-Cre (F) | Y | Y | Y | Y | Y | Y |
| MMTV-Cre (Muller) | Y | Y | Y | NR | NR | NR |
| MMTV-NIC | Y | Y | Y | NR | NR | NR |
| MMTV-Cre (Kemler) | Y | NR | NR | NR | NR | NR |
| MMTV-Cre (Lane) | Y | Y | NR | NR | NR | NR |
| WAP-Cre (Ludwig) | Y | N | NR | N | N | N |
| WAP-Cre (Wagner) | Y | N | Y | N | N | N |
| WAPiCre | Y | NR | NR | N | N | N |
| BLG-Cre | Y | NR | NR | NR | N | N |
N - no expression
NR - not reported
Y - expression
Cell-specificity
While the BLG-Cre and WAP-Cre mice exhibit an exquisite mammary alveolar specificity, MMTV-Cre transgenes are expressed, at least temporally, in a number of other cell types. MMTV-Cre-mediated gene deletion is detected not only in mammary epithelium but also in other epithelia and hematopoietic lineages. In particular, the expression of MMTV-LTR in salivary epithelium, prostate and skin has been used to investigate development and carcinogenesis in these organs (see supplementary references). In general, deletion in the immune system is not as extensive as in mammary epithelium. However, if loss of the gene in question in immune cells or any of the other off target cells affects mammary epithelium, it would be necessary to transplant the gene knock-out mammary epithelium to demonstrate the epithelial cell intrinsic nature of the gene loss.
Conclusion
The purpose of this report is to inform researchers about physiological differences observed in three lines of transgenic mice expressing an identical Cre transgene targeted to the mammary gland. This knowledge should be considered in choosing a specific line for gene deletion in mammary epithelium and interpreting data that indicate impairment in mammary function. The transgenic expression of CRE can cause cellular toxicity as demonstrated in various systems before [10; 11; 12] and requires “Cre-only” controls. Moreover, mosaic deletion activity can lead to artifacts such as selection for only undeleted cells depending on the specific effect the absence of a gene elicits. It also calls for careful monitoring of the deletion on a cellular level. Having said this, mosaic deletion also provides a means to elegantly study wild type and knock-out cells in close proximity within the same tissue by microscopic analyses.
Methods
Preparation of histology samples
Mature females 2 to 3 months of age were mated and inspected for vaginal plugs daily. On the morning after delivering pups animals were anesthetized and one of the inguinal glands was collected by biopsy. Mammary samples were fixed in neutral buffered formalin and processed for paraffin embedding or whole mount staining. At least 3 animals per transgenic line were sampled. All procedures were approved by the Animal Care and Use Committee of NIDDK. The animals were treated humanely in agreement with NIH regulations.
Detection of gene deletion by -glactosidase staining
Tissues of two-month old bitransgenic MMTV-Cre/ROSA26-lox-Stop-lox-LacZ virgins were spread on a glass slide and fixed for one hour for X-gal staining as previously described (Wagner et al., 2001). After color development they were fixed for 2 hours in neutral buffered formalin, dehydrated and mounted with Permount.
Determination of pup growth
Pups from at least seven litters from the different Cre lines were counted and weighed each day. Weight (g) per pup vs pup age (days) was plotted and compared by Fisher’s t test to pups nursed by non-transgenic littermates (4), which served as controls. The average number of pups per litter was between 7 and 10.
Supplementary Material
Acknowledgement
Dr. Oksana Gavrilova is gratefully acknowledged for help with statistical analyses. This work was supported by the Intramural Research Program of NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 3.Wintermantel TM, Mayer AK, Schutz G, Greiner EF. Targeting mammary epithelial cells using a bacterial artificial chromosome. Genesis. 2002;33:125–130. doi: 10.1002/gene.10097. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig T, Fisher P, Murty V, Efstratiadis A. Development of mammary adenocarcinomas by tissue-specific knockout of Brca2 in mice. Oncogene. 2001;20:3937–3948. doi: 10.1038/sj.onc.1204512. [DOI] [PubMed] [Google Scholar]
- 5.Selbert S, Bentley DJ, Melton DW, Rannie D, Lourenco P, Watson CJ, Clarke AR. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 1998;7:387–396. doi: 10.1023/a:1008848304391. [DOI] [PubMed] [Google Scholar]
- 6.Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci U S A. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Robinson GW, Lesche R, Martinez-Diaz H, Jiang Z, Rozengurt N, Wagner KU, Wu DC, Lane TF, Liu X, Hennighausen L, Wu H. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development. 2002;129:4159–4170. doi: 10.1242/dev.129.17.4159. [DOI] [PubMed] [Google Scholar]
- 8.Lin SC, Lee KF, Nikitin AY, Hilsenbeck SG, Cardiff RD, Li A, Kang KW, Frank SA, Lee WH, Lee EY. Somatic mutation of p53 leads to estrogen receptor alpha-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res. 2004;64:3525–3532. doi: 10.1158/0008-5472.CAN-03-3524. [DOI] [PubMed] [Google Scholar]
- 9.Boussadia O, Kutsch S, Hierholzer A, Delmas V, Kemler R. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech Dev. 2002;115:53–62. doi: 10.1016/s0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- 10.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 11.Lee JY, Ristow M, Lin X, White MF, Magnuson MA, Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J Biol Chem. 2006;281:2649–2653. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 12.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 14.Buono KD, Robinson GW, Martin C, Shi S, Stanley P, Tanigaki K, Honjo T, Hennighausen L. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 15.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu P, Ewald AJ, Martin GR, Werb Z. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol. 2008;321:77–87. doi: 10.1016/j.ydbio.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 18.Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ursini-Siegel J, Hardy WR, Zuo D, Lam SH, Sanguin-Gendreau V, Cardiff RD, Pawson T, Muller WJ. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J. 2008;27:910–920. doi: 10.1038/emboj.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.