Abstract
LIS1 is a critical regulator of dynein function during mitosis and organelle transport. Here, we investigated how Pac1, the budding yeast LIS1 homologue, regulates dynein targeting and activity during nuclear migration. We show that Pac1 and Dyn1 (dynein heavy chain) are dependent upon each other and upon Bik1 (budding yeast CLIP-170 homologue) for plus end localization, whereas Bik1 is independent of either. Dyn1, Pac1 and Bik1 interact in vivo at the plus ends, where an excess amount of Bik1 recruits approximately equal amounts of Pac1 and Dyn1. Overexpression of Pac1 enhanced plus end targeting of Dyn1 and vice versa, while affinity-purification of Dyn1 revealed that it exists in a complex with Pac1 in the absence of Bik1, leading us to conclude that the Pac1-Dyn1 complex preassembles in the cytoplasm prior to loading onto Bik1-decorated plus ends. Strikingly, we found that Pac1-overexpression augments cortical dynein activity through a mechanism distinct from loss of She1, a negative regulator of dynein-dynactin association. While Pac1-overexpression enhances the frequency of cortical targeting for dynein and dynactin, the stoichiometry of these complexes remains relatively unchanged at the plus ends compared to that in wild-type cells (~3 dynein to 1 dynactin). Loss of She1, however, enhances dynein-dynactin association at the plus ends and the cell cortex, resulting in an apparent 1:1 stoichiometry. Our results reveal differential regulation of cortical dynein activity by She1 and Pac1, and provide a potentially new regulatory step in the off-loading model for dynein function.
Keywords: LIS1, CLIP-170, Pac1, Bik1, She1, dynein, dynactin
Introduction
LIS1 is a highly conserved protein that was originally identified as the target for sporadic mutations causing human type I lissencephaly (Reiner et al. 1993), a brain developmental disease that arises from defects affecting both the division and migration of neuronal precursor cells (Tsai et al. 2005). Although highly enriched in the brain (Smith et al. 2000), LIS1 function is not restricted to the nervous system. Elegant genetic and cellular studies (Faulkner et al. 2000; Smith et al. 2000; Xiang et al. 1995) have revealed that LIS1 plays a major role in non-neuronal cells as a regulator of cytoplasmic dynein, a minus end-directed microtubule (MT) motor that participates in numerous aspects of mitosis (Howell et al. 2001; O’Connell and Wang 2000; Rusan et al. 2002; Salina et al. 2002), intracellular transport (Burakov et al. 2003; Paschal et al. 1987; Vaughan et al. 1999), and directed cell migration (Dujardin et al. 2003).
In dividing cultured vertebrate cells, LIS1 localizes prominently to prometaphase kinetochores and the cell cortex (Coquelle et al. 2002; Faulkner et al. 2000), known sites of action for cytoplasmic dynein and its accessory complex dynactin. Several lines of evidence indicate that kinetochore LIS1 plays a role in dynein motor regulation rather than dynein motor recruitment. Most notably, targeting of LIS1 to kinetochores is dependent on dynein and dynactin, but not vice versa (Coquelle et al. 2002; Tai et al. 2002). Overexpression of LIS1 displaces CLIP-170 from the kinetochore without affecting dynein targeting, suggesting that LIS1 mediates the linkage between CLIP-170 and the motor protein complexes at this site (Tai et al. 2002). Additionally, overexpression of LIS1 or injection of an anti-LIS1 antibody delays anaphase onset and interferes with chromosome congression to the metaphase plate (Faulkner et al. 2000), a known activity of dynein, further implicating a role for LIS1 in dynein motor regulation at the kinetochore.
In contrast, the role for LIS1 at the cortex of mitotic cells is poorly understood. A defect in spindle orientation was reported in LIS1-overexpressing polarized MDCK cells, supporting the notion that LIS1 regulates dynein-dependent interactions between astral MTs and the cell cortex (Faulkner et al. 2000). LIS1 has been observed to colocalize with CLIP-170 at punctate cortical sites and the distal tips of astral MTs in mitotic HeLa cells (Coquelle et al. 2002), although the functional significance of this colocalization with regard to dynein-mediated spindle orientation is not known. Dynein and dynactin have also been reported to localize to punctate cortical sites and the distal tips of astral MTs in several mammalian cell lines (Busson et al. 1998; Faulkner et al. 2000; O’Connell and Wang 2000), but the extent to which the distribution of dynein and dynactin overlaps with that of LIS1 has not been examined. Additionally, overexpression of LIS1 is known to displace dynactin from MT plus ends in interphase cells, and disrupt cortical punctates of dynactin in mitotic mammalian cells (Faulkner et al. 2000); however, LIS1-overexpression significantly alters the MT organization in these cells (Faulkner et al. 2000; Smith et al. 2000), making it unclear how LIS1 affects dynein and dynactin targeting to the cortex of mitotic cells.
In fungi, LIS1 orthologues participate in dynein-mediated nuclear migration and spindle orientation (Morris 2003; Xiang 2003; Yamamoto and Hiraoka 2003). Budding yeast LIS1, known as Pac1, is required for dynein targeting to astral MT plus ends (Lee et al. 2003; Sheeman et al. 2003), a prerequisite for dynein targeting to the cell cortex (Markus et al. 2009). In budding yeast, the only known site of dynein action is the cell cortex, but how Pac1 affects cortical dynein activity is unclear since it is not found at the cell cortex. In the filamentous fungus Aspergillus nidulans, however, the LIS1 orthologue NUDF depends on dynein and dynactin for targeting to MT plus ends (Zhang et al. 2003), a requirement reminiscent of LIS1 targeting to kinetochores in mammalian cells. Deletion of NUDF results in an accumulation of dynein and dynactin at MT plus ends found within the hyphal tip of this organism (Zhang et al. 2003), suggesting that NUDF is important for retrograde movement of dynein. Recent studies indicate that this NUDF-mediated function is important for retrograde endosome transport rather than nuclear migration (Lenz et al. 2006). The discrepancy in the requirement of LIS1 for dynein plus end-targeting in budding yeast versus filamentous fungi might be due to functional divergence, or the existence of redundant mechanisms for dynein interaction with plus ends in filamentous fungi. A detailed understanding of how LIS1 interacts with dynein at the astral MT plus ends, and how this ultimately regulates cortical dynein activity for spindle orientation, are not well understood in any organism.
In budding yeast cells, dynein at the cortex functions to draw the mitotic spindle and nucleus into the bud neck by pulling on astral MTs emanating from the spindle pole bodies (SPBs) (Adames and Cooper 2000; Carminati and Stearns 1997). This activity is regulated by numerous factors, including dynactin, which has been shown to affect dynein targeting and force production at the cell cortex (Kardon et al. 2009; Moore et al. 2008; Moore et al. 2009a), and She1, which was recently shown to affect the association between dynein and dynactin at MT plus ends (Woodruff et al. 2009). Although we previously proposed a model whereby dynein is off-loaded from the plus ends of astral MTs to the cell cortex (Lee et al. 2005), the molecular details of this process remain unclear. Furthermore, neither the mechanism underlying the loading of dynein on to MT plus ends, nor how its activity is regulated following delivery to the cortex are understood. We now provide evidence that dynein targeting begins with the preassembly of Pac1 and Dyn1 in the cytoplasm, most likely as a 1:1 complex. The Pac1-Dyn1 complex is then targeted to MT plus ends in a manner that requires Bik1, the yeast homologue of CLIP-170, but not Bim1, the yeast homologue of EB1. Next, dynactin is recruited in substoichiometric amounts, in a She1-regulated manner, to mediate off-loading of dynein to the cell cortex. Our data also suggest that She1 plays a novel role in promoting the dissociation of dynactin from cortical dynein following the off-loading process. Furthermore, our analysis of dynein and dynactin quantities at MT plus ends and the cell cortex in Pac1-overexpressing and she1Δ cells reveals distinct steps of regulation within the dynein pathway. Most notably, Pac1 and She1 differentially regulate motor activity by tuning either dynein targeting or its association with dynactin at the cell cortex.
Materials and Methods
Plasmid construction
We constructed a family of tagging vectors designed to integrate VN (amino acid 1-172 of Venus) or VC (amino acid 155-238 of Venus) at the 3′ end of any chromosomal locus. Briefly, a PCR fragment containing VN was amplified from pKT103 (Sheff and Thorn 2004) using a forward primer flanked with a XhoI site followed by a linker sequence encoding AlaCysLysIleProAsnGluLeuLysGlyLysValMetAsnHis, as described (Shyu et al. 2008), and a reverse primer flanked with a stop codon and a NotI site. The PCR product was digested with XhoI and NotI and ligated into pRS305 digested with XhoI and NotI to yield pRS305:VN. Similarly, a PCR product containing VC was amplified from pKT103 using primers that resulted in a 5′ XhoI site followed by the same linker sequence as above, and a 3′ stop codon followed by a NotI site. The PCR product was digested with XhoI and NotI and ligated into pRS305 digested with XhoI and NotI to yield pRS305:VC. These plasmids were verified by sequencing. XhoI-NotI VN or VC fragments were subcloned from pRS305 into pRS303, pRS304 and pRS306 digested with XhoI and NotI, yielding pRS303:VN, pRS303:VC, pRS304:VN, pRS304:VC, pRS306:VN and pRS306:VC. These plasmids were used as templates for PCR-mediated tagging of DYN1, PAC1, and BIK1.
To express full-length or truncated Pac1 constructs, PCR products corresponding to PAC1, PAC1CC (nucleotides 1-429), or PAC1WD40 (nucleotides 430-1482) flanked 5′ by SalI and BglII sites and 3′ by a SalI site were digested with SalI and ligated into pRS315:3mCherry digested with SalI, yielding pRS315:PAC1-3mCherry, pRS315:PAC1CC-3mCherry, and pRS315:PAC1WD40-3mCherry. Next, an engineered ApaI-BglII fragment containing the MET3 promoter (MET3p) was amplified by PCR from pRS315:MET3p (Markus et al. 2009), digested with ApaI and BglII, and ligated into the ApaI and BglII sites upstream of the PAC1, PAC1CC, and PAC1WD40 sequences, yielding pRS315-MET3p:PAC1-3mCherry, pRS315-MET3p:PAC1CC-3mCherry, and pRS315-MET3p:PAC1WD40-3mCherry. To produce 13myc-tagged variants of the Pac1 constructs, a SalI-BglII fragment containing 13myc was cut from pFA6a-13myc-kanMX6 (Longtine et al. 1998), blunted with Blunt Enzyme Mix (New England Biolabs), and ligated into pRS315-MET3p:PAC1-3mCherry, pRS315-MET3p:PAC1CC-3mCherry, and pRS315-MET3p:PAC1WD40-3mCherry that had been digested with HindIII and NotI (to remove 3mCherry) and blunted with Blunt Enzyme Mix. Orientation of the 13myc was confirmed by restriction digestions, yielding pRS315-MET3p:Pac1-13myc, pRS315-MET3p:Pac1CC-13myc and pRS315-MET3p:Pac1WD40-13myc.
To construct pRS306-MET3p:mCherry-TUB1, an engineered SalI fragment containing the MET3 promoter was amplified by PCR, digested with SalI, and ligated into pAK011 (Khmelinskii et al. 2007) digested with XhoI.
A plasmid was constructed to tag the endogenous BIK1 locus with 3CFP. To do so, we amplified the 3′ end of the BIK1 gene (nucleotides 983-1320) from genomic DNA using a forward primer flanked with a BamHI site followed by a linker encoding GlyAlaGlyAlaGlyAla, and a reverse primer flanked with a BamHI site. The PCR fragment was digested with BamHI and ligated into pBS-3CFP::URA3 (Lee et al. 2005) digested with BamHI, yielding pBS-BIK1983-1320-3CFP::URA3.
To construct pRS315-MET3p:VN and pRS315-MET3p:VC (used as negative controls for BiFC experiments), PCR products corresponding to VN or VC flanked 5′ by a BglII site and 3′ by a NotI site were digested with BglII and NotI and ligated into pRS315-MET3p:PAC1-3mCherry digested with BglII and NotI, yielding pRS315-MET3p:VN and pRS315-MET3p:VC.
Media and Strain Construction
All strains (Table S1) are derived from YWL36 or YWL37 (Vorvis et al. 2008), or the protease-deficient background YWL29 (BJ5457; Jones 1990). We transformed yeast strains using the lithium acetate method (Knop et al. 1999). Strains carrying null mutations or tagged components were constructed by PCR product-mediated transformation (Longtine et al. 1998) or by mating followed by tetrad dissection. Transformants were clonally purified by streaking to individual colonies on selective media. Proper tagging was confirmed by PCR. At least two independent transformants were chosen from each tagging and disruption procedure for subsequent localization studies. Yeast synthetic defined (SD) media was obtained from Sunrise Science Products (San Diego, CA).
To label MTs, strains were transformed with HindIII-digested pRS305-MET3p:mCherry-TUB1 (MET3p:mCherry-TUB1::LEU2) (Markus et al. 2009), ApaI-digested pRS306-MET3p:mCherry-TUB1 (MET3p:mCherry-TUB1::URA3), StuI-digested pAFS125C (CFP-TUB1::URA) (Moore et al. 2008), StuI-digested pBJ1333 (GFP-TUB1::URA3) (Bloom et al. 1999), or undigested pBJ1351 (GFP-TUB1::LEU2) (Song and Lee 2001). Leu+ or Ura+ transformants were selected and examined for mCherry-Tub1, CFP-Tub1, or GFP-Tub1 fluorescence by microscopy.
To tag chromosomal loci with yEGFP, mCherry or 3mCherry, we PCR amplified from the tagging vectors pBJ776 (Cormack et al. 1997), pRS303:mCherry, or pRS303:3mCherry (Markus et al. 2009), respectively. To tag the endogenous BIK1 locus with 3CFP, pBS-BIK1983-1320-3CFP::URA3 was linearized with NsiI (after a partial digest) before transformation and selection on Uracil-lacking plates. To integrate the KANR::GAL1p-TAP cassette (containing an S-tag and a TEV protease recognition site followed by two IgG-binding motifs, termed ZZ) at the 5′ end of the DYN1 locus, we used the pFA6a-kanMX6-PGAL1-TAP-PCN plasmid as a template for PCR.
Image Acquisition
Yeast cultures were grown to mid-log phase at 30°C and analyzed on an agarose pad containing nonfluorescent SD media or 50 mM potassium phosphate buffer, pH 7. Wide-field fluorescence images were collected using a 1.49 NA 100X objective on a Nikon 80i upright microscope equipped with piezo Z-control (Physik Instrumente), electronically controlled SmartShutter (Sutter Instrument), motorized filter cube turret, and a cooled EM-CCD Cascade II:512 camera (Photometrics). The microscope system was controlled by NIS-Elements software (Nikon). A step size of 1 μm was used to acquire Z-stack images 2 μm thick. Sputtered/ET filter cube sets (Chroma Technology) were used for imaging CFP (49001), GFP (49002), YFP (49003), and mCherry (49008) fluorescence.
Ratiometric Quantification and Statistical Analysis
Cse4-standardized ratiometric measurements were performed as described (Joglekar et al. 2008; Markus et al. 2009). Strains expressing fluorescently-tagged Cse4 or dynein pathway components were imaged using identical imaging conditions and with 1×1 binning of the camera chip (Cascade II:512). We used ImageJ to draw a circle encompassing 3 × 3 pixels to quantify the intensity of individual Cse4, or the relevant plus end or cortical dynein pathway foci. With a 100× magnification objective and a camera pixel size of 16 μm (Cascade II:512), each pixel corresponds to 160 nm in the object plane, a value which we verified using a micrometer. The mean intensity of Cse4 (Fig. S3A–C) was assigned a value of 32 molecules (Joglekar et al. 2006; Meluh et al. 1998) and was used to normalize fluorescence intensity values (A.U.) to number of molecules. We used R and the model-based clustering algorithm Mclust (Fraley and Raftery 2007) to determine whether each data set was a single normal or a mixture of multiple normal distributions, and to determine the mean and standard deviation for each component. The Mclust package for R uses an expectation-maximization algorithm to determine the number of components within a given data set by maximizing the Bayesian information criterion (BIC) (Schwarz 1978) (see Fig. S4I for example).
Cell Lysis, Protein Isolation and Immunoblotting
For purification of TAP-Dyn1-GFP (Fig. 2E), yeast cultures were grown to OD600 ~2–3 in YPA supplemented with 2% galactose, harvested, washed with cold water, resuspended in 0.5 volume of ice cold 2X lysis buffer (1X buffer: 30 mM HEPES pH 7.4, 50 mM potassium acetate, 2 mM magnesium acetate, 0.2 mM EGTA, 0.2% Triton X-100, 1 mM DTT, 0.1 mM Mg-ATP, 0.4 mM PMSF, 0.5 mM Pefabloc SC, 0.7 μg/ml Pepstatin) and then lysed by bead beating in round bottom glass tubes four times for 1 minute each, with 1 minute on ice between each beating. Crude lysate was clarified at 21,000 × g for 20 min and the supernatant was then bound to S-protein agarose (EMD Chemicals) for 45 minutes at 4°C. The S-protein agarose was subsequently washed three times in wash buffer (30 mM HEPES pH 7.4, 50 mM potassium acetate, 2 mM magnesium acetate, 0.2 mM EGTA, 300 mM KCl, 0.1% Triton X-100, 10% glycerol, 1 mM DTT, 0.1 mM Mg-ATP, 0.4 mM PMSF, 0.5 mM Pefabloc SC, 0.7 μg/ml Pepstatin), twice in TEV buffer (10 mM Tris pH 8.0, 150 mM KCl, 0.1% Triton X-100, 10% glycerol, 1 mM DTT, 0.1 mM Mg-ATP, 0.5 mM Pefabloc SC) and then incubated in TEV buffer supplemented with AcTEV protease (Invitrogen) for 1 hour at 16°C. The resulting eluates were mixed with 5X sample buffer and analyzed by immunoblotting as described below.
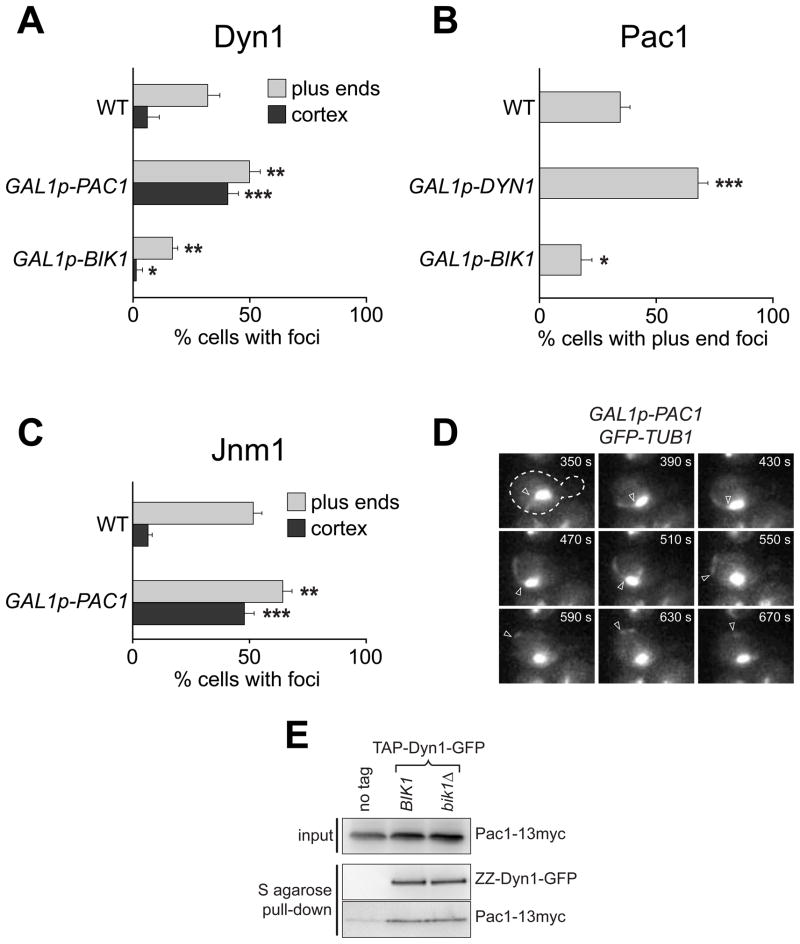
Figure 2. Overexpression of Pac1 increases cortical dynein targeting and activity.
(A–C) The percentage of cells that exhibit plus end or cortically associated (A) Dyn1, (B) Pac1, or (C) Jnm1 foci is plotted for indicated strains. The GAL1 promoter (GAL1p) was inserted at the 5′ end of the chromosomal DYN1, BIK1 or PAC1 locus, replacing the endogenous promoter. GAL1p-carrying strains were imaged after growth to mid-log in synthetic defined media supplemented with 2% galactose. A 2 μm Z-stack of images was collected at 5 s intervals for Dyn1-3mCherry, Pac1-3mCherry or Jnm1-3mCherry with CFP-Tub1 or GFP-Tub1. Stationary cortical or dynamic plus end foci were identified in two-color movies and scored accordingly. Error bars represent standard error of proportion (n ≥ 67 cells; *p < 0.03; **p < 0.01; ***p < 0.0001). (D) Movie frames of GAL1p-PAC1 cells expressing GFP-Tub1 depicting an astral MT undergoing dynein-mediated sliding at the cell cortex and subsequently being pulled out from the SPB (at frame 550 s). Each image is a maximum intensity projection of a 2 μm Z-stack of wide-field images. The time elapsed in seconds is indicated. Open arrowhead indicates approximate position of MT minus end (see Video S1). (E) TAP-tagged Dyn1-GFP pulls down equivalent amounts of Pac1-13myc in the presence (BIK1) and absence (bik1Δ) of Bik1. Equal amounts of cell lysate were incubated with S-protein agarose. Bound proteins were released by TEV protease digestion and analyzed by immunoblotting for ZZ (IgG) and 13myc (anti-c-myc).
To prepare lysates for immunoblotting (Figs. S1B and S2E), yeast cultures were grown to mid-log phase in 3 ml SD media with either glucose or galactose as the sole carbon source (as indicated in figure legends) and harvested. Equal numbers of cells were pelleted and resuspended in 0.2 ml of 0.1 M NaOH and incubated for 5 minutes at room temperature as described (Kushnirov 2000). Following centrifugation, the resulting cell pellet was resuspended in sample buffer.
Crude lysates or TEV eluates were separated on a 4% (for Dyn1-13myc or ZZ-Dyn1-GFP), 8.75% (for Pac1-13myc, Pac11-13myc, and Bik1-13myc), or 12.5% (for Pac1CC-13myc, Pac1WD40-13myc and Pac1-13myc) SDS-PAGE. Proteins were electroblotted to PVDF in 25 mM Tris, 192 mM glycine supplemented with 0.05% SDS and 10% methanol for 30–40 minutes. Rabbit anti-GFP (for CFP-Tub1 loading controls; Invitrogen), anti-PGK1/2 (Santa Cruz Biotechnology), IgG (MP Biomedical), anti-c-Myc (GenScript), and HRP-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories) were used at 1:500, 1:1000, 1:1000, 1:1000 and 1:3333 dilutions, respectively. Chemiluminescence signal was acquired and imaged using a G:BOX Chemi HR16 (Syngene) equipped with a 16-bit CCD camera (Sony ICX285AL; pixel size of 6.45 μm × 6.45 μm). Immunoblots were exposed (durations ranged from 10 s to 10 min) without saturating the camera’s pixels.
Results
Pac1 and Dyn1 are Co-Dependent for Plus End Targeting
How Pac1 localizes to and tracks with the MT plus ends and how it functions in concert with Bik1 for plus end-targeting of dynein are not well understood. Pac1 has a predicted coiled-coil domain in the N-terminus and a WD40 repeat domain in the C-terminus (Fig. S1A). To examine the domain requirement for plus end localization of Pac1, we fused either the N-terminal coiled-coil domain (Pac1CC; amino acids 1-143) or the C-terminal WD40 repeat domain (Pac1WD40; amino acids 144-494) to 3mCherry, and expressed them from plasmids in a pac1Δ mutant expressing CFP-labeled MTs (CFP-Tub1; Fig. S1A). Two-color time-lapse imaging revealed that both fusion proteins exhibited diffuse cytoplasmic fluorescence, with no detectable localization at the MT plus ends or along the length of cytoplasmic MTs (Fig. S1C). This observation contrasts with a previous study that found that LIS1 binds directly to MTs through its N-terminal region containing the coiled-coil domain (Sapir et al. 1997). This discrepancy in MT association is not due to instability of the fusion proteins, as evidenced by western blotting (Fig. S1B), but may reflect a divergence between Pac1 and LIS1 proteins. Alternatively the presence of the C-terminal 3mCherry tag may interfere with MT association of our fusion constructs. However, since full-length Pac1 tagged with 3mCherry is a functional fusion protein (Fig. S2A) and localizes to cytoplasmic MT plus ends (Figs. S1C and S2B), our observations of the fusion constructs suggest that neither the coiled-coil nor the WD40 repeat domain alone is sufficient for MT binding or +TIP activity in vivo. Instead, they suggest that both domains are necessary for these activities.
We next determined if Pac1 localization to MT plus ends requires the zinc knuckle domain of Bik1. A previous study showed that Pac1 interacts with the C-terminal zinc knuckle domain of Bik1 in a two-hybrid assay (Sheeman et al. 2003). We examined Pac1-3mCherry localization in a bik1CTΔ39 mutant, which expresses Bik1 lacking the last 39 amino acids encompassing the zinc knuckle domain. We observed a complete loss of Pac1-3mCherry plus end foci in bik1CTΔ39 cells (Fig. S1D and E), a result consistent with previous observations of Pac1-3GFP (Li et al. 2005). Loss of Pac1-3mCherry plus end foci was similarly observed in bik1Δ cells (Fig. 1A and S2B). These data indicate that the C-terminal zinc knuckle domain of Bik1 is required for Pac1 plus end localization.
Figure 1. Pac1 and Dyn1 are dependent on each other for plus end targeting, whereas Bik1 is independent of either.
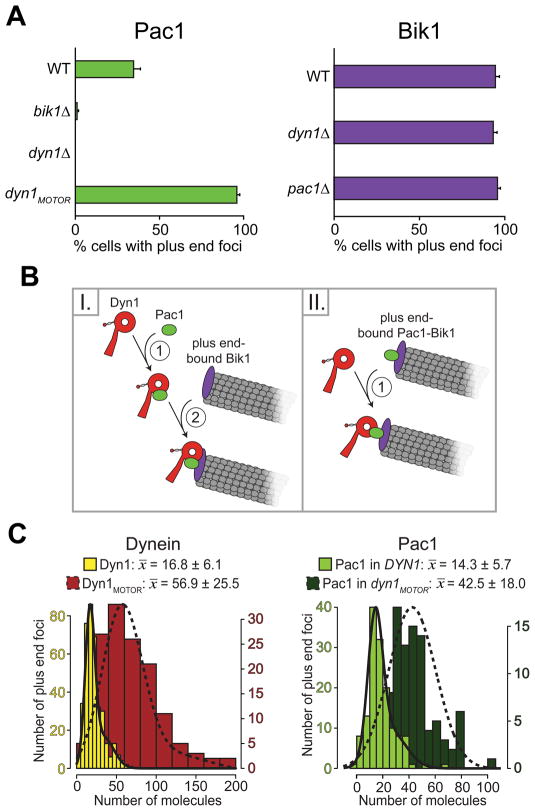
(A) Plus end localization of Pac1 or Bik1 in wild-type or mutant backgrounds. A 2 μm Z-stack of images was collected at 5 s intervals for CFP-Tub1 and either Pac1-3mCherry (left) or Bik1-3mCherry (right). The percentage of cells that display plus end-associated Pac1-3mCherry or Bik1-3mCherry foci is plotted for each indicated strain background (n ≥ 92 cells). Error bars represent standard error of proportion. (B) Potential mechanisms for dynein recruitment to a MT plus end. Left schematic depicts co-dependence of Pac1 and Dyn1 for recruitment to the plus end. Right schematic depicts Pac1 and Bik1 recruited to the plus end independently of Dyn1. (C) Histograms of the number of molecules per plus end focus, shown together with Gaussian fits (see Materials and Methods and Supplementary Note 1), for Dyn1-3mCherry (left plot, left axis; yellow; n = 380 foci), Dyn1MOTOR (left plot, right axis; red; n = 137 foci), Pac1-3mCherry in DYN1 (right plot, left axis; light green; n = 143 foci), or Pac1-3mCherry in dyn1MOTOR-3YFP background (right plot, right axis; dark green; n = 98 foci). Mean values for each data set ± standard deviation are shown. In the case of bimodal distributions, the mean of the first major component is indicated.
Conversely, Bik1 plus end localization (examined using a functional Bik1-3mCherry fusion; Fig. S2A) was unaffected in cells lacking Pac1 (Fig. 1A and S2B). Notably, Bik1 plus end localization is also unaffected by the loss of Bim1 (yeast EB1) (Carvalho et al. 2004), which contrasts with recent findings demonstrating that mammalian CLIP-170 requires EB1 for +TIP activity (Bieling et al. 2008; Dixit et al. 2009). We reasoned that if Bik1 tracks MT plus ends autonomously and independently of Bim1, then Bim1 should also be dispensable for Pac1 plus end localization. We quantified Pac1 plus end localization in bim1Δ cells and found that it was unaffected by the loss of Bim1 compared to wild type cells (Fig. S1D and E). Taken together, our data indicate that, like Dyn1 (Sheeman et al. 2003), the in vivo +TIP activity of Pac1 is mediated by Bik1.
Next, we asked whether Pac1 is targeted to plus ends independently of dynein. We recently proposed that Pac1 binds tightly to a motor domain fragment of Dyn1 (Dyn1MOTOR) in the cytoplasm (Markus et al. 2009). This suggests that plus end-associated Bik1 may recruit a preassembled Pac1-Dyn1 complex to the plus end (see model I in Fig. 1B). However, previous studies (Lee et al. 2003) suggested that Pac1 could be recruited to plus ends in the absence of dynein (see model II in Fig. 1B). Since these previous observations of Pac1 localization in dyn1Δ cells were performed without fluorescently-labeled MTs, it was unclear whether the observed cytoplasmic Pac1 foci were associated with the plus ends. We reexamined Pac1 localization in dyn1Δ cells expressing CFP-labeled MTs (CFP-Tub1) and noted a complete loss of Pac1-3mCherry plus end foci (Fig. 1A, left; Fig. S2B). Furthermore, we found that Bik1 plus end localization was unaffected by the loss of Dyn1 (Fig. 1A, right). Our findings support the schematic depicted in model I (Fig. 1B), that Dyn1 and Pac1 are co-dependent for +TIP activity, while Bik1 associates with plus ends independently of the other two.
We next asked whether there is any correlation between dynein and Pac1 targeting to the MT plus end, as predicted if Pac1 and Dyn1 are co-dependent for plus end localization. We previously showed that Dyn1MOTOR exhibits enhanced plus end targeting compared with Dyn1, based on frequency and intensity measurements at MT plus ends (Markus et al. 2009). We examined Pac1-3mCherry localization in dyn1MOTOR-3YFP cells, which express Dyn1MOTOR-3YFP as the only source of dynein. Compared to a wild-type DYN1 background, we observed that Pac1-3mCherry exhibited enhanced plus end-targeting in the dyn1MOTOR-3YFP background, both in the foci-counting assay (2.8 fold; Fig. 1A, left) and the molecule-counting assay (3.0 fold; Fig. 1C; see below). These results indicate a clear positive correlation for Pac1 and Dyn1 targeting to MT plus ends. Additionally, as reported (Lee et al. 2003), we observed that Pac1-3mCherry accumulated in the nucleus with a diffuse distribution in the DYN1 strain, but this nuclear localization was absent in the dyn1MOTOR-3YFP strain, suggesting that the observed enhancement of Pac1-3mCherry at plus ends was due to redistribution of the protein from the nucleus to the cytoplasm (compare Fig. 3C to Fig. S1C, top row). The function of Pac1 in the nucleus is unknown at this point.
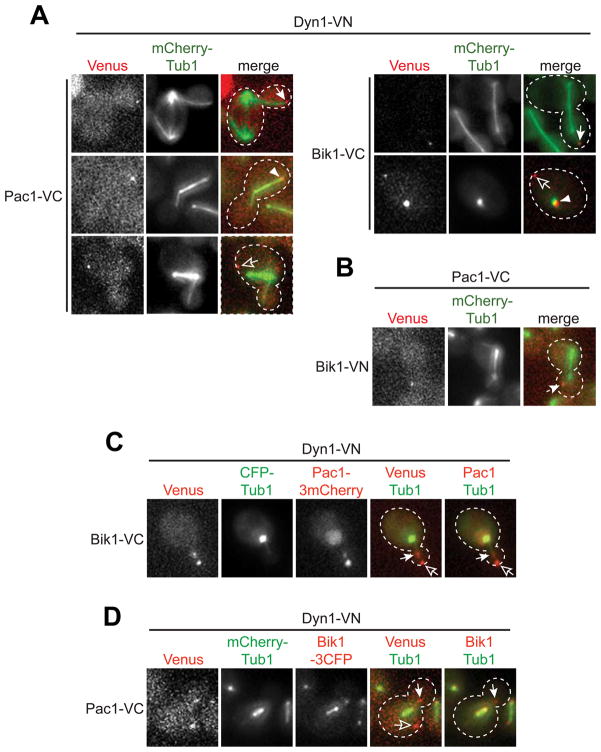
Figure 3. In vivo interactions of Pac1 with Dyn1 and Bik1 in Bimolecular Fluorescence Complementation (BiFC) assay.
(A and B) The N- and C-terminal fragments of Venus, VN and VC, respectively, were fused to the C-terminus of Pac1, Bik1, or Dyn1, as indicated. Fluorescence due to reconstitution of the Venus fluorophore was acquired in the YFP channel. Merged images show Venus in red and MTs (mCherry-Tub1) in green. (C) BIK1-VC DYN1-VN strain coexpressing Pac1-3mCherry and CFP-Tub1. Merged images show CFP-Tub1 in green and either Venus or Pac1-3mCherry in red. (D) PAC1-VC DYN1-VN strain coexpressing Bik1-3CFP and mCherry-Tub1. Merged images show mCherry-Tub1 in green and either Venus or Bik1-3CFP in red. A 2 μm Z-stack of wide-field images was collected at 5 s intervals (maximum intensity projections are shown) to identify foci associated with SPB (closed arrowheads), plus end (closed arrows), or cell cortex (open arrows).
Molecule Counting Reveals Similar Levels of Dyn1 and Pac1 but Substoichiometric Amounts of Dynactin at MT Plus Ends
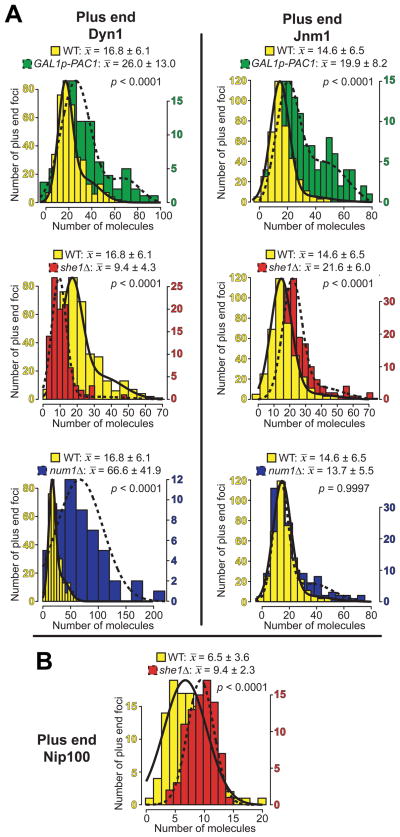
We asked whether Pac1 and Dyn1 exist at the same level at the plus end, as would be expected if they are recruited together to this site. To determine the amount of the dynein-Pac1 complex, we used quantitative ratiometric methods to measure the number of molecules of Pac1 and individual dynein complex subunits (Fig. S4; see Materials and Methods). Each protein was fluorescently-tagged at its native locus in wild-type cells and confirmed to be functional by a spindle position assay (Fig. S2A). Two-color movies of the tagged protein and CFP-Tub1 were collected to confirm the localization of each fluorescent focus at the MT plus end. Cse4, a stable centromeric protein present at two molecules per chromosome (Joglekar et al. 2006; Meluh et al. 1998), was used as a standard for the intensity measurements, as previously described (Joglekar et al. 2008; Markus et al. 2009). We measured a large number of foci for each tagged protein (Fig. S3 and S4) and fit each intensity data set with either a single normal distribution, or a mixture of two normal distributions (see Materials and Methods and Supplementary Note 1), in order to obtain an accurate mean number of molecules per plus end focus. Many of the data sets were bimodal (i.e., Dyn1, Dyn3, Pac11, Pac1 and Jnm1), exhibiting one major peak plus a second minor peak encompassing foci that were approximately twice the fluorescence intensity value of those within the first peak. In all cases, the second distribution constituted only a small subset of each bimodal data set (from 12% to 36%). Although unclear at this point, we reasoned that these brighter plus end foci may represent overlapping plus ends foci (see Supplementary Note 1). For the purpose of comparison and analysis, the mean values obtained from the first normal distribution (x̄1) were used in each case.
We found an average of 14.3 ± 5.7 molecules of Pac1 per plus end, compared to 16.8 ± 6.1 molecules of Dyn1 (Table 1; Figure 1C). Since Pac1 and Dyn1 are dimers (Kardon et al. 2009; Li et al. 2005; Tarricone et al. 2004), these numbers correspond to approximately 7 Pac1 dimers and 8 dynein complexes per plus end, a stoichiometry that is close to 1:1 (see Discussion). Additionally, in each dynein complex, we found that the relative number of molecules of Dyn1 (heavy chain) to Pac11 (intermediate chain) to Dyn3 (light-intermediate chain) is approximately 2 to 4 to 3 (Table 1; Fig. S4A – C), a stoichiometry similar to that documented for mammalian dynein (King et al. 2002; Kini and Collins 2001; Paschal et al. 1987; Vallee et al. 1988). Dyn2 (light chain) was omitted from our counting analysis due to its association and involvement with other cellular structures (Stelter et al. 2007) (Fig. S6A). We also observed an incomplete dependence of Dyn1 and Pac11 plus end localization on Dyn2 (Fig. S6B – C and E), and vice versa (Fig. S6A, bottom). As a result, we could not reliably estimate the number of Dyn2 molecules within each dynein complex.
Table 1. Mean number of molecules per plus end focus or stationary cortical focus for various dynein pathway components.
Summary of molecule counting results. Mean values for each data set ± standard deviation is shown for indicated dynein pathway component. In the case of bimodal distributions, the mean of the first major component (x̄1 from Figs. S4 and S5) is shown.
| Mean number of molecules | Ratio at plus end | Ratio at cortex | |||
|---|---|---|---|---|---|
| Plus end | Cortex | X: 2 Dyn1 | X: 2 Nip100 | X: 2 Dyn1 | |
| Dyn1 | 16.8 ± 6.1 | 6.3 ± 2.7 | 2.0 | 2.0 | |
| Dyn3 | 25.2 ± 10.9 | n/a* | 3.0 | n/a* | |
| Pac11 | 31.2 ± 16.4 | 8.4 ± 3.5 | 3.7 | 2.7 | |
| Jnm1 | 14.6 ± 6.5 | 8.2 ± 3.8 | 4.5 | ||
| Nip100 | 6.5 ± 3.6 | 2.0 | |||
| Arp10 | 3.5 ± 2.0 | 1.1 | |||
| Ldb18 | 6.7 ± 2.4 | 2.1 | |||
| Pac1 | 14.3 ± 5.7 | 1.7 | |||
| Bik1 | 42.3 ± 17.0 | 5.0 | |||
| Kip2 | 22.7 ± 10.1 | 2.7 | |||
n/a = not applicable since Dyn3 is not found at the cortex in wild-type cells (see Fig. 6).
We next determined the molecular ratio of the dynein-Pac1 complex to various components required for plus end-targeting and off-loading of dynein (Table 1). We found an average of 42.3 ± 17.0 molecules of Bik1 per plus end (Fig. S3D), or ~21 Bik1 dimers, which corresponds to >2-fold excess compared to the number of dynein-Pac1 complexes. Kip2, a kinesin that transports Bik1 to the plus end (Carvalho et al. 2004), was present at approximately 22.7 ± 10.1 molecules (Fig. S3E) or ~11 dimers per plus end, less than the number of Bik1 dimers (see Fig. 6). Ndl1, the yeast homologue of NudEL, a protein that acts transiently to stabilize Pac1 at the plus end (Li et al. 2005), was not included in our measurements since we observed Ndl1 present at only a small fraction of plus ends, as reported (Li et al. 2005). The dynactin complex, which relies on dynein for plus end-targeting (Moore et al. 2008), is required for the off-loading of dynein to the cell cortex (Lee et al. 2003; Moore et al. 2009b). We found an average of 6.5 ± 3.6 molecules of Nip100 (p150Glued) per plus end (Table 1; Fig. S4F), indicating that dynactin is present at ~3 complexes per plus end (since p150Glued is a dimer). Additionally, we found the relative number of molecules of Nip100 (p150Glued) to Jnm1 (p50 dynamitin) to Arp10 (Arp11) to Ldb18 (p24) to be approximately 2 to 5 to 1 to 2 (Table 1; Fig. S4D – G and K), a stoichiometry strikingly similar to that documented for mammalian dynactin (Eckley et al. 1999; Kini and Collins 2001; Schafer et al. 1994), suggesting conservation throughout evolution. Taken together, our data are consistent with an initial Bik1-dependent plus end loading of the dynein-Pac1 complex, followed by recruitment of the dynactin complex. Given the stoichiometry, only ~1 dynactin complex is recruited after ~3 dynein-Pac1 complexes are loaded on to the plus ends (see Discussion).
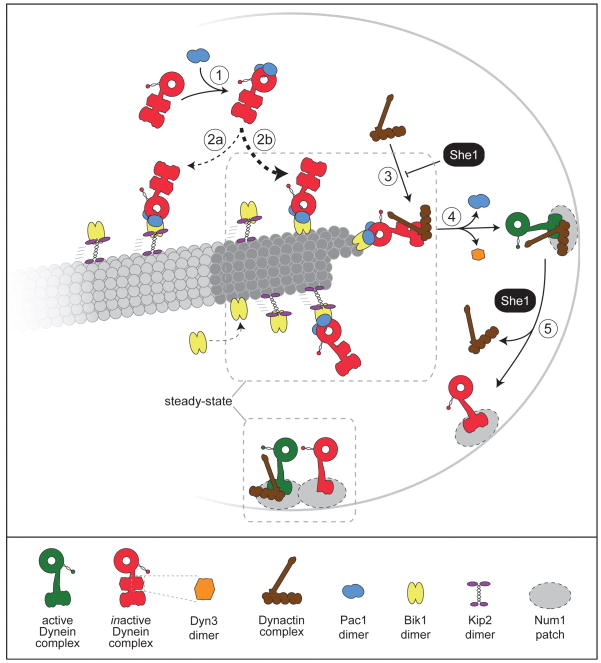
Figure 6. Schematic of the pathway for dynein and dynactin targeting to the plus end and cell cortex.
Our data indicate that the dynein complex associates with cytoplasmic Pac1 in a 1:1 ratio (step 1) prior to interacting with plus end-bound Bik1 (step 2). Previous studies have established that Bik1/CLIP-170 is targeted to the plus end either by direct recruitment from the cytoplasm (Bieling et al. 2008; Dixit et al. 2009; Dragestein et al. 2008; Folker et al. 2005) or via a Kip2 kinesin-dependent mechanism (Carvalho et al. 2004). Our previous data (Markus et al. 2009) showed that dynein plus end targeting is predominantly independent of Kip2 (reflected by the thickness of the dashed arrows for step 2a versus 2b). A previous study in budding yeast demonstrated that dynein then recruits dynactin to the plus end (Moore et al. 2008). Our data here indicate that only 1 dynactin complex is recruited for every 3 plus end dynein complexes. Association of dynein with dynactin (step 3), which is negatively regulated by She1 (Woodruff et al. 2009) (Fig. 4; Table 3), is a requisite for the offloading of dynein-dynactin to Num1 patches at the cell cortex (step 4) (Lee et al. 2003; Moore et al. 2008; Sheeman et al. 2003). Each cortical Num1 patch consists of ~14 molecules of Num1 (Tang et al. 2009) (not all molecules are drawn). Since Pac1 and Dyn3 are not found at the cell cortex, they likely dissociate from the dynein complex during step 4 (Lee et al. 2005; Lee et al. 2003). Based on our observations in this study, we propose that She1 mediates the dissociation of dynactin from cortically anchored dynein (step 5) (see Discussion), thus maintaining a steady-state ratio of 1 dynactin per 2 dynein complexes. Our data indicate that assembled cortical dynein-dynactin complexes (green) are active, while cortical dynein without dynactin is inactive (red). Dashed gray boxes outline a diagrammatic summary of the stoichiometry our counting data has revealed for MT plus ends and the cell cortex. See Figure S4 regarding plus end Dyn3 and Figure S5 regarding cortical Pac11.
Pac1 and Dyn1 Overexpression Enhance Each Other’s Plus End Targeting
The observed 1:1 stoichiometry and interdependence between dynein and Pac1 for +TIP activity suggest that they might associate in the cytoplasm prior to loading on to MT plus ends. We reasoned that this association would follow a second-order reaction, which predicts that increasing the cytoplasmic concentration of either component would result in an increased level of dynein-Pac1 complex competent for loading on to the plus ends. To test this, we integrated the galactose-inducible GAL1 promoter (GAL1p) at the 5′ end of the PAC1 or DYN1 chromosomal locus and assayed for plus end foci of Dyn1-3mCherry or Pac1-3mCherry, respectively. Immunoblot analysis revealed that GAL1p drove overexpression of Pac1 and Dyn1 (Fig. S2E). As predicted, Pac1-overexpression increased the frequency of finding Dyn1-3mCherry foci at plus ends by 1.6 fold (from 32.1% to 50.0%; Fig. 2A). Similarly, Dyn1-overexpression increased the frequency with which Pac1-3mCherry was observed at plus ends by ~2 fold (from 34.6% to 68.0%; Fig. 2B). In contrast, we found that GAL1p-driven overexpression of Bik1 (Fig. S2E) did not target more Dyn1 or Pac1 to the plus ends. Instead, the frequency of finding Dyn1-3mCherry or Pac1-3mCherry at the plus ends was decreased by a moderate amount (Fig. 2A and 2B), which could be due to sequestration of dynein-Pac1 by excess Bik1 in the cytoplasm. When overexpressed, Bik1-3CFP was enhanced at the plus ends and found along the length of cytoplasmic MTs (Fig S2C and D); however, no Dyn1-3mCherry or Pac1-3mCherry fluorescence was observed along the MT length (not shown). Thus, while overexpression of Bik1 alone was sufficient to enhance its own association with MTs, this increased MT-associated pool of Bik1 was not sufficient to enhance the targeting of the dynein-Pac1 complex.
To confirm that Bik1 and Pac1 have discrete roles with regard to their dynein plus end-targeting function (as suggested from above), we asked whether overexpression of Pac1 could compensate for the loss of Bik1. We found that, although Pac1 overexpression could drive more Dyn1-3mCherry to plus ends in wild-type cells (as described above), it could not restore the loss of Dyn1-3mCherry at the plus ends in a bik1Δ mutant (not shown).
Our data suggest that Dyn1 and Pac1 form a complex in the cytoplasm prior to association with Bik1 at the plus end. To test this, we performed a biochemical pull-down assay to assess whether Pac1 and Dyn1 exists in a complex in the absence of Bik1. We found that affinity purification of TAP-tagged Dyn1 with S-protein agarose specifically precipitated myc-tagged Pac1 from wild-type and bik1Δ cell extracts (Fig. 2E). The relative amount of Pac1-13myc that coprecipitated with TAP-Dyn1 was equivalent whether it was purified in the presence or absence of Bik1. Only a small amount of Pac1-13myc was precipitated from a control extract in which Dyn1 was not tagged, owing to non-specific binding to S-protein agarose. These results indicate that Dyn1 and Pac1 form a complex in the cytoplasm in a Bik1-independent manner. Taken together with our results from above, this complex is likely subsequently recruited on to Bik1-coated MT plus ends as depicted in model I in Figure 1B (see also Fig. 6).
Interactions of Pac1, Dyn1 and Bik1 at Plus Ends In Vivo
To confirm that Pac1 and dynein interact at MT plus ends in vivo, we made use of the bimolecular fluorescence complementation (BiFC) assay (Shyu et al. 2008). Pac1 was fused to the C-terminal half of Venus (termed VC), while Dyn1 was fused to the N-terminal half of Venus (termed VN). The Venus chromophore is only restored if VC and VN are brought together by interaction of the two proteins fused to the two respective halves. In wild-type cells, Dyn1 is observed as foci at SPBs, MT plus ends, and the cell cortex (Grava et al. 2006; Lee et al. 2005; Markus et al. 2009; Moore et al. 2008; Sheeman et al. 2003), whereas Pac1 is observed in the nucleus and at MT plus ends, but not the cell cortex (Lee et al. 2003). As shown in Figure 3A, coexpression of Pac1-VC and Dyn1-VN restored Venus fluorescence at SPBs, MT plus ends, and the cell cortex. The observed fluorescence was due to Pac1-Dyn1 interaction as no Venus signal was detected in cells expressing either Pac1-VC or Dyn1-VN alone, or in combination with VN or VC, respectively (not shown). We and others have previously proposed that dynein is delivered by plus ends to the cell cortex, where it becomes anchored to generate forces for pulling the spindle (Lee et al. 2003; Markus et al. 2009; Sheeman et al. 2003). Additionally, we recently showed that plus end targeting of dynein is a prerequisite for cortical targeting (Markus et al. 2009). Thus, our finding of cortical Venus fluorescence suggests that Pac1-VC and Dyn1-VN are off-loaded together from the plus ends to the cell cortex. Whether this occurs in wild-type cells, as suggested (Lee et al. 2003), or whether this is a result of the irreversible intermolecular association due to reconstitution of the Venus chromophore is unclear at this point (Hu et al. 2002).
We next asked whether Dyn1 interacts with Bik1 in vivo, as would be expected if interaction between Dyn1 and Pac1 forms a binding site for Bik1. Previous studies have shown that Bik1 is present at the plus ends, as discontinuous foci along cytoplasmic MTs, and on spindle MTs (see Fig. S2B and D) (Lin et al. 2001). In cells coexpressing Bik1-VC and Dyn1-VN, we found Venus fluorescence at SPBs, plus ends, and cell cortex (Fig. 3A). Bik1-VC alone did not exhibit any Venus signal (not shown). Thus, this finding indicates that dynein interacts only with a subset of cellular Bik1. Since Bik1 is absent from the cortex in wild-type cells, the observed cortical fluorescence suggests that Bik1-VC and Dyn1-VN were off-loaded together from the plus ends, perhaps due to the restored Venus chromophore.
We next asked whether Pac1 associates with the cortical Dyn1-Bik1 Venus foci, as would be expected if dynein only interacts with Bik1 in the presence of Pac1. Localization of Pac1-3mCherry and CFP-Tub1 in a strain coexpressing Bik1-VC and Dyn1-VN showed that all cortical Venus foci contained Pac1-3mCherry fluorescence (Fig. 3C; n = 19 foci). In contrast, examination of Bik1-3CFP in a strain coexpressing Pac1-VC and Dyn1-VN showed that approximately half of the cortical Venus foci did not contain Bik1-3CFP signal (5 of 8; Fig. 3D). Thus, Pac1 is always present when Dyn1 interacts with Bik1, a result consistent with the co-dependence of Dyn1 and Pac1 for loading on to Bik1-coated MT plus ends.
Pac1 interacts with Bik1 in a yeast two-hybrid assay (Sheeman et al. 2003). We confirmed this interaction in cells coexpressing Pac1-VC and Bik1-VN (Fig. 3B). The reconstituted Venus signal was observed only at the plus ends, a finding consistent with the loading of the dynein-Pac1 complex at this site.
In yeast, loss of dynein activity results in spindle misorientation defects. We asked whether cortical association of dynein with Pac1 or Bik1 – crosslinked by the Venus chromophore – affects dynein pathway function. A strain coexpressing Dyn1-VN and Pac1-VC exhibited a degree of spindle misorientation similar to that of a dynein null strain (dyn1Δ) in a cold spindle position assay (50.2% versus 40.8%; Fig. S2A, right), indicating that crosslinked Dyn1-Pac1 is non-functional. In contrast, a strain coexpressing Dyn1-VN and Bik1-VC exhibited a degree of spindle misorientation that was significantly lower than dyn1Δ (16.0% versus 40.8%; Fig. S2A, right), indicating that crosslinked Dyn1-Bik1 is partially functional. As controls, strains carrying DYN1-VN, PAC1-VC, or BIK1-VC alone did not exhibit any spindle misorientation defects (Fig. S2A, middle).
Pac1 Overexpression Enhances Targeting and Activity of Cortical Dynein
We investigated how Pac1-mediated targeting of dynein to plus ends affects the targeting and activity of cortical dynein. We quantified stationary cortical foci of Dyn1 in Pac1-overexpressing cells by analyzing time-lapse two-color images of Dyn1-3mCherry with CFP-Tub1 or GFP-Tub1 (cortical foci are defined as those that were not associated with MT plus ends and remained stationary at the cell cortex for at least 3 consecutive frames). We found that overexpression of Pac1 dramatically enhanced cortical targeting of Dyn1-3mCherry, both in the foci-counting assay (from 6.2% to 40.7% cells; Fig. 2A) and the molecule-counting assay (from 3 to ~9 dynein complexes per cortical focus; Fig. 5, top left). Immunoblots showed that Dyn1 protein levels were unaffected by Pac1 overexpression, indicating that the enhanced targeting was not due to an increase in protein expression or stability (Fig. S2E). Furthermore, we observed a 1.6-fold increase in cortical MT sliding events and a 6-fold increase in the frequency of observing MT detachment from the SPB (Fig. 2D; Video S1; Table 2). Thus, Pac1 overexpression enhances the targeting and activity of cortical dynein.
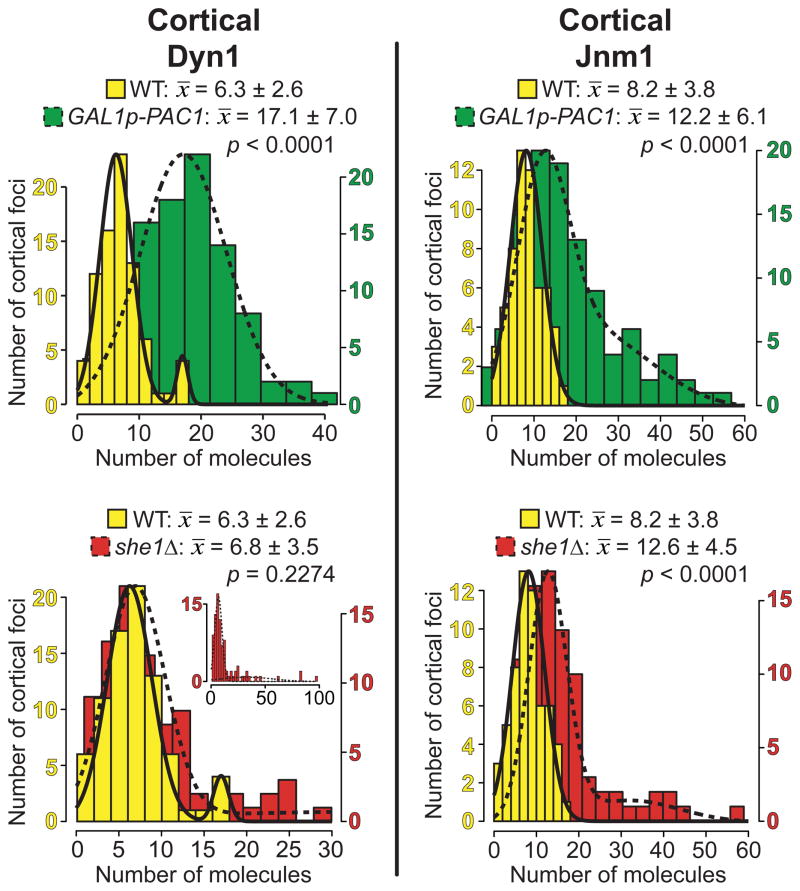
Figure 5. GAL1p-PAC1 and she1Δ differentially affect the recruitment of dynein and dynactin to the cell cortex.
Histogram of the number of molecules per stationary cortical focus, shown together with Gaussian fits (see Materials and Methods and Supplementary Note 1), for Dyn1 or Jnm1 in a wild-type (nDyn1 = 80 foci; nJnm1 = 58 foci), GAL1p-PAC1 (nDyn1 = 96 foci; nJnm1 = 107 foci), or she1Δ strain (nDyn1 = 88 foci; nJnm1 = 82 foci). Mean values for the first major component (i.e., x̄1 from Fig. S5) ± standard deviation are shown. P values between wild-type and mutant are indicated. For 2-component data sets, only those values probabilistically determined to occupy the first component were used to calculate P values. Inset in bottom left panel depicts entire data set for cortical Dyn1 molecule number in she1Δ cells.
Table 2. Quantitation of dynein-mediated spindle movement events.
Indicated strains were imaged after growth to mid-log in synthetic defined media supplemented with either 2% glucose (wild-type and she1Δ strains) or galactose (GAL1p-PAC1 strain). A 2 μm Z-stack of images was collected at 10 s intervals for GFP-Tub1 or mCherry-Tub1. Dynein-mediated MT sliding events were identified and scored accordingly (see Fig. 2D, Videos S1 and S2). Mean values ± standard errors are shown (n ≥ 146 cells and t ≥ 40 min per strain). Numbers in boldface indicate statistical significance from wild-type (p < 0.003).
| sliding events (cell−1 min−1) | mean displacement (μm) | mean velocity (μm/min) | MTs pulled out due to dynein (cell−1 min−1) | |
|---|---|---|---|---|
| Wild-type | 0.0040 ± 0.0004 | 1.22 ± 0.14 | 2.46 ± 0.18 | 0.0002 ± 0.0001 |
| GAL1p-PAC1 | 0.0063 ± 0.0006 | 1.18 ± 0.15 | 2.25 ± 0.20 | 0.0012 ± 0.0004 |
| she1Δ | 0.0175 ± 0.0011 | 1.54 ± 0.25 | 3.03 ± 0.40 | 0.0029 ± 0.0007 |
In Pac1-overexpressing cells, we noted that the enhancement of cortical dynein was more dramatic than the enhancement of plus end dynein (Fig. 2A). We observed similar effects on the targeting of dynactin in Pac1-overexpressing cells: a moderate increase at plus ends, and a dramatic increase at the cell cortex, based on counting Jnm1-3mCherry (p50 dynamitin) foci (Fig. 2C). We next wanted to investigate the mechanism by which cortical dynein activity was enhanced by Pac1 overexpression. We considered the possibility that dynein and dynactin interaction was enhanced at the cortex, given that a recent study demonstrated that dynein activity is likely regulated via interaction with dynactin (Woodruff et al. 2009). Thus, we determined the dynein:dynactin stoichiometry by counting the relative number of molecules of Dyn1-3mCherry and Jnm1-3mCherry at the cell cortex. In wild-type cells, we found that each cortical focus has an average of 6.3 ± 2.6 molecules of Dyn1 and 8.2 ± 3.8 molecules of Jnm1 (Fig. S5A and C; Table 3). In Pac1-overexpressing cells, each cortical focus has an average of 17.1 ± 7.0 molecules of Dyn1 and 12.2 ± 6.1 molecules of Jnm1 (Fig. 5, top), while each plus end focus has approximately 26.0 ± 13.0 molecules of Dyn1 and 19.9 ± 8.2 molecules of Jnm1 (Fig. 4A, top). Thus, although the levels of dynein and dynactin per plus end and cortical focus were elevated by Pac1 overexpression, the resulting ratio of dynein to dynactin complexes indicated that dynactin was still limiting with respect to dynein at both sites (see Table 3), suggesting that the enhanced cortical activity was not due to an enhancement in dynein-dynactin association per se.
Table 3. Loss of She1 but not overexpression of Pac1 significantly increases the ratio of dynactin to dynein at plus ends and the cell cortex.
Mean values for each data set ± standard deviation are shown for the number of Dyn1 or Jnm1 molecules per plus end or stationary cortical focus in indicated strains. In the case of bimodal distributions, the mean of the first major component (x̄1 from Figs. 4, 5, S4 and S5) is shown. Cells were imaged after growth to mid-log in synthetic defined media supplemented with either 2% glucose (wild-type, num1Δ, she1Δ strains) or galactose (GAL1p-PAC1 strain). n/a = not applicable. Dynein:dynactin ratios (boldface) assume two molecules of Dyn1 per dynein complex and five Jnm1 per dynactin complex (see Table 1), the latter of which is also supported by Kini and Collins (Kini and Collins 2001) and Schafer et al., (Schafer et al. 1994). Asterisks (*) in the last two columns indicate a statistically significant difference from wild-type (p < 0.04).
| Mean number of molecules | Dynein: Dynactin ratio | ||||
|---|---|---|---|---|---|
| plus end | cortex | plus end | cortex | ||
| Wild-type | Dyn1 | 16.8 ± 6.1 | 6.3 ± 2.7 | 2.9 | 1.9 |
| Jnm1 | 14.6 ± 6.5 | 8.2 ± 3.8 | |||
| GAL1p- PAC1 | Dyn1 | 26.0 ± 13.0 | 17.1 ± 7.0 | 3.3 | 3.5* |
| Jnm1 | 19.9 ± 8.2 | 12.2 ± 6.1 | |||
| she1Δ | Dyn1 | 9.4 ± 4.3 | 6.8 ± 3.5 | 1.1* | 1.3* |
| Jnm1 | 21.6 ± 6.0 | 12.6 ± 4.5 | |||
| num1Δ | Dyn1 | 66.6 ± 41.9 | n/a | 12.2* | n/a |
| Jnm1 | 13.7 ± 5.5 | n/a | |||
Figure 4. GAL1p-PAC1, she1Δ, and num1Δ differentially affect the recruitment of dynein and dynactin to the MT plus end.
Histograms of the number of molecules per plus end focus, shown together with Gaussian fits (see Materials and Methods and Supplementary Note 1), for (A) Dyn1 or Jnm1 in a wild-type (nDyn1 = 380 foci; nJnm1 = 385 foci), GAL1p-PAC1 strain (nDyn1 = 80 foci; nJnm1 = 124 foci), she1Δ (nDyn1 = 100 foci; nJnm1 = 170 foci), or num1Δ (nDyn1 = 52 foci; nJnm1 = 153 foci) strain, or (B) Nip100 in a wild-type or she1Δ strain (nSHE1 = 108 foci; nshe1Δ = 83 foci). Mean values for the first major component (i.e., x̄1 from Fig. S4) ± standard deviation are shown. P values between wild-type and mutant are indicated. For 2-component data sets, only those values probabilistically determined to occupy the first component were used to calculate P values.
She1 Restricts Dynein Activity by Regulating Dynein Association with Dynactin both at MT Plus Ends and the Cell Cortex
She1 was recently demonstrated to be a negative regulator of the dynein pathway in budding yeast (Woodruff et al. 2009). It was proposed that She1 modulates dynein pathway activity by inhibiting the association between dynein and dynactin at MT plus ends. However, a quantitative assessment of dynein-dynactin MT plus end targeting has not been performed in she1Δ cells, nor was it known what effect She1 deletion has on cortical dynein and dynactin targeting. To investigate whether the dynein-dynactin ratio is affected by loss of She1 and how the ratio compares to wild-type and GAL1p-PAC1 cells, we quantitated the number of molecules of Dyn1 and Jnm1 in she1Δ cells both at MT plus ends and the cell cortex.
In a she1Δ strain, we observed no change in the frequency of finding cells exhibiting Dyn1-3mCherry or Jnm1-3mCherry at plus ends or the cell cortex as compared to wild-type cells (not shown). However, as reported (Woodruff et al. 2009), we confirmed that loss of She1 dramatically enhanced the association between dynein and dynactin. Our molecule counting assay showed that, in she1Δ cells, each plus end focus has an average of 9.4 ± 4.3 molecules of Dyn1 and 21.6 ± 6.0 molecules of Jnm1 (Fig. 4A, middle), while each cortical focus has an average of 6.8 ± 3.5 molecules of Dyn1 and 12.6 ± 4.5 molecules of Jnm1 (Fig. 5, bottom row). Thus the resulting stoichiometry of dynein to dynactin was reduced significantly to approximately 1:1 at plus ends and cell cortex (Table 3), a result consistent with an enhancement in dynein-dynactin association at both sites in she1Δ cells. We confirmed the change in dynein:dynactin stoichiometry at MT plus ends and the cell cortex in she1Δ cells by quantitating the number of Nip100 molecules (9.4 ± 2.3 per plus end and 4.9 ± 1.5 per cortical focus; Fig. 4B and data not shown). In contrast, in num1Δ cells, where Dyn1 fails to off-load and is enhanced at plus ends (Lee et al. 2003; Markus et al. 2009; Sheeman et al. 2003), we found approximately 33 dynein and 3 dynactin complexes per plus end (Fig. 4A, bottom; Table 3), a 10-fold increase in the dynein-dynactin stoichiometry. This finding confirms that the recruitment of dynactin to the plus ends is not limited by the number of dynein molecules, but is negatively regulated by She1 (Woodruff et al. 2009). Like Pac1-overexpressing cells, she1Δ cells exhibited enhanced cortical dynein activity (Video S2; Table 2), despite a lack of change in the frequency of cortical dynein targeting. Taken together, these results demonstrate that the activity of cortical dynein in budding yeast cells is regulated via two mechanisms: by fine-tuning the frequency of targeting without affecting dynein-dynactin association (a mechanism sensitive to Pac1 levels), or by modulating dynein-dynactin association without affecting the frequency of targeting (a mechanism sensitive to She1 levels).
Discussion
In summary, we have provided evidence that Pac1 and Dyn1 preassemble in the cytoplasm prior to association with Bik1 at MT plus ends. Our measurements provide the first quantitative inventory of dynein pathway components at MT plus ends and the cell cortex, and reveal that Pac1 and dynein are likely recruited to plus ends as a 1:1 complex. The number of molecules and the stoichiometry of dynein to dynactin at plus ends suggest that the association between dynein and dynactin is a limiting step in the off-loading process (Fig. 6). Furthermore, our measurements of cortical dynein and dynactin in GAL1p-PAC1 and she1Δ strains indicate different ratios of dynein to dynactin in these two strains, and reveal a new regulatory step that may involve a She1-mediated dissociation of dynactin from cortically-anchored dynein (Fig. 6; discussed below).
A Pathway for Dynein from the Cytoplasm to MT Plus Ends
In budding yeast, we propose that dynein is targeted to the plus end, then off-loaded to the cortex, where it is anchored and activated to power the sliding of astral MTs. Our observations (i.e., ~1:1 stoichiometry, co-dependence of targeting, enhancement of targeting by overexpression of partner, and biochemical pull-downs) support the notion that dynein preassembles with Pac1 in the cytoplasm (Fig. 6, step 1) prior to binding to plus end-associated Bik1 (Fig. 6, step 2). We propose that the interaction between dynein and Pac1 results in the formation of a binding site for Bik1, as suggested by data from our split-Venus (BiFC) assays. An alternate hypothesis may be that a transient Pac1-Bik1 or dynein-Bik1 complex exists along MTs or at the plus ends, which may be subsequently stabilized by the binding of either dynein or Pac1, respectively. Since we observed no such localization of either Pac1-3mCherry or Dyn1-3mCherry in the dyn1Δ or pac1Δ strains, respectively, we favor the former model. Given that there are more than twice as many molecules of Bik1 than Pac1 or dynein complex per plus end, we reason that Bik1 is not a limiting component for dynein-Pac1 recruitment. Our results revealed a slightly higher number of dynein complexes than Pac1 dimers (~8 to ~7) per MT plus end. These differences may be due to error inherent to the molecule counting assay, and thus, in the cell, the ratio of dynein to Pac1 at plus ends may indeed be 1:1. Alternatively, it is possible that following the Pac1-dependent recruitment of dynein to plus end-bound Bik1, dynein may be ‘handed off’ directly to the MT lattice or plus end, as was recently proposed by Caudron et al (Caudron et al. 2008). We observed a similar discrepancy in the Dyn1MOTOR background (~57 Dyn1MOTOR monomers: ~22 Pac1 dimers) that may be partly explained by each Pac1 dimer binding to two Dyn1MOTOR molecules (yielding an apparent ratio of ~29 Dyn1MOTOR dimers: ~22 Pac1 dimers), while the excess Dyn1MOTOR molecules may be bound directly to MT plus end as a result of the proposed ‘hand off’.
Based on work from other labs (Bieling et al. 2008; Dixit et al. 2009; Dragestein et al. 2008; Folker et al. 2005; Gupta et al. 2009; Lin et al. 2001), we speculate that the dynein-Pac1 complex may interact with Bik1 that is bound directly to the plus end through its N-terminal MT-binding CAP-Gly domain (Fig. 6, step 2b). Alternatively, the dynein-Pac1 complex may interact with Bik1 that is being transported to the plus end by the kinesin Kip2 (Fig. 6, step 2a) (Carvalho et al. 2004). We previously showed that the Kip2-mediated mechanism accounts for only ~30% of plus end-associated dynein (Markus et al. 2009), suggesting that the former mechanism is the major pathway for plus end targeting of the dynein-Pac1 complex. Our measurements of Bik1 molecules, however, did not distinguish between Bik1 that was bound directly to tubulin from those associated with Kip2 at the plus end. The presence of a two-fold excess of Bik1 versus Kip2 (Fig. S3D and E; Table 1), along with the observation that plus end-tracking of Bik1 is not abolished in kip2Δ cells (Carvalho et al. 2004), suggest that a population of Bik1 is associated with the plus end independently of Kip2. It is therefore likely that some Bik1 molecules are directly recruited from the cytoplasm to plus ends, which is consistent with the observed fast turnover rate of mammalian CLIP-170 on MT plus ends (Dragestein et al. 2008). Although not mutually exclusive, it is also possible that some Bik1 molecules are ‘handed off’ by Kip2 to the MT plus end upon its arrival there. Whether the dynein-Pac1 complex interacts differentially with these two pools of Bik1 remains to be determined.
Dynein and Dynactin Targeting by LIS1 proteins
Although the players involved in plus end-targeting of dynein appear to be similar among species (such as Pac1/NUDF/LIS1, Bik1/CLIPA/CLIP-170, Ndl1/NUDE/NudE, and dynactin), the molecular details and the requirements for plus end-targeting of dynein are not identical and appear to differ considerably. For instance, dynactin is required for the plus end accumulation of dynein in A. nidulans (Xiang et al. 2000; Zhang et al. 2003), U. maydis (Lenz et al. 2006), and cultured vertebrate cells (Vaughan et al. 1999); however, the converse is true in S. cerevisiae (Moore et al. 2008). Additionally, in contrast to S. cerevisiae Pac1, A. nidulans NUDF is not required for the localization of dynein and dynactin to the distal ends of cytoplasmic MTs at hyphal tips (Zhang et al. 2002). Rather, loss of NUDF resulted in an enhancement of dynein and dynactin at the hyphal tips (Zhang et al. 2003), suggesting an involvement of NUDF in the retrograde movement but not the plus end-targeting of dynein in this organism (Lenz et al. 2006) (see Introduction). In cultured vertebrate cells, although the role of LIS1 in dynein targeting is currently unclear (see Introduction), overexpression of LIS1 resulted in the displacement of dynactin from astral MT plus ends (Faulkner et al. 2000), a finding that contrasts with our current study in budding yeast. However, our findings for Bik1 appear to be similar with those for mammalian CLIP-170. Lansbergen et al., (2004) showed that CLIP-170 plays an important role in the targeting of LIS1, dynein, and dynactin to MT plus ends in cultured mammalian cells. Nonetheless, LIS1 and p150Glued compete for binding to CLIP-170, which contrasts with the findings in budding yeast where dynactin relies on dynein (Moore et al. 2008), which in turn co-depends with Pac1 for binding to Bik1 and tip-tracking (Lee et al. 2003; Sheeman et al. 2003) (current study). These differences and similarities among vertebrates, A. nidulans, and S. cerevisiae may reflect a loss or gain of particular LIS1-ligand interactions subsequent to the divergence of these organisms, as evidenced by the weak sequence similarity between vertebrate and yeast CLIP-170s (Lin et al. 2001), NUDEs (Li et al. 2005), and dynein/dynactin components (Lee et al. 2005).
Interestingly, a recent biophysical study (McKenney et al. 2010) revealed that LIS1 could prolong the interaction of dynein with MTs, specifically when subjected to high loads in an optical trap assay. The same study also demonstrated a role for NudE in stabilizing the dynein-LIS1 interaction, consistent with a model previously proposed from genetic studies in budding yeast (Li et al. 2005). Since Pac1 and Ndl1 are not found at the site of dynein activity in budding yeast (i.e., the cell cortex), it raises the possibility that Pac1 may facilitate dynein targeting by affecting its MT-binding and/or motor properties at the MT plus end.
A Pathway for Dynein from MT Plus Ends to the Cell Cortex
We observed that, in wild-type cells, only one dynactin complex is targeted to the plus end per ~3 dynein-Pac1 complexes. Previous studies have shown that dynactin targeting is dependent on dynein, and have implicated dynactin in the off-loading of dynein from plus ends to the cell cortex (Lee et al. 2005; Lee et al. 2003; Moore et al. 2008; Sheeman et al. 2003). A recent study demonstrated that, when purified separately from budding yeast and mixed together in vitro, dynein and dynactin associate in a 1:1 ratio while moving along MTs (Kardon et al. 2009). Furthermore, following isolation from vertebrate cells, species of dynein and dynactin that co-migrate on a sucrose density gradient are present in a 1:1 ratio (Kini and Collins 2001). Based on these functional, biophysical, and biochemical studies, we reason that the association of a single dynactin complex with a single dynein complex at the plus end is required for their subsequent off-loading to the cell cortex (Fig. 6, step 3). Because the dynactin complex is limiting relative to the dynein-Pac1 complex at the plus end, we hypothesize that this association is the crucial step in the regulation of the offloading process (Fig. 6, step 4).
Interestingly, we observed that, in cells lacking She1, the increase in dynactin association with the plus ends is accompanied by a significant decrease in the number of plus end dynein complexes (from ~17 to ~9 copies of Dyn1; Fig. 4A, middle; Table 3; p < 0.0001). Furthermore, we observed that she1Δ cells exhibited a wide range of cortical Dyn1 values (up to 98 for she1Δ versus up to ~18 for wild-type; Fig. 5, bottom left panel), 21.5% of which were greater than the maximum value observed in wild-type cells, suggesting that an enhancement of dynactin plus end-targeting leads to a concomitant increase in dynein off-loading. Since we and others (Woodruff et al. 2009) did not observe an increased frequency of cortical dynein targeting in she1Δ cells relative to wild-type (SHE1) cells, we speculate that the enhanced association of plus end dynein with dynactin enables more dynein to be off-loaded to the cell cortex per off-loading event. The extent of dynein plus end-targeting in she1Δ cells is likely limited by the level of cellular Pac1; thus, we speculate that only in rare cases where there are very high numbers of dynein (and dynactin) molecules at the plus end will there be a concomitant off-loading of large numbers of dynein-dynactin complexes.
We previously proposed a model whereby the association between dynein and Num1 at the cell cortex is inhibited due to a ‘masking’ of the N-terminal cortical association domain of Dyn1 (Markus et al. 2009). We proposed that this domain is ‘unmasked’ upon association of Dyn1 with the plus end (omitted from the schematic in Fig. 6 for simplicity), thus accounting for the need of dynein to associate with plus ends prior to cortical anchoring. Our current data suggest that the association of dynactin with dynein at MT plus ends may trigger this unmasking event.
A New Regulatory Step for the Dynein Pathway at the Cell Cortex
Although overexpression of Pac1 leads to robust cortical targeting of dynein (Fig. 2A) and dynactin (Fig. 2B) and an increase in cortical dynein activity (Fig. 2D and Table 2), these cells still exhibited a relatively high dynein:dynactin ratio at the cell cortex (3.5 to 1; Table 3). Since dynein and dynactin are likely together in a 1:1 ratio at the moment of off-loading (as discussed above), we reason that there must be a mechanism that induces the dissociation of dynactin from dynein at the cortex after the off-loading event, thus working to maintain a steady-state ratio of 1 dynactin to ~3–4 dynein in Pac1-overexpressing cells, and 1 dynactin to ~2 dynein in wild-type cells (Table 3; see Fig. 6). In contrast, loss of She1 leads to an enhancement of cortical dynein activity (more so than Pac1-overexpression) by altering the ratio to ~1:1 at the cell cortex. Thus, we propose that, in addition to its role at plus ends, She1 functions at the cortex in a wild-type cell to mediate the dissociation of dynactin from cortically-anchored dynein (Fig. 6, step 5), thereby regulating cortical dynein activity. This function of She1 may be important to prevent hyperactivity of cortical dynein or to enable cell cycle dependent activation of dynein activity, as suggested by Woodruff et al., (Woodruff et al. 2009).
Supplementary Material
Acknowledgments
We thank Drs. Jeffrey Moore and John Cooper for sending a yeast strain expressing Ldb18-3GFP. We also thank Dr. Michael Lavine in the Department of Mathematics and Statistics at the University of Massachusetts Amherst for his assistance with statistical methods and R. We are grateful to Drs. Xianying Tang, Elizabeth Collins, and Patricia Wadsworth for valuable discussions throughout this study. This work was supported by an HHMI Academic Research Internship, a University of Massachusetts Amherst Biology Department Junior Fellowship and American Heart Association Undergraduate Summer Fellowship to J. P., and an NIH/NIGMS grant (1R01GM076094) to W.-L. L.
Abbreviations
- MT
microtubule
- SPB
spindle pole body
- +TIP
plus end-tracking protein
- WT
wild-type
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149(4):863–74. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183(7):1223–33. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom KS, Beach DL, Maddox P, Shaw SL, Yeh E, Salmon ED. Using green fluorescent protein fusion proteins to quantitate microtubule and spindle dynamics in budding yeast. Methods Cell Biol. 1999;61:369–83. doi: 10.1016/s0091-679x(08)61990-1. [DOI] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162(6):963–9. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8(9):541–4. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138(3):629–41. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6(6):815–29. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Caudron F, Andrieux A, Job D, Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J Cell Sci. 2008;121(Pt 9):1506–13. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, et al. LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22(9):3089–102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143 ( Pt 2):303–11. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106(2):492–7. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragestein KA, van Cappellen WA, van Haren J, Tsibidis GD, Akhmanova A, Knoch TA, Grosveld F, Galjart N. Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J Cell Biol. 2008;180(4):729–37. doi: 10.1083/jcb.200707203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163(6):1205–11. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999;147(2):307–20. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2(11):784–91. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Folker ES, Baker BM, Goodson HV. Interactions between CLIP-170, tubulin, and microtubules: implications for the mechanism of Clip-170 plus-end tracking behavior. Mol Biol Cell. 2005;16(11):5373–84. doi: 10.1091/mbc.E04-12-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C, Raftery AE. Bayesian Regularization for Normal Mixture Estimation and Model-Based Clustering. Journal of Classification. 2007;24:155–181. [Google Scholar]
- Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10(4):425–39. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Gupta KK, Paulson BA, Folker ES, Charlebois B, Hunt AJ, Goodson HV. Minimal plus-end tracking unit of the cytoplasmic linker protein CLIP-170. J Biol Chem. 2009;284(11):6735–42. doi: 10.1074/jbc.M807675200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155(7):1159–72. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–98. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8(6):581–5. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Salmon ED, Bloom KS. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 2008;85:127–51. doi: 10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Vacuolar proteases in yeast Saccharomyces cerevisiae. Methods Enzymol. 1990;185:372–86. doi: 10.1016/0076-6879(90)85033-k. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci U S A. 2009;106(14):5669–74. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177(6):981–93. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Bonilla M, Rodgers ME, Schroer TA. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 2002;11(5):1239–50. doi: 10.1110/ps.2520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini AR, Collins CA. Modulation of cytoplasmic dynein ATPase activity by the accessory subunits. Cell Motil Cytoskeleton. 2001;48(1):52–60. doi: 10.1002/1097-0169(200101)48:1<52::AID-CM5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15(10B):963–72. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–60. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lansbergen G, Komarova Y, Modesti M, Wyman C, Hoogenraad CC, Goodson HV, Lemaitre RP, Drechsel DN, van Munster E, Gadella TW, Jr, et al. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol. 2004;166(7):1003–14. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168(2):201–7. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160(3):355–64. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25(11):2275–86. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7(7):686–90. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155(7):1173–84. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee WL. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19(3):196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141(2):304–14. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94(5):607–13. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9(4):510–27. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci U S A. 2009a;106(13):5147–52. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009b;66(8):546–55. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr Opin Cell Biol. 2003;15(1):54–9. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11(5):1765–74. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105(3):1273–82. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364(6439):717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Tulu US, Fagerstrom C, Wadsworth P. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol. 2002;158(6):997–1003. doi: 10.1083/jcb.200204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108(1):97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16(23):6977–84. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126(2):403–12. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. Estimating the Dimension of a Model. The Annals of Statistics. 1978;6(2):461–464. [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13(5):364–72. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21(8):661–70. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shyu YJ, Hiatt SM, Duren HM, Ellis RE, Kerppola TK, Hu CD. Visualization of protein interactions in living Caenorhabditis elegans using bimolecular fluorescence complementation analysis. Nat Protoc. 2008;3(4):588–96. doi: 10.1038/nprot.2008.16. [DOI] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2(11):767–75. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Song S, Lee KS. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol. 2001;152(3):451–69. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Kunze R, Flemming D, Hopfner D, Diepholz M, Philippsen P, Bottcher B, Hurt E. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol. 2007;9(7):788–96. doi: 10.1038/ncb1604. [DOI] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156(6):959–68. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Punch JJ, Lee WL. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 2009;8(19) doi: 10.4161/cc.8.19.9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim MH, Derewenda ZS, Knapp S, Tsai LH, Musacchio A. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44(5):809–21. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170(6):935–45. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Wall JS, Paschal BM, Shpetner HS. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 1988;332(6164):561–3. doi: 10.1038/332561a0. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112 ( Pt 10):1437–47. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- Vorvis C, Markus SM, Lee WL. Photoactivatable GFP tagging cassettes for protein-tracking studies in the budding yeast Saccharomyces cerevisiae. Yeast. 2008;25(9):651–9. doi: 10.1002/yea.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell. 2009;20(13):3003–11. doi: 10.1091/mbc.E09-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X. LIS1 at the microtubule plus end and its role in dynein-mediated nuclear migration. J Cell Biol. 2003;160(3):289–90. doi: 10.1083/jcb.200212168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol. 2000;10(10):603–6. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6(3):297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci. 2003;116(Pt 22):4501–12. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
- Zhang J, Han G, Xiang X. Cytoplasmic dynein intermediate chain and heavy chain are dependent upon each other for microtubule end localization in Aspergillus nidulans. Mol Microbiol. 2002;44(2):381–92. doi: 10.1046/j.1365-2958.2002.02900.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14(4):1479–88. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.