Summary
The Drosophila Apaf-1 related killer (Dark) forms an apoptosome in the intrinsic cell death pathway. In this study, we show that Dark forms a single-ring when initiator procaspases are bound. The resulting Dark-Dronc complex cleaves DrICE efficiently; hence, a single-ring represents the Drosophila apoptosome. We then determined the 3D structure of a double-ring at ~6.9Å resolution and created a model of the apoptosome. Subunit interactions in the Dark complex are similar to those in Apaf-1 and CED-4 apoptosomes, but there are also significant differences. In particular, Dark has “lost” a loop in the nucleotide binding pocket, which opens a path for possible dATP exchange in the apoptosome. In addition, caspase recruitment domains (CARDs) form a crown on the central hub of the Dark apoptosome. This CARD geometry suggests that conformational changes will be required to form active Dark-Dronc complexes. When taken together, these data provide novel insights into apoptosome structure, function and evolution.
Introduction
The Drosophila Apaf-1 related killer (Dark) assembles into an apoptosome that functions within the intrinsic cell death pathway. This complex is essential for most developmental cell death and stress-induced apoptosis (Rodriguez et al., 1999; Zhou et al., 1999; Kanuka et al., 1999; Mills et al., 2006; Srivastava et al., 2007). Apoptosomes are formed by NOD family members, including Apaf-1 and CED-4, and these platforms activate procaspases. In addition, Dark, Apaf-1 and CED-4 are members of the AAA+ ATPase superfamily (Danot et al., 2009). A characteristic feature of these proteins is the presence of a nucleotide binding and oligomerizaton domain (NOD), which mediates platform assembly in the presence of nucleotide (Yuan et al., 2010; Qi et al., 2010; Inohara and Nunez, 2003; Acehan et al., 2002). By analogy with Apaf-1 and CED-4 (Riedl et al., 2005; Yan et al., 2005), Dark contains an N-terminal caspase recruitment domain (CARD) followed by the NOD, which is comprised of a nucleotide binding domain (NBD), a small helical domain (HD1) and a winged-helix domain (WHD). The NOD of Dark, like Apaf-1, is followed by a second helical domain (HD2) and WD-40 repeats which form two β-propellers (Yu et al., 2006; Yuan et al., 2010).
During apoptosis, the initiator caspase Dronc binds to the Dark apoptosome through CARD-CARD interactions (Dorstyn et al., 1999). This activates Dronc so that it can cleave DrICE, the executioner procaspase (Dorstyn & Kumar, 2008; Kumar 2007), and a cascade of proteolytic events then culminates in cell death. Initially, it was thought that cytochrome c may serve as an activator of Dark assembly (Rodriguez et al., 1999; Kanuka et al., 1999). More recent data suggests that cytochrome c is not required for Dark assembly or Dronc activation (Dorstyn et al., 2002, 2004; Dorstyn and Kumar, 2008; Yu et al., 2006). However, a role for cytochrome c in apoptosis has been demonstrated in some tissues (Arama et al., 2006; Mendes et al., 2006; Kornbluth and White, 2005).
In healthy cells, Dronc activation is suppressed by the Drosophila inhibitor of apoptosis protein (DIAP1) through complex formation between the DIAP1 BIR2 domain and a 12-residue linker region of Dronc (Chai et al., 2003). During apoptosis, Reaper, Hid and Grim proteins (Grether et al., 1995) bind to the DIAP1 BIR2 domain and relieve DIAP-mediated inhibition of Dronc (Wu et al., 2001; Hawkins et al., 2000). In addition, RGH proteins interact with the BIR1 domain of DIAP1 through their inhibitor of apoptosis (IAP) binding motifs. This releases DrICE from an inhibited complex with DIAP1 (Yan et al., 2004; Wang et al., 1999; Kaiser et al., 1998). Hence, activation of this death pathway by pro-apoptotic signals may involve a number of steps including DIAP1 removal, Dark assembly and Dronc activation. Previously, a 3D structure of Dark complexes at 18.8Å resolution revealed a novel double-ring with D8 symmetry. However, the nature of the physiological apoptosome remained unclear. In addition, molecular details of Dronc activation are not known, although Dronc undergoes proximity-induced dimerization like other initiator procaspases (Dorstyn & Kumar, 2008; Snipas et al, 2008; Boatright et al., 2003; Renatus et al., 2001).
In this paper, we show that Dark single-rings are stable in physiological salt when Dronc is bound. The Dark-Dronc single-ring complex is active in cleaving DrICE and thus, may represent the physiological apoptosome. We then determined a 3D structure of the double-ring at ~6.9Å resolution. This map was used to create a model of the central hub and extended arms in the apoptosome. In addition, the putative regulatory region was modeled as tandem 7- and 8-blade β-propellers, which are connected to the central hub by the HD2 arm. Interactions between adjacent subunits in the central hub are used to assemble an NBD ring and an outer HD1-WHD ring. Three helices in the NBD, including α12, α13 and linker helix α8, may play important roles in apoptosome assembly. Intriguingly, an extended loop that interacts with nucleotide in other apoptosomes has been lost in Dark. Hence, an open path to bulk solution has been created for bound nucleotides in the Dark apoptosome. We also find that Dark CARDs form an octagonal crown on the central hub. In contrast, CED-4 CARDs form a two-layered "disk" on the CED-4 apoptosome (Qi et al., 2010), while Apaf-1 CARDs are flexibly-linked to the human apoptosome (Yuan et al., 2010). This unexpected disparity in how CARDs are presented on apoptosomes from flies, nematodes and humans suggests that certain aspects of procaspase activation must differ in these organisms.
Results and Discussion
Stability of Dark single- and double-rings
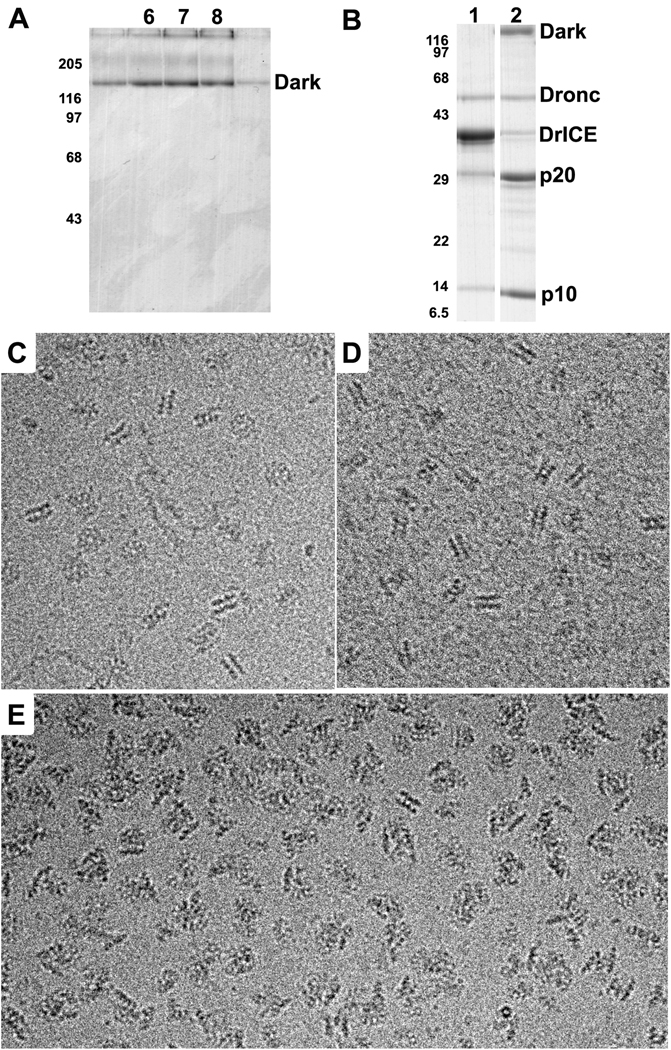
In a previous study, double-rings were assembled from Dark in low salt buffer, using EDTA to promote nucleotide exchange with dATP (Yu et al., 2006). Thus, we wondered whether the double-ring would be stable in a physiological buffer. To test stability, we assembled Dark in low salt buffer (Experimental Procedures) and fractionated the sample on a 10–40% glycerol gradient in buffer supplemented with 90 mM KCl (PSB). In this condition, Dark ran as a smaller complex in fractions 6–8 (Figure 1A) relative to double-ring complexes, which typically run in fractions 9–11 (see Yu et al., 2006). The mobility of the smaller Dark complex is similar to the Apaf-1 apoptosome, which has a calculated molecular mass of ~1 MDa (Yu et al., 2005). This suggested that the Dark complex may form a single-ring in 100 mM KCl.
Figure 1. Stability and activity of Dark rings.
A. Glycerol gradient profile of Dark complexes assembled and run in PSB. The peak occurs in fractions 6–8, similar to the migration of the Apaf-1 apoptosome (not shown). Also note that Dark aggregates after heating in SDS gel loading buffer to give a triplet instead of a single band. The positions of molecular weight markers (in kDa) are shown on the left.
B. Proteolysis of DrICE in the absence and presence of Dark complexes. Lane 1: Dronc + DrICE in PSB; Lane 2: co-assembled Dark-Dronc complex + DrICE in PSB; p20 and p10 are cleavage products of DrICE.
C. Frozen-hydrated Dark double-rings at 3 mg/ml in PSB imaged over a hole in the carbon support film.
D. Dark double-rings assembled at 0.5 mg/ml in PSB and imaged on a thin carbon film
E. Dark-Dronc complexes were assembled in PSB and imaged over holes at 3 mg/ml. The images show mostly single-ring aggregates as evidenced by the lack of typical side views for the double-ring and the presence of single-ring edge views.
We then imaged Dark complexes in PSB over holes in a carbon support film using electron cryo-microscopy. For this experiment, the sample was concentrated to ~3 mg/ml in the presence of NP40 to obtain sufficient particles in the holes after freezing. Unexpectedly, defocused images showed top and edge views consistent with double-rings (Figure 1C). In addition, when Dark complexes were imaged on a carbon film after assembly at 0.5 mg/ml in PSB, they also appeared as double-rings (Figure 1D). Hence, interactions in the double-ring are stable in 100 mM KCl when the protein concentration is greater than or equal to 0.5 mg/ml (~3 uM). However, only putative single-rings were observed when Dark complexes were run on a glycerol gradient, which resulted in a dilution of the protein concentration to ~0.3–0.5 uM (Figure 1A). These data suggest that single- and double-rings are in a facile equilibrium that is dependent upon Dark concentration. Hence, single-rings may be favored at the much lower protein concentration that is present in cells, but this remains to be investigated.
DrICE cleavage by Dark apoptosomes
Dronc is the initiator procaspase for the Dark apoptosome (Dorstyn et al., 1999; Chew et al., 2004; Daish et al. 2004). Thus, we asked whether Dronc could be incorporated into complexes when co-assembled with Dark. For these experiments, we mutated internal-cleavage sites in Dronc to alanine (E143A and E352A) to minimize proteoysis during over-expression in bacteria. Interestingly, we found that Dronc co-migrated on glycerol gradients with Dark complexes when they were co-assembled in either PSB or low salt buffer (not shown). We then imaged Dronc-Dark complexes over holes in a carbon film after freezing the sample at ~3 mg/ml in PSB. Under these conditions, Dark-Dronc complexes formed a mixture of single- and double-rings with single-rings greatly predominating, as shown by typical edge views (>95%; Figure 1E). Unfortunately, single rings with bound Dronc readily formed aggregates under these conditions.
We then ascertained whether recombinant Dark-Dronc complexes are active. To monitor activity we expressed and purified DrICE, a Dronc substrate. An active site mutation (C211A) was made in DrICE to prevent feedback cleavage of both Dark and Dronc by activated DrICE. After co-assembly of the Dark-Dronc complex in low salt buffer, the sample was diluted in PSB to a protein concentration of ~0.2 uM. DrICE cleavage reactions were carried out at room temperature with a final DrICE concentration of ~2 µM, and after 3 hrs, reactions were stopped and visualized on a 10–18% gradient gel by SDS-PAGE. As expected, we found very little DrICE cleavage by Dronc in the absence of Dark (Figure 1B, lane 1). However, Dark complexes were active in cleaving DrICE to form the p20 and p10 subunits (Figure 1B, lane 2). Based on the data, we suggest that Dark single-rings bind Dronc to form the active apoptosome.
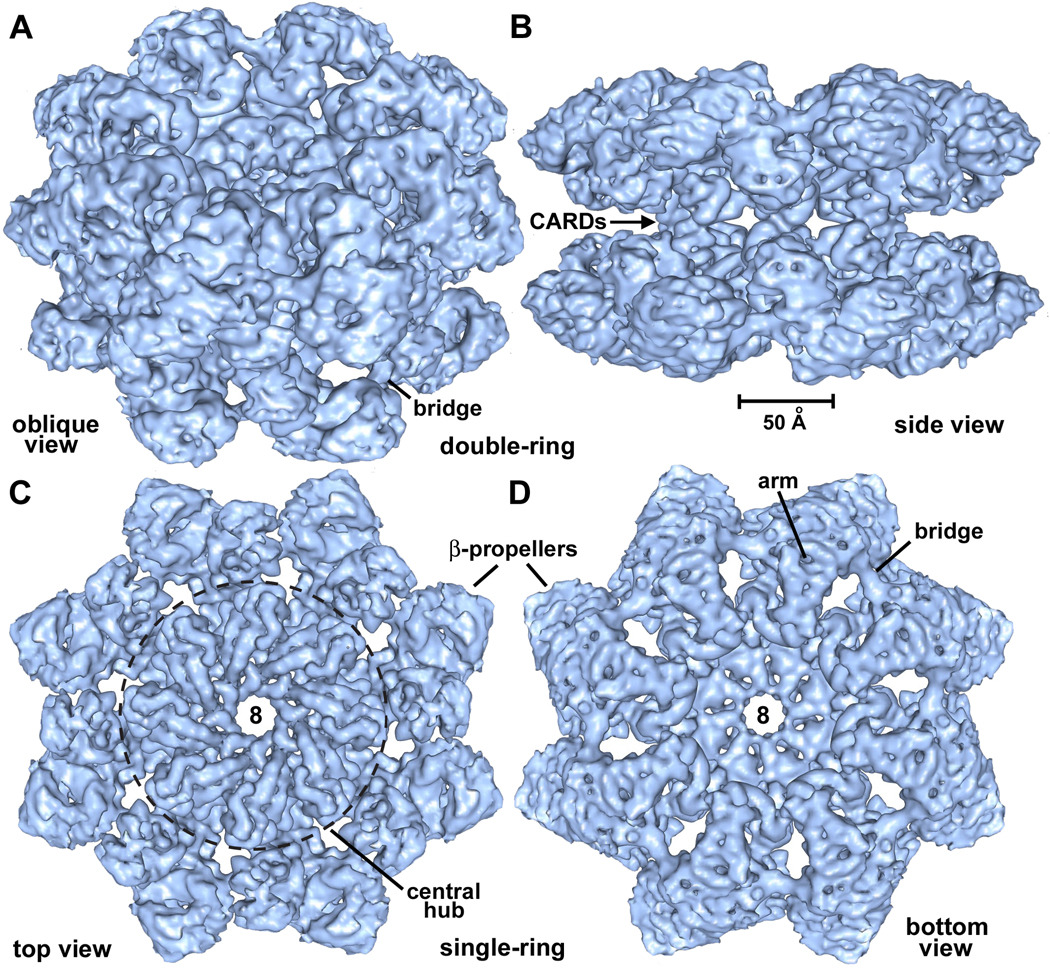
Structure of Dark rings
Dark single- and double-rings appear to be closely-related, with the two rings being linked by a single set of connections (Yu et al., 2006; this work). Hence, a structure of the double-ring should provide information on the architecture of a single-ring apoptosome. To this end, we imaged double-rings (~2.5 MDa) on a thin carbon support film in vitreous ice. We adjusted grid surface conditions such that ~30% of the complexes were imaged edge-on, to give a range of views about the 8-fold axis. Refinements were done in EMAN2 with D8 symmetry (Tang et al., 2007) and the final 3D map had a resolution of ~6.9Å (FSC0.5; Figure S1). The accuracy of the 3D alignment is demonstrated by a comparison of projections from the symmetry-enforced model with their corresponding class averages (Figure S2). The resulting map was of sufficient quality to visualize α-helices, β-sheets and ordered linkers. Surface representations of the double-ring are shown in oblique top and side views (Figures 2A, 2B). A single connection is present between opposing subunits in the two rings and is formed by opposing CARDs (see next section). Segmentation tools in Chimera were used to remove one ring and provide unobstructed views of the apoptosome (Figures 2C, 2D). The central hub is dominated by 8–10Å diameter rods consistent with the high α-helical content of this region. In addition, density at high radius forms a bridging connection between β-propellers in adjacent subunits.
Figure 2. Surface views of Dark double- and single-rings.
A. An oblique view is shown of the Dark double-ring, which has been surface rendered in blue. This and subsequent molecular figures were made with Chimera (Goddard et al., 2005).
B. The Dark double-ring is viewed edge-on along the 2-fold axis. The CARD-CARD connections are visible and a scale bar is provided.
C. A top view is shown of a Dark single-ring extracted from the 3D map. In this view, the central hub within the dashed circle is dominated by ~8–10Å diameter rods.
D. A bottom view of the Dark single-ring reveals the arm which connects to the β-propellers and a bridge between propellers in adjacent subunits.
See also Figures S1 and S2.
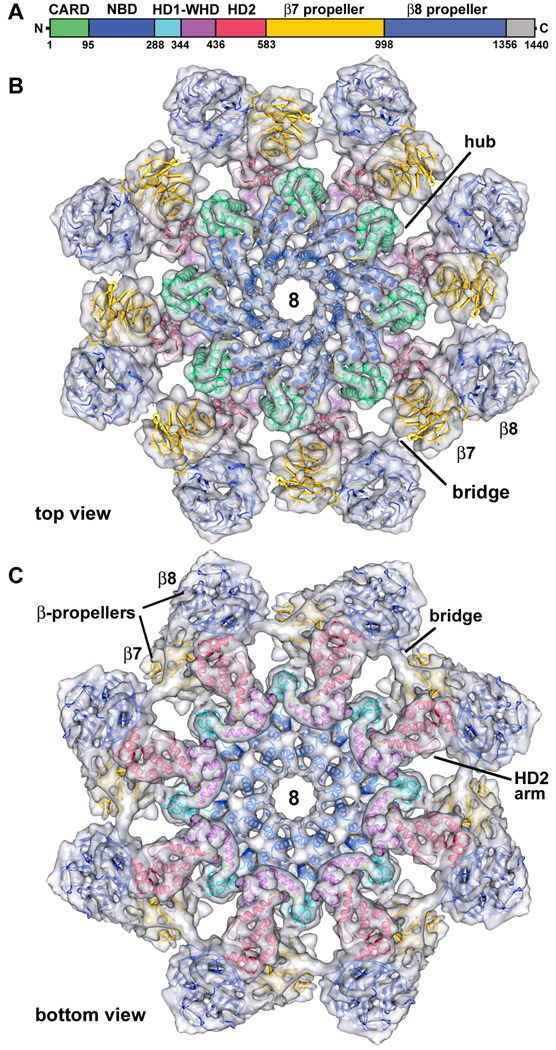
A color-coded diagram of seven Dark domains is shown in Figure 3A, along with a C-terminal region of unknown function. Since no crystal structures were available for Dark, we created homology models of the five N-terminal domains with Modeller (Sali and Blundell, 1993), based on their counterparts in Apaf1-591 (1Z6T, Riedl et al., 2005) and CED4 (2A5Y, Yan et al., 2005). The modeling was aided by the ability to accurately position α-helices and β-sheets within the map. Sequence alignments for Dark, Apaf-1 and CED-4 were made with TCoffee (Notredame et al., 2000). Structure-based models were then displayed in Chimera along with the sequences (Goddard et al., 2005; Figure S3), to verify that the domains were modeled as accurately as possible. We also modeled 15 predicted WD40 repeats in the C-terminal domain as tandem 7- and 8-blade propellers (Supplemental Methods).
Figure 3. Homology model of the Dark apoptosome.
A. A linear diagram of Dark is shown with color-coded domains.
B. A top view is shown of the final Dark model docked within a semi-transparent single-ring. Important features are labeled (see text for details).
C. A bottom view of the final Dark model shows the excellent fit within the density map.
See also Figures S3 and S4.
The final model for the Dark apoptosome is shown within a semi-transparent map of the single-ring, in top and bottom views (Figures 3B, 3C). Clearly, the overall fit of the model is excellent. The wheel-like complex is comprised of a hub formed in part by a central ring of eight NBDs. This architecture is reminiscent of the subunit packing of other AAA+ ATPase rings (Donot et al., 2009). However, important differences are present, which include an inner ring of NBD helices that line the octagonal pore and an outer ring comprised of alternating HD1s and WHDs. Similar features are also present in Apaf-1 and CED-4 apoptosomes (Yuan et al., 2010; Qi et al., 2010). Finally, the HD2 domain in each Dark subunit forms an extended arm that provides a base for the regulatory region, which is formed by 7- and 8-blade β-propellers. An overview of the modeled double-ring is shown in Figure S4.
The resulting model of the Dark apoptosome differs significantly from a previous analysis at 18.8Å resolution (Yu et al., 2006). In brief, the first model was obtained at a much lower resolution and thus, was dependent on structural homology with the human apoptosome (Yu et al., 2005). In these early models, CARDs were placed at the center of the hub since there was biochemical data to support the idea that CARDs interact with each other (Yu et al., 2005; 2006). However, it is now apparent that CARD-CARD interactions occur in the active state of the Apaf-1 apoptosome, rather than in the ground state (Yuan et al., 2010). Moreover, current 3D maps of Dark and Apaf-1 apoptosomes show unequivocally that NBDs are located at the center of the hub (Yuan et al., 2010; this work), in good agreement with a crystal structure of the smaller CED-4 apoptosome (Qi et al., 2010).
Dark domains
Although we used a double-ring for our structure determination, it is clear that Dark single-rings are closely-related to Apaf-1 and CED-4 apoptosomes (Yuan et al., 2010; Qi et al., 2010). Domains within the Dark apoptosome will be described in the next sections, starting with the central hub.
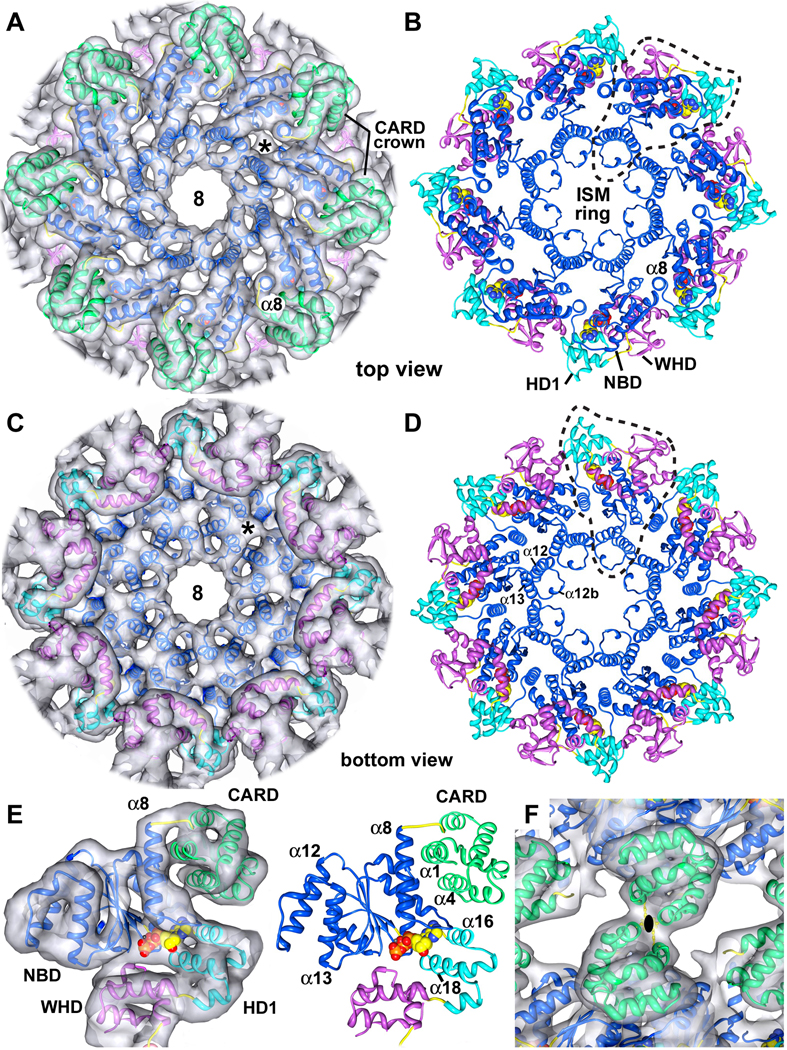
CARDs
The CARDs are well ordered in the central hub of the Drosophila apoptosome (shown in green, Figure 4A), and have a helical fold that is similar to other CARDs, although helix α2 is strongly bent. The CARDs interact with lateral surfaces of their respective NBDs (Figure 4E, left and right panels). This interface involves helix α1 in the CARD and helices α8 and α11 of the NBD. In addition, helix α4 in the CARD may interact with helix α9 and the α8–α9 loop in the NBD, which are also involved in nucleotide binding (Figure S5). The extensive nature of the CARD-NBD interface suggests that a similar arrangement may occur within single-ring apoptosomes. In the double-ring, homotypic interactions between opposing Dark CARDs are responsible for forming the larger complex (Figures 4F, S4C). The interface formed by oppositely facing CARDs in the double-ring may be stabilized by hydrophobic interactions between Phe19 and Phe87. In addition, Lys86 and Glu20/Asp21 may also be involved this interface. The summation of CARD-NBD and CARD-CARD interactions over 8 subunit pairs promotes the formation of a stable double-ring at higher protein concentrations.
Figure 4. Central hub of the Dark apoptosome.
A. The CARD crown is shown in a top view of the central hub, with the final model docked within the density map. An asterisk marks un-modeled density that may represent a bound small molecule.
B. A top view is shown of the central hub with the CARDs omitted for clarity. NBD, HD1 and WHD from a single monomer are encircled with a dashed line and the ISM ring is indicated.
C. A bottom view is shown of the central hub without the CARDs.
D. A bottom view of the central hub is shown without the 3D map to clearly resolve the HD1-WHD ring.
E. (left) Each CARD is linked to helix 8 and interacts with the lateral surface of its respective NBD. The model is shown within the density map. (right) Molecular interactions between a CARD (helices α1 and α4) and the NBD are shown.
F. Homotypic interactions between CARDs in opposing rings mediate double-ring formation. This view is along a 2-fold axis.
See also Figures S5, S6 and S7.
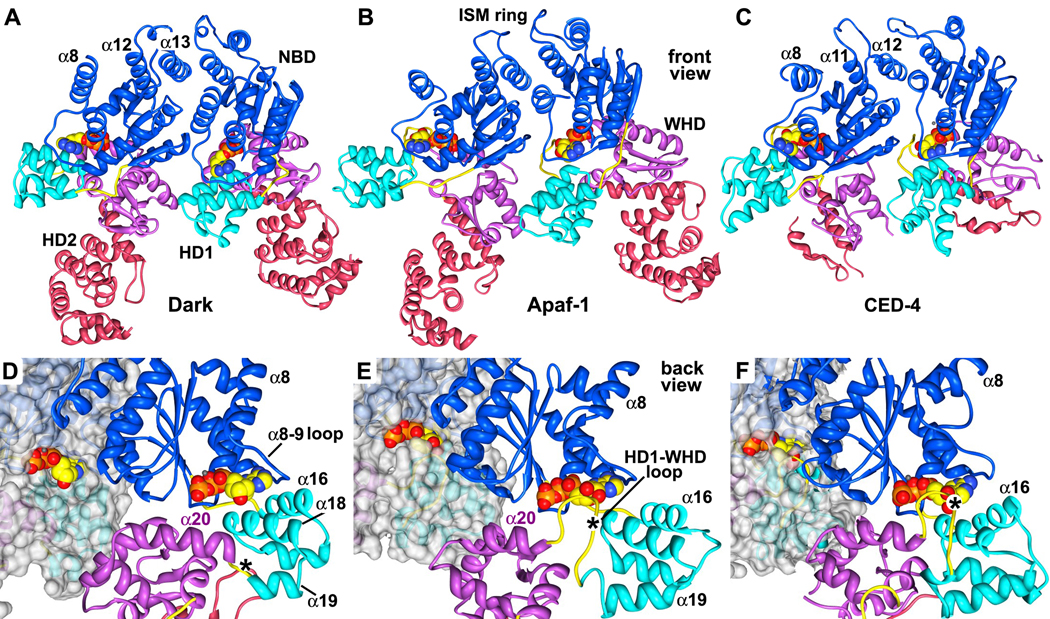
NBD and ISM ring
The nucleotide binding domain forms an inner ring within the central hub. Density for the NBD is excellent, as shown by the fit of a central β-sheet, which is resolved as a thin, flattened density that accommodates 5 parallel β-strands (Figures S5A, S5C, S6). The nucleotide binding pocket is formed by the NBD, linker helix α8 and HD1 (Figure 4E). This pocket is large enough to hold an NTP molecule, which was modeled as dATP. The loop connecting helices α8 and α9 cradles the adenine ring, while helices α16 and α18 in HD1 form one wall of the pocket (Figures 4E and S5A).
The initiator specific motif (ISM) corresponds to an α-helical insert in the canonical NBD fold that is present in the initiator clade of AAA+ ATPases (Danot et al., 2009). In Dark, this helix (α12) forms a helix-loop-helix pair with helix α13, and a short helix (α12b) is present in the intervening loop. These paired helices (α12 and α13) are aligned almost vertically along the cylindrical axis (Figures 4A–4D). Together with equivalent pairs from 7 other subunits, these helices form a cylindrical picket fence that lines the central pore (the ISM ring). Interestingly, a short helix is also observed in a CED-4 monomer between equivalent helices α11 and α12 (Yan et al., 2005). However, this short helix is not observed in the CED-4 apoptosome (Qi et al., 2010).
HD1-WHD
Helix domain 1 and the WHD in adjacent subunits interact to form a second ring in the central hub (Figures 4B–4D). This observation is consistent with mutagenesis studies, which show that the loss of HD1 and/or WHD inactivates Dark-mediated apoptosis (Srivastava et al., 2007). The interface between HD1 and WHD is formed by contacts between helices α18 and α23, and similar interactions are present in Apaf-1 and CED-4 apoptosomes (Yuan et al., 2010; Qi et al., 2010). Remarkably, only three residues form a short HD1-WHD loop that connects helices α19 and α20 (Figure S4A; residues 344–346), whereas this loop is much longer in Apaf-1 and CED-4 (Figure S3). Finally, subunit interactions in the central hub of the Dark apoptosome are highlighted in Figure S7.
WHD-HD2 arm and β-propellers
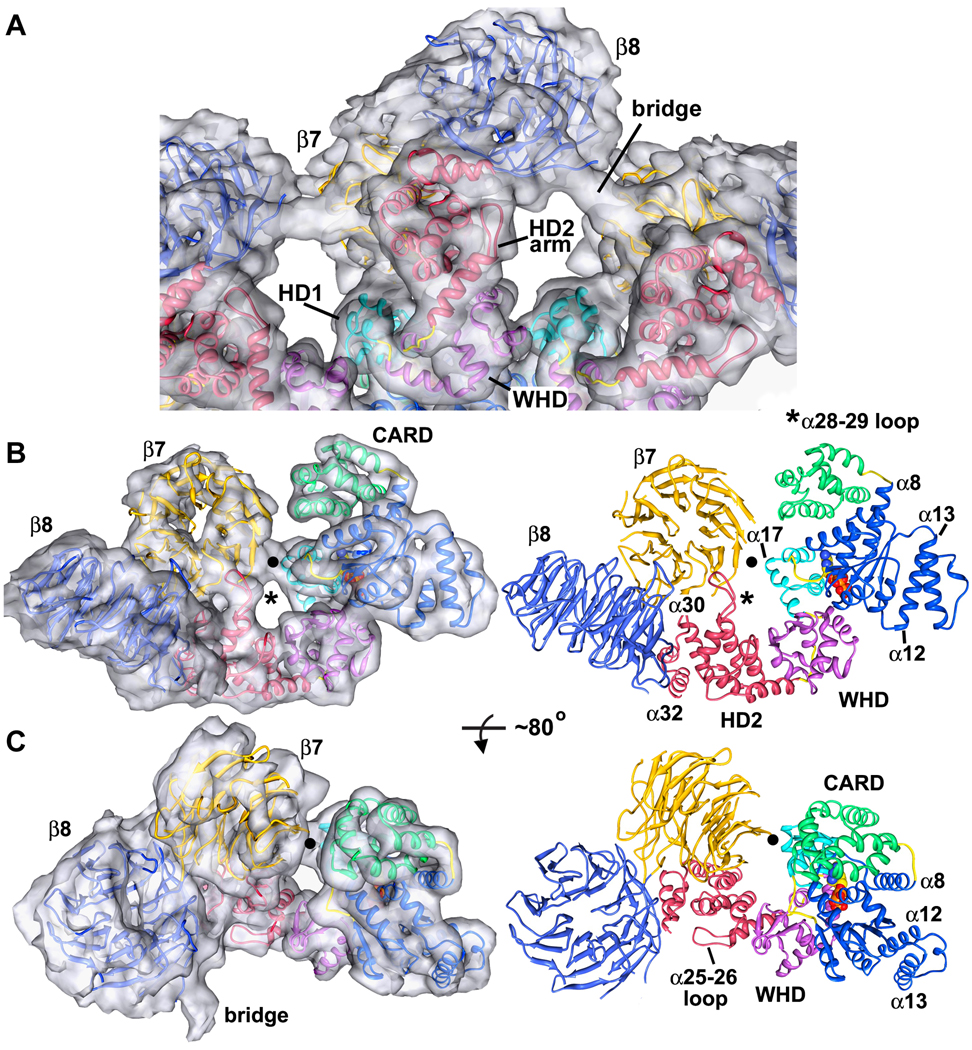
The WHD is closely-associated with HD2 and positions this α-helical domain to form a rigid arm, which extends from the central hub and supports the two β-propellers. An overview of this region is shown in Figure 5A, as viewed from the bottom of the ring. The overall topology of the HD2 is similar to its counterpart in Apaf-1, with two exceptions. First, a long loop is inserted between helices α28 and α29, which extends upwards to interact with the β7 propeller (Figure 5B, see asterisk). Second, a loop between α25 and α26 is reoriented to form part of the bottom surface of the HD2 arm (Figures 5A, 5C). As seen previously in the Apaf-1 apoptosome, three helices in HD2 (α30–α32) form a base upon which the V-shaped regulatory domain rests (Figures 5B, 5C; Yuan et al., 2010). At a reasonable threshold, there is also a connection between the central hub and the α17-blade propeller, which involves helix 17 in HD1 (see black dot in Figures 5B, 5C).
Figure 5. Molecular interactions in the arm and regulatory region.
A. A bottom view is shown of the HD2 arm and regulatory region. The final model is docked within the semi-transparent 3D map and relevant features are labeled.
B. (left) A side view is shown of the entire Dark subunit with a clear view of the HD2 arm and β-propellers, docked within the density map (right) The final model is shown in the same orientation. A molecular contact between the β7 propeller and HD1 is indicated with a black dot and the novel α28–29 loop is marked with an asterisk.
C. (left) A tilted view of the Dark subunit is shown, after rotation about the horizontal axis by ~80°. (right) Features within the monomer are labeled including the α25–26 loop which forms part of the HD2 arm. The bridge density has not been modeled, though its appearance is suggestive of an extended β-hairpin that connects β8 and β7 propellers in adjacent subunits.
Based on sequence analysis, 15 WD40 repeats in the regulatory region were modeled with tandem 7- and 8-blade propellers. The local fit of propeller blades in high density regions of these cylindrical features was excellent (Figures 5B, 5C; left panels). The resulting V-shaped model is similar to a pair of β-propellers in actin interacting protein 1 (Voegtli et al., 2003; 1PGU), except that the second 7-blade propeller in Aip1 has been replaced with an 8-blade propeller in Dark. However, an accurate assignment of WD40 repeats within the map was difficult at this resolution. We therefore modeled β-propeller sequences with alanines in our model. Hence, this region is less well defined than the central hub and arm (Supplemental Methods).
After modeling the two β-propellers, there are roughly 200 residues at the Dark C-terminus that remain unaccounted for. Many of these extra residues may be located in extended loops between β-strands in the β-propeller and are probably quite flexible. In addition, ~75 residues are predicted to form a small helical domain at the C-terminus, following the 8-blade propeller (not shown). However, these residues are difficult to assign into the density map and may also be disordered. In addition, density in one blade of the 7-blade propeller is weak due to flexibility in this region. We also note that the V-shaped, tandem β-propellers are tilted both circumferentially and upwards, relative to their positions in the Apaf-1 apoptosome (Figure S8). This reflects small cumulative changes in the arm α-helices of Dark that facilitate a much closer approach between propellers from adjacent subunits (Yuan et al., 2010). Finally, there is a strong bridging density between adjacent regulatory regions that has not been modeled. Due to its shape, this density is likely formed by a rigid β-hairpin that extends between adjacent β-propellers. The function of this unique bridging density is not known, but it may stabilize the extended conformation of the V-shaped regulatory region in the absence of a bound activator. This stabilization may be essential for Dark assembly, as recent mutagenesis studies showed that loss of WD40s in Dark suppressed HID-induced apoptosis (Srivastava et al., 2007).
A conserved yet adaptable architecture for apoptosomes
A recent analysis suggests that apoptotic pathways and their proteins have undergone a complicated evolutionary process, presumably due to differences in selection and generation times (Zmasek et al., 2007). Hence, assembly of Apaf-1 like molecules into signaling platforms and the activation of initiator procaspases may differ significantly in arthropods, chordates and nematodes (Danial and Korsmeyer, 2004). With this in mind, it was instructive to compare available structures of these apoptosomes to understand their similarities and differences.
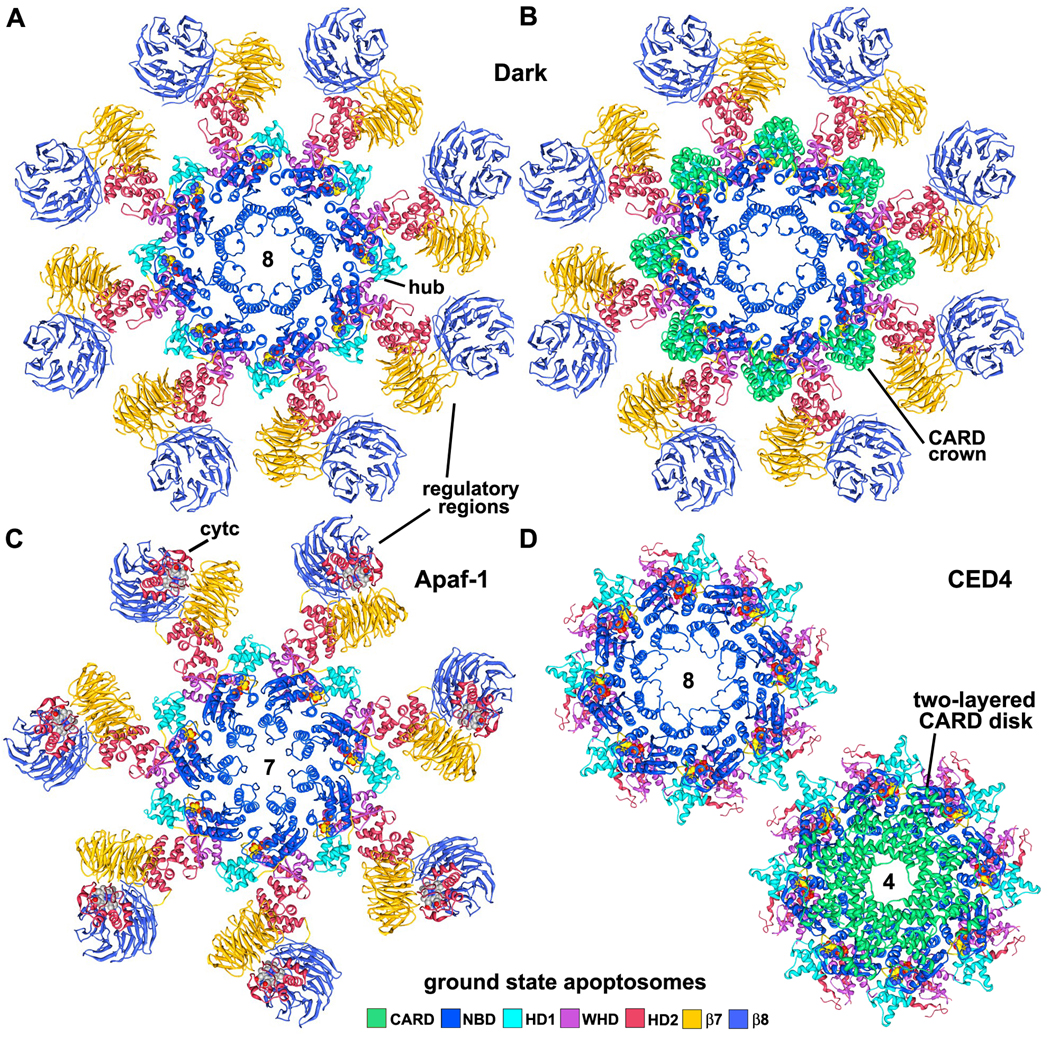
Apoptosomes from D. melanogaster, H. sapiens and C. elegans are shown as molecular ribbons in Figures 6A–6D, as viewed along their symmetry axes. All three apoptosomes use similar, though not identical interactions between subunits to form activation platforms (Figures 7A–7C). This is remarkable because they have different rotational symmetries (8, 7 and quasi-8 fold respectively). Lateral contacts between HD1 and WHD in adjacent subunits generate an outer ring in the central hub, in which adjacent domains are staggered in an up (HD1) and down (WHD) pattern. The HD1 and WHDs in Dark and Apaf-1 apoptosomes are structurally homologous to each other and form similar rings. Conversely, HD1 and WHDs in the CED-4 apoptosome are more divergent in structure, perhaps because a significant region of HD2 and the regulatory domains have been lost during evolution. Interactions between the NBD and HD1-WHD pairs in adjacent subunits promote the formation of similar lateral dimers in Dark, Apaf-1 and CED-4 apoptosomes (Figures 7A–7C), which demonstrates the remarkable plasticity of the NOD.
Figure 6. Models of Dark, Apaf-1 and CED-4 apoptosomes.
A. The platform region of the octagonal Dark apoptosome is shown as a molecular ribbon diagram, viewed along the 8-fold axis in a top view.
B. The Dark apoptosome is shown in a top view with the CARDs, to highlight the formation of a CARD crown on the central hub.
C. The heptameric Apaf-1 apoptosome is shown in a top view with bound cytochrome c (although the position of the latter is still being refined; Yuan et al., 2010).
D. (top left) A top view is shown of the octameric CED-4 apoptosome, with CARDs omitted to provide an unobstructed view of the central hub. (bottom right) The CED-4 apoptosome is shown with the double-layered CARD disk, which has 4-fold symmetry (Qi et al., 2010).
See also Figure S8.
Figure 7. A comparison of lateral dimers and nucleotide binding sites in Dark, Apaf-1 and CED-4 apoptosomes.
A. A lateral dimer is shown from the Dark apoptosome. Structures in panels A–C have been aligned on the leftmost NBD.
B. A lateral dimer from the Apaf-1 apoptosome is shown. Differences in the packing of helix α8 are apparent.
C. The CED-4 lateral dimer is shown with its truncated HD2. The conformation of the α8 helix varies in the two subunits due to differences in packing of their respective CARDs in the disk (Qi et al., 2010).
D. A close-up is shown of the nucleotide binding region of Dark within the context of a lateral dimer. This view is from the bottom of the Dark single-ring. The subunit on the left has been rendered as a molecular surface to show the exposed nature of dATP. In panels D–F the lateral dimers are aligned on the rightmost NBD in each pair. The dATP binding pocket, as seen on the right, has a much shorter HD1-WHD loop than in the other apoptosomes (\marked with an asterisk).
E. A close-up of the same region in an Apaf-1 lateral dimer shows a more obscured path to bound ATP on the left, and the presence of a modeled HD1-WHD loop on the right, which embraces the nucleotide from the inward surface.
F. A similar close-up is shown for the atomic structure of the CED-4 dimer within the apoptosome. In this case, bound ATP is almost totally occluded within its binding pocket.
See also Figure S9.
There are also intriguing structural differences in the central hub of these apoptosomes. In Dark, the ISM helix (α12 in Dark/Apaf-1, α11 in CED-4) is part of a helix-loop-helix motif that lines the central pore to form an α-helical picket fence. Interactions of the helical ISM motifs and helix α8 are probably critical in directing assembly of the NBD ring in all apoptosomes (Figures 6 and 7). Intriguingly, the ISM ring in each of these apoptosomes has a different diameter due to changes in the local geometry of the paired helices. The different diameters of the ISM rings also reflect altered interactions of helices equivalent to α8, α12 and α13 with the conserved core of their respective NBDs. In general, there are many local changes even when the two complexes being compared have the same number of subunits per ring. We conclude that the multi-domain nature of these NOD proteins has allowed compensatory changes in domain interfaces during evolution, resulting in the construction of rings with similar hubs yet different symmetries. Hence, modeling other NOD platforms such as inflammasomes will require judicious use of the toolkit provided by crystal structures of Apaf1-591 (Riedl et al., 2005) and the CED-4 apoptosome (Qi et al., 2010), along with hybrid models for Apaf-1 and Dark apoptosomes (Yuan et al., 2010; this work).
The local environment for bound dATP also differs in Dark because the HD1-WHD linker is much shorter than in Apaf-1 and CED-4. This is important because the HD1-WHD linker makes contacts with ATP in the CED-4 apoptosome (Figure 7F; Qi et al., 2010). The TPYSY motif in this CED-4 loop is also present in Apaf-1 (SSYDY; Figure S3). This allowed us to explicitly model the HD1-WHD linker of Apaf-1 using our published 3D map of the human apoptosome (Yuan et al., 2010). Thus, the HD1-WHD linker reaches up within the subunit-subunit interface to interact with bound nucleotide in both the CED-4 and Apaf-1 apoptosomes (Figures 7E, 7F). The conserved Ser/Thr residue in this motif hydrodgen bonds with the ribose ring, while the first tyrosine forms a hydrogen bond with a phosphate (Qi et al., 2010). In humans, the HD1-WHD linker undergoes a large conformational change, coupled with movements of the WHD-HD2 pair during apoptosome assembly (Yuan et al., 2010; Figure S9).
In Drosophila, a much shorter HD1-WHD linker creates an open path for possible exchange of bound dATP in the apoptosome. This path is readily visible on the underside of the Dark central hub (Figure 7D). Conversely, bound NTPs are screened from solvent by side chains in the HD1-WHD loop of Apaf-1 and CED-4 apoptosomes (Figures 7E, 7F). Loss of the longer HD1-WHD loop in Dark may have resulted in a local rearrangement of helices α16 and α18 in HD1, which are rotated in towards the nucleotide binding pocket. At the same time, helix α20 in the WHD has become markedly longer and now may interact with helix α18 of HD1 (Figure 7D). These changes may create new interactions between Dark and bound dATP involving Arg322 and Ser325 on helix α18 (Figure S3). This idea remains to be tested. Currently, it is not clear why a more accessible dATP binding site is required in the Dark apoptosome. However, increased accessibility of the nucleotide may reflect differences in the assembly mechanism (see below) or alternatively, Dark apoptosomes might disassemble if dATP levels are diminished during apoptosis. More experiments are needed to follow up on these ideas.
Dark assembly
Cytochrome c is essential for Apaf-1 asembly (Liu et al., 1996; Li et al, 1997; Acehan et al., 2002), but its role as an activator in Drosophila is controversial (Dorstyn et al. 2002, 2004; Arama et al., 2006; Rodriguez et al., 1999; Kanuka et al., 1999; Kornbluth and White, 2005; Yu et al., 2006; Mendes et al., 2006). At the same time, Apaf-1 assembly requires nucleotide exchange with either dATP or ATP as cofactors, although the nature of the NTP and the requirement for hydrolysis prior to exchange is being debated (Reubold et al., 2009; Jiang and Wang, 2000; Kim et al., 2005; Riedl et al., 2005). For Dark, ring assembly requires dATP in the absence of an activator, rather than ATP (Yu et al., 2006; this work). Moreover, Dark is not expected to be an efficient ATPase because adjacent aspartates in the Walker B motif have been replaced by a leucine-asparagine pair (L245-N246; Figure S3). Thus, nucleotide exchange may provide the driving force for in vitro assembly of Dark. In our hands, efficient assembly required the use of high concentrations of EDTA. This treatment may have facilitated nucleotide exchange on nonproductive Dark monomers that are formed during purification, by chelating Mg+2 ions.
Given the significant divergence in platform assembly mechanisms for Apaf-1 and CED-4, it is possible that Dark may not follow the Apaf-1 paradigm, even though these NOD proteins are similar in many respects. In one scenario, Dark may spontaneously assemble into a single-ring in vivo, when newly synthesized monomers bind dATP. This would be similar to the lateral association of subunits that occurs in the CED-4 dimer, but instead of being inhibited by a functional homolog of CED-9 (Yan et al., 2005), Dark dimers would complete their assembly into an apoptosome. In addition, local crowding in the cytoplasm might favor the formation of double-rings from single-rings, which would sequester active site CARDs. Hence, the major regulatory step in Dronc activation would be down-regulation of DIAP1 (Chai et al., 2003; Wu et al., 2001; Hawkins et al., 2000), coupled with Dronc synthesis, which in turn would allow the formation of single-ring Dark-Dronc complexes. In this model, conserved β-propellers might stabilize the Dark apoptosome through circumferential interactions or create a binding site for other factors. Cytochrome c may also play a role in Dark activation in certain tissues, in a manner that has yet to be determined (Arama et al., 2006; Mendes et al., 2006; Kornbluth and White, 2005).
CARD presentation
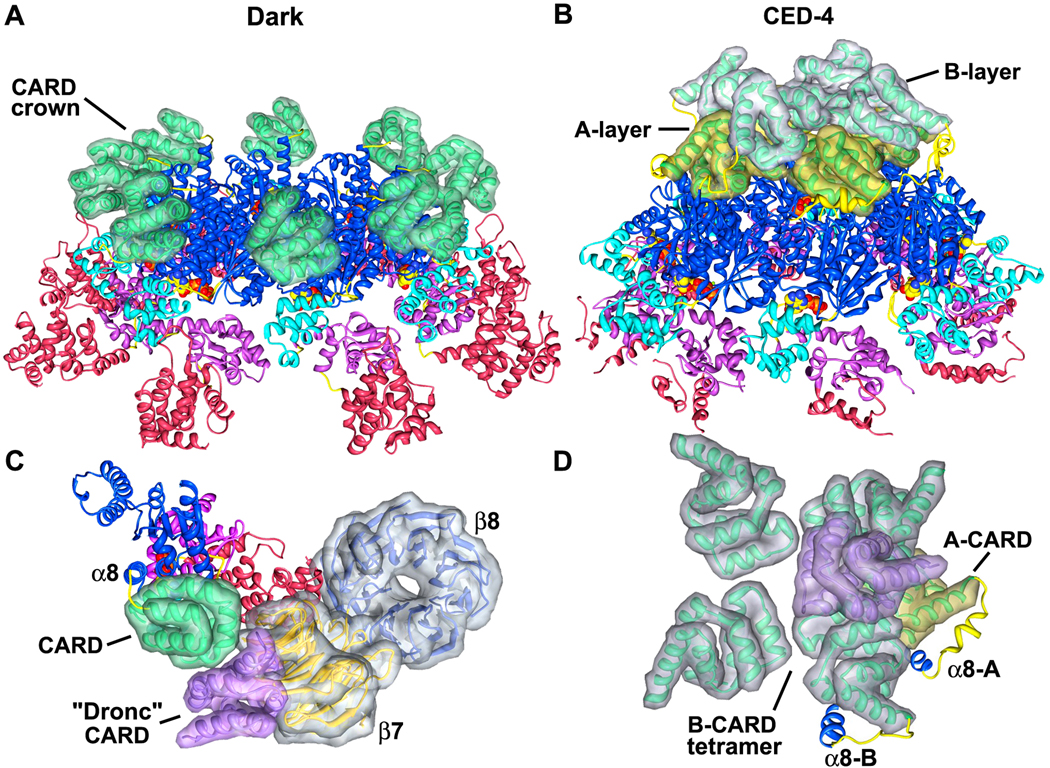
Although the basic architecture of the central hub is similar in Dark, Apaf-1 and CED-4 apoptosomes, the way in which these platforms display their N-terminal CARDs is quite distinctive. In the Dark apoptosome, eight CARDs are bound to lateral surfaces of their cognate NBDs through an extensive interface. This suggests that CARD-NBD interactions in our model may be physiologically relevant. The positioning of CARDs on the central hub creates a crown, that when viewed from the top, sits directly above the HD1s. In addition, neighboring CARDs do not interact with each other (Figures 6B, 8A). Importantly, the Dark CARD is positioned on the hub so that it can bind the Dronc CARD. This interaction was modeled using a crystal structure of an Apaf-1 CARD-pc-9 CARD heterodimer (Qin et al., 1999). Based on the docking, we find that the Dronc CARD has clear access to its presumed binding site on the Dark CARD, but heterodimer formation would create a significant clash at higher radius with the 7-blade propeller (Figure 8C). This clash could be eliminated by a local rearrangement of the Dark CARD on the central hub or alternatively, by release of the Dark CARD from the hub. In the latter case, the Dark CARD would remain tethered to the NBD by the 8 linker, as occurs in the Apaf-1 apoptosome (Yuan et al., 2010). Indeed, Apaf-1 CARDs are flexibly-linked to the central hub in the ground state and thus, are not visible in a 3D map (Figure 6C; Yuan et al., 2010). However, an Apaf-1 CARD-pc-9 CARD “disk” may be formed when pc-9 is bound and this disk is flexibly-linked to the apoptosome (Yuan et al., 2010).
Figure 8. The presentation of CARDs on Dark and CED-4 apoptosomes.
A. A tilted view is shown of the central Dark hub. The 8 CARDs in the crown are shown within calculated surfaces to provide better depth perception.
B. A tilted view is shown of the CED-4 apoptosome with CARDs from the A- and B-subunits displayed within calculated surfaces. The tetrameric A-layer is shown in gold and B-layer is in silver.
C. A strong clash exists between the bound CARD of the initiator procaspase (in purple) and the β-7 propeller of Dark. A monomer is shown and domains are displayed within calculated surfaces.
D. This panel shows the relative orientations of A- and B-CARDs in two layers on the CED-4 apoptosome. Only CARDs and α8 helices are shown for clarity. The modeled binding of an initiator CARD (in purple) by a CED-4 CARD in the B-layer would preclude the binding of other initiator CARDs and shows significant clashes.
In the CED-4 apoptosome, two non-equivalent CARDs in each of the four lateral CED-4 dimers combine to form a two-layered disk (Qi et al., 2010; Figures 6D, 8B). In this case, CED-4 CARDs from the A-subunits form a tetramer that sits directly on the central hub, while CARDs from B-subunits form a similar tetramer that sits above the A-layer with a rotational offset of roughly 45° (Figure 8D). Both A- and B-CARDs are rotated ~170° relative to Dark CARDs and are slightly tilted, when their respective subunits are aligned on the NBD (not shown). In addition, CED-4 CARDs are located much closer to the center of the hub and thus, obscure the ISM ring helices when the complex is viewed from above (Figure 6D, bottom right). Intriguingly, this orients CED-4 CARDs so that binding sites for CED-3 CARDs point in towards the particle axis and laterally towards an adjacent subunit. In modeling this interaction, it is clear that binding of a single CED-3 CARD to a B-CARD is sterically hindered and more importantly, multiple CED-3 CARDs could not bind simultaneously to the CED-4 CARD disk. Hence, major rearrangements may be required for CED-3 binding to the CED-4 apoptosome. It also remains to be determined if crystal packing interactions may have led to an artificially-ordered array of CARDs on the CED-4 apoptosome. Remarkably, the CED-4 apoptosome is thought to bind only two CED-3 molecules (Qi et al., 2010). However, all CARDs are equally accessible in Drosophila and human apoptosomes, which suggests that these platforms may bind 8 and 7 initiator procaspases, respectively (Yuan et al., 2010 and this work).
Summary
Our data are consistent with the idea that single-ring complexes represent the physiological Dark apoptosome. However, double-rings are formed by Dark assembly in vitro at higher protein concentrations and could play a role in the cell. A 3D structure of the double-ring at ~6.9Å resolution allowed us to create a detailed blueprint of the Dark apoptosome. A comparison of Drosophila, human and C. elegans apoptosomes revealed many similarities, even though subunit number (and size) differs in each of these cell death platforms. These organisms span the animal kingdom; hence, their structures provide snapshots of divergent features, including differences in their nucleotide binding pockets. In addition, active site CARDs are displayed in distinctive ways on these apoptosomes. Since initiator CARD binding may be blocked in Dark and CED-4 apoptosomes, we suggest that procaspase activation will require local conformational changes of the CARD crown and double-layered disk. Finally, it remains to be determined whether a CARD-CARD activation disk is formed when initiator procaspases bind to Dark and CED-4 apoptosomes, in a manner similar to the Apaf-1-pc-9 complex (Yuan et al., 2010). In any case, the observed plasticity of the CARD-NBD linker may play an important role in procaspase activation.
Experimental Procedures
Preparation of Dronc and DrICE
Full-length Dronc (1–450) was cloned into a pET32a vector (Novagen) between the Nde I and EcoRI sites, with a C-terminal 6x-His tag and an intervening TEV site. The construct contains two point mutations (E143A, E352A) to prevent zymogen self-cleavage during expression and purification. For protein expression, 6 liters of BL21 (DE3) cells at OD600=1.0 were induced with 0.3 mM IPTG for 4 hrs at 24°C. Pelleted cells were lysed as described (Dutta et al., 2001). The soluble high speed supernatant (100,000×g) was purified with a Ni-NTA column (3 ml resin packed in a 20 ml column (Qiagen) and eluted with 250 mM imidazole in 25 mM Tris buffer (pH 7.5).
The Ni-NTA column eluate was passed through a DEAE DE52 column (2 ml resin; Whatman) and a CM52 column (2 ml; Whatman). Flow-through from the CM52 column was further purified with a hydroxyapatite column (1 ml, Biorad) and Dronc was eluted with a potassium phosphate (pH 7.5) gradient from 20 mM to 1 M. Purified Dronc was dialyzed into low salt buffer (LSB; 20 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT) with protease inhibitor cocktail (1 mM PMSF, 1 mM benzamidine, 20 µM E64) at 4°C. After dialysis, protein (~1 mg/ml) was aliquoted and flash frozen with liquid nitrogen. All protein concentrations were determined with calculated UV A280 extinction coefficients (ExPASy).
Full-length DrICE (1–339) was cloned into a pET32a vector. The active site cysteine was mutated to alanine (C211A) to prevent self-cleavage and feedback cleavage of Dark/Dronc on the Drosophila apoptosome. In total, 6 liters of BL21 (DE3) cells at OD600=0.6 were induced with 1 mM IPTG for 3 hrs at 37°C. Cells were harvested and lysed as before. The high speed supernatant was purified with a Ni-NTA column (3 ml), followed by a Hi-trap Q column (1 ml, GE Healthcare). Purified protein was dialyzed into LSB supplemented with 90 mM KCl at room temperature, aliquoted and flash frozen.
Dark complex assembly and glycerol gradients
A doubly His-tagged, full length Dark (1–1440; with N-terminal 9x-His and C-terminal 8x-His) was expressed in baculovirus infected, sf-21 insect cells and purified as described (Yu et al., 2006). For Dark assembly, 50 µg of Dark (~0.5 mg/ml in LSB) was incubated with 10 mM EDTA and 10 mM dATP at 37°C for 30 min. The sample was then incubated at room temperature overnight to allow complete assembly. Glycerol gradients were used to evaluate Dark assembly and complex formation between Dark and Dronc. A 10–40% glycerol gradient (~2.2 ml total volume) was made in LSB or in this buffer supplemented with 90 mM KCl (PSB), by manually overlaying 7 step solutions between 40% and 10%. The gradient was equilibrated for ~16 hrs at 4°C before use. Protein samples were loaded onto the top and centrifuged in an RP55S swinging bucket rotor with an RCM100 Micro Ultracentrifuge (Sorvall) for 2.5 hrs at 55,000 rpm and 17°C. Fractions of 150 µl were harvested from the top and analyzed by SDS-PAGE on 8–18% gradient gels.
Structure determination and modeling
Dark double-rings were prepared for cryo-electron microscopy and imaged on thin carbon films to provided optimal contrast for single particle alignments (Yuan et al., 2010; Yu et al., 2006). Particles in 4k × 4k CCD frames (TVIPS) were selected and processed in EMAN2 (Tang et al., 2007). Modeling was done as described previously (Yuan et al., 2010) and additional details are provided in Supplemental Methods. Final figures were made with Chimera (Goddard et al., 2005) and Adobe Photoshop.
Supplementary Material
Acknowledgements
We thank Drs. Lai Wang and Xiaodong Wang for providing Dark baculovirus. Coordinates for apoptosome models (Dark: 3IZ8; Apaf-1: 3IYT) and the Dark density map (EMD-5235) have been deposited. The Topf laboratory is supported by the Human Frontier Science Program (RGY0079/2009-C) and an MRC Career Development Award (G0600084). S. Kumar and L. Dorstyn are supported by the National Health and Medical Research Council, Australia. Ludtke and Akey laboratories are supported by NIH grants (S.J.L. R01GM080139; C.W.A. R01GM63834).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding and activation. Mol. Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nature Struct. Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev. Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase Dronc is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insights into signaling by STAND proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Dorstyn L, Kumar S. A biochemical analysis of the activation ofthe Drosophila caspase DRONC. Cell Death Differ. 2008;15:461–470. doi: 10.1038/sj.cdd.4402288. [DOI] [PubMed] [Google Scholar]
- Dorstyn L, Mills K, Lazebnik Y, Kumar S. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 2004;167:405–410. doi: 10.1083/jcb.200408054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 2002;156:1089–1098. doi: 10.1083/jcb.200111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes and Develop. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to apaf-1. J. Biol. Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Kim H-E, Du F, Fang M, Wang X. Formation of an apoptosome is initiated by cytochrome c induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. U SA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis DIAP1 suppresses cell death induced by the caspase drICE. FEBS Letters. 1998;440:243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1 a Drosophila Apaf-1/CED-4 related caspase activator. Mol. Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- Kim H-E, Du F, Fang M, Wang X. Formation of an apoptosome is initiated by cytochrome c induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. (U SA) 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;13:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B. Cytochrome c–d regulates developmental apoptosis in the Drosophila retina. EMBO reports. 2006;7:933–939. doi: 10.1038/sj.embor.7400773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K, Daish T, Harvey KF, Pfleger CM, Hariharan IK, Kumar S. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J. Cell Biol. 2006;172:809–815. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol. Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, Luo G, Li H, Wang J, Yan N, Shi Y. Crystal Structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J. Biol. Chem. 2009;284:32717–32724. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase-9. Proc. Natl. Acad. Sci. (USA) 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nature Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Snipas SJ, Drag M, Stennicke HR, Salvesen GS. Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ. 2008;15:938–945. doi: 10.1038/cdd.2008.23. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, Bergmann A. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by CryoEM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli WC, Madrona AY, Wilson DK. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J. Biol. Chem. 2003;278:34373–34379. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nature Struct. Molec. Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, Xue D, Shi Y. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure. 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An Apaf-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.