Abstract
Background
Recent genome-wide association studies have identified common variants associated with high-density lipoprotein cholesterol (HDL-C). Whether these associations are modified by physical activity, which increases HDL-C levels and reduces the risk of cardiovascular disease (CVD), is uncertain.
Methods and Results
In a prospective cohort study of 22,939 apparently healthy Caucasian US women, we selected 58 single nucleotide polymorphisms (SNPs) in 9 genes that demonstrated genome-wide association (P<5×10−8) with HDL-C levels and sought evidence of effect modification according to levels of physical activity (PA). PA modified the effects on HDL-C of 7 SNPs at 3 loci, and the strongest evidence of effect was observed for rs10096633 at LPL, rs1800588 at LIPC and rs1532624 at CETP (each P-interaction <0.05). The per-minor-allele increase in HDL-C for rs1800588 at LIPC and rs1532624 at CETP was greater in active than inactive women, whereas the reverse was observed for rs10096633 at LPL. Minor-allele carrier status at the LPL SNP was associated with a reduced risk of MI in active (Hazard Ratio [HR] 0.42, 95% Confidence Interval [CI] 0.23–0.76) but not amongst inactive women (HR 1.10, 95% CI 0.83–1.44; P-interaction=0.007). By contrast, carrier status at the CETP SNP was associated with a reduced risk of MI regardless of activity level (HR 0.72, 95% CI 0.57–0.92; P-interaction=0.71). No association between LIPC SNP carrier status and MI risk was noted
Conclusions
The effects of common variants in the LPL, LIPC and CETP genes on HDL-C levels are modified by PA. For a common variant in LPL, the impact on MI varied by activity level, while the effects of a common variant in CETP on MI risk did not.
Keywords: genomic studies, HDL cholesterol, myocardial infarction, exercise
Introduction
Prospective cohort studies demonstrate a strong inverse relationship between high density lipoprotein cholesterol (HDL-C) and risk of cardiovascular disease.1 HDL-C levels are highly heritable and recent genome wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in at least 9 genes that alter HDL-C levels.2–6 While some studies have demonstrated an association between SNPs associated with HDL-C levels and the risk of future cardiovascular events, others have not.6–12
Environmental factors also contribute to the variation observed in HDL-C levels. Physical activity is associated with higher levels of HDL-C in several epidemiologic studies,13–15 and variation in the CETP, LIPC, APOA1, LIPG, APOE and LPL genes have been reported to contribute to inter-individual variability in the HDL-C response to exercise.16–23 The extent to which regular physical activity modifies the effect of recently discovered SNPs on HDL-C levels, or their effect on risk of myocardial infarction (MI) has not been well studied, particularly among women.
We sought to determine whether associations between common SNPs and HDL-C levels are modified by physical activity in a large cohort of healthy US Caucasian women. Additionally, to examine the clinical implications of our findings, we investigated whether favorable genotypes at significant loci are associated with lowered risk of MI, and if this risk is modified by level of physical activity.
Methods
Study participants
Participants were from the Women’s Genome Health Study (WGHS), a prospective genetic evaluation of women in the Women's Heath Study (WHS).24 Study participants in the WHS were female health professionals aged 45 years and older at the time of enrollment who were free of any major chronic disease including cancer and cardiovascular diseased (CVD) at study entry (1992–95). Information on baseline variables including race/ethnicity, demographic characteristics, medical history, medications, and dietary and lifestyle facts were reported on questionnaires. The WGHS includes 23,294 Caucasian women who consented to ongoing analyses using genetic data and for whom we have baseline plasma and DNA. We used all 23,294 women in our initial SNP screening step, and then restricted our sample to the 22,939 women for whom we also had information on physical activity and HDL-C. The study was approved by the institutional review board of the Brigham and Women's Hospital (Boston, Massachusetts).
Genotyping and SNP Selection
DNA samples were genotyped with the Infinium II technology from Illumina (Human HAP300 panel) as previously described.24 All samples were required to have successful genotyping using the BeadStudio v. 3.3 software (Illumina, San Diego, CA) for at least 98% of the SNPs. SNPs with call rates <90% and with a minor allele frequency <1% in Caucasians were removed from the analysis. SNPs were evaluated for deviation from Hardy-Weinberg equilibrium using an exact method and were excluded when the P-value was lower than 10−6. After quality control, a total of 339,596 SNPs were left for the analysis. Genotyping success at SNPs used for our analysis ranged from 97% at rs1532624 to >99.99% at rs10096633 and rs1800588.
Physical activity
At baseline, each participant was asked to report her approximate average time per week during the previous year spent on 8 groups of recreational activities (walking/hiking, jogging, running, bicycling, aerobic exercise/dance, lap swimming, tennis/squash/racquetball, and lower-intensity exercise) and the number of flights of stairs climbed on a questionnaire previously shown to be both valid and reliable.25, 26 A metabolic equivalent (MET) score was assigned to each activity based on the energy cost of that activity and the energy expended on each activity was then estimated by multiplying its MET score with hours/week, and summed across all activities (MET-hours/week). The United States Government recommends at least 150 minutes per week of moderate-intensity aerobic activity (e.g., brisk walking), the equivalent of ≥ 7.5 MET-hrs per week.27
Lipid Assessment
EDTA blood samples were obtained at the time of enrollment in the WHS and stored in vapor-phase liquid nitrogen (−170°C). Study participants had baseline blood samples assayed for HDL-C, ApoA1 and triglyceride (TG) levels in a core laboratory certified by the National Heart Lung and Blood Institute/Centers for Disease Control and Prevention Lipid Standardization Program and had low coefficients of variance.4 Among the 21,806 women for whom we have information on fasting status, 16562 (76%) were fasting for at least 8 hours prior to blood collection.
Ascertainment of Cardiovascular Events
Criteria for endpoint assessment have been reported previously.28 All study participants were followed for myocardial infarction through February 2008. An end-points committee of physicians reviewed medical records for all participants reporting myocardial infarction. Events were confirmed if symptoms met World Health Organization Criteria and if the event was associated with abnormal levels of cardiac enzymes or diagnostic electrocardiographic criteria. Only confirmed end-points were included in this analysis.
Statistical Analysis
We identified the common SNPs in 9 previously described genes3, 6 influencing HDL-C levels at a genome-wide level of statistical significance (P<5×10−8) in the 23,294 women for whom we have genotype information by performing linear regression in PLINK. In total 58 SNPs achieved genome wide significance for HDL-C (Supplementary Table 1). All 58 SNPs were screened for statistically significant evidence of effect modification by physical activity across the median in our cohort (8.8 MET-hours/week), using the unadjusted quantitative trait interaction procedure available in PLINK.29 An a priori threshold of P<0.05 was used to select SNPs for further analysis. A sensitivity analysis using physical activity adjusted HDL-C levels identified 5 additional SNPs, two of which were in a tenth gene (LCAT) that was not identified in the primary analysis but which has been identified previously as a determinant of HDL-C.6 These 5 SNPs were also tested for evidence of effect modification.
After restricting the sample to those with complete information for HDL-C and MET-hours of physical activity (N=22,939), characteristics of the study population were computed across the physical activity median in our cohort (8.8 MET-hours/week), and compared using two-sided t-tests for continuous variables and χ2 tests for categorical variables. We performed a multivariable linear regression analyses for each significant SNP, assuming an additive model of inheritance, to test the association with HDL-C levels after adjustment for age, BMI, alcohol intake, hypertension, diabetes, and hormone use status. We calculated the effect estimates and mean levels of HDL-C for each genotype at each selected SNP, stratified by median activity levels. Tests for interaction were performed using multivariable linear regression models that included the above covariates, number of copies of the minor allele at each SNP (0, 1, or 2), physical activity below (0) and above (1) the median, and an interaction term (SNP*physical activity). We tested the significant SNPs for evidence of effect modification of ApoA1 levels, the major apolipoprotein on the HDL-C molecule, in similarly adjusted models. As a sensitivity analysis, we used a P-value threshold of 0.0056, which represents a P-value threshold of 0.05 corrected for the 9 genes tested for evidence of interaction.
We created three separate genotype scores, one for each SNP (rs10096633 in LPL, rs1800588 in LIPG, and rs1532624 in CETP). At each SNP, we used a dominant model to categorize women into two groups: those who were homozygous for the major allele and those who were carriers of a minor allele. At rs10096633, rs1800588, and rs1532624, women homozygous for the major allele had lower HDL-C levels than women carrying at least one copy of the minor allele, and thus were defined as having a “deleterious” genotype. Minor allele carrier status was defined as “beneficial” for each of these three SNPs on the assumption that any impact on MI risk would be consistent with the observed effect on HDL-C. We used multivariable Cox proportional hazard models, adjusting for age, BMI, blood pressure (Framingham categories), history of diabetes, smoking, and total cholesterol, to estimate the hazard ratio (HR) of incident MI in the entire cohort associated with each genotype score. Next, we performed a stratified analysis, according to genotype score, to estimate the HR of incident MI associated with increased level physical activity, using inactive women (≤8.8MET-hrs/week, the median in our cohort) as the reference group.
All analyses were carried out using SAS/Genetics 9.1 package (SAS Institute Inc, Cary, NC) and PLINK.29
Results
Women who reported physical activity levels above the median of 8.8 MET-hours/wk had, on average, a lower baseline body mass index (BMI), lower TG levels, higher levels of HDL-C and ApoA1, and a lower prevalence of self-reported diabetes and hypertension (Table 1). Post menopausal hormone use and moderate alcohol use was also more common amongst more active women.
Table 1.
Characteristics of Study Participants According to Physical Activity Levels
| ≤8.8MET-hours/week N=11,446 |
> 8.8MET-hours/week N=11,493 |
|
|---|---|---|
| Age, y | 52.0 (48.0–58.0) | 52.0 (48.0–59.0) |
| BMI, kg/m2 | 25.7 (22.9–29.6) | 24.1 (21.9–27.0) |
| Cholesterol treatment, % | 3.2 | 3.2 |
| Diabetes, % | 3.0 | 2.1 |
| Hypertension, % | 27.1 | 22.0 |
| Metabolic syndrome, % | 28.4 | 17.8 |
| Paternal history of MI, % | 13.2 | 12.7 |
| Post-menopausal hormone use, % | 42.2 | 45.5 |
| Alcohol consumption, % | ||
| Rarely | 48.2 | 38.4 |
| 1–3 drinks/mo | 13.4 | 13.2 |
| 1–6 drinks/wk | 28.8 | 36.7 |
| ≥ drinks/d | 9.7 | 11.8 |
| TG, mg/dL | 127.0 (88.0–184.0) | 114.0 (79.0–167.0) |
| HDL-C, mg/dL | 50.1 (41.9–60.4) | 53.7 (44.7–63.9) |
| ApoA1, mg/dL | 146.8 (130.5–165.4) | 151.6 (134.7–170.4) |
Values shown for continuous variables are median (IQR).
HDL denotes high-density cholesterol, ApoA1 denotes Apolipoprotein A1 and TG denotes Triglycerides.
The SNPs associated with HDL-C levels at a genome-wide level of statistical significance and their unadjusted P-values for interaction with the median level of physical activity (8.8 MET-hrs/week) are displayed in Supplementary Table 1, sorted by chromosomal position. Evidence of effect modification was observed for 8 SNPs in 4 genes, including 2 SNPs at LPL (8q21), 3 at LIPC (15q22), 2 at CETP (16q13), and 1 at LIPG (18q45). The SNPs with the smallest interaction P-value at each locus were rs10096633 (LPL; P-interaction=0.006), rs1800588 (LIPC; P-interaction=0.01), rs1532624 (CETP; P-interaction=0.008) and rs4939883 (LIPG; P-interaction=0.03). The absolute difference (SE) in HDL-C level per copy of each significant allele ranged from 1.098 (0.193) for rs4939883 in LIPG (minor allele frequency (MAF) = 0.17) to 3.067 (0.146) for rs1532624 in CETP (MAF = 0.43). We did not observe evidence of effect modification for other SNPs with similar MAF and effect sizes (Supplementary Table 1). None of the 5 SNPs identified in a sensitivity analysis using physical activity adjusted HDL-C met the pre-specified criteria for evidence of effect modification (Supplementary Table 2).
Mean (±SD) HDL-C by genotype and increase in HDL-C per copy of the minor alleles at significant SNPs within the LPL, LIPC, CETP and LIPG genes in all women and in women stratified by median levels of physical activity are displayed in Table 2. As shown, we observed a smaller per-allele increase in HDL-C for rs10096633 at LPL among active women (1.0 mg/dL per copy) than among inactive women (2.1 mg/dL per copy, P-interaction=0.004). By contrast, for rs1532624 at CETP and rs1800588 at LIPC, we observed a larger increase in HDL-C levels per copy of the minor allele in active women than in inactive women (P-interaction= 0.04 and 0.02, respectively). While our initial test for interaction suggested that the effect of rs4939883 at LIPG on HDL-C levels was modified by physical activity, this effect was no longer statistically significant in the fully adjusted model. Similar results were seen for the SNPs in LPL, LIPC, and CETP when using an alternative physical activity cutpoint (Supplementary Table 3). The distribution of HDL-C levels stratified by genotype and physical activity level are presented in the Supplementary Figure. The proportion of variance in HDL-C explained by the physical activity-genotype interaction was 0.03% for rs10096633 in LPL, 0.01% for rs1800588 in LIPC, and 0.03% for rs1532624 in CETP. By comparison, adding each of SNP individually to adjusted models explained an additional 0.27% (rs10096633), 0.58% (rs1800588), or 1.99% (rs1532624) of the variance in HDL-C. In sensitivity analyses using a P-value threshold corrected for the number of genes analyzed (P<0.0056), evidence of interaction remained statistically significant only for rs10096633 in LPL. When we restricted our analysis to women who had been fasting for at least 8 hours at the time of blood collection our observations did not differ substantially.
Table 2.
Mean HDL-C (mg/dl) Levels per copy of the Minor Allele at Significant SNPs in the Entire Cohort and Across Median levels of Physical Activity
| Mean (SD) HDL-C per allele, copy mg/dl | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome, Gene | N | 0 | 1 | 2 | β (SE) | P | |
| rs10096633 | 8p21, LPL | All women | 22,938 | 53.3 (15.0) | 55.0 (15.3) | 55.9 (15.2) | 1.6 (0.2) | <0.0001 |
| ≤8.8MET-hours/week | 11,445 | 51.5 (14.4) | 53.9 (14.9) | 54.1 (14.6) | 2.1 (0.3) | <0.0001 | ||
| >8.8MET-hours/week | 11,493 | 55.2 (15.2) | 56.1 (15.6) | 57.7 (15.6) | 1.0 (0.3) | <0.001 | ||
| Test for interaction | 0.004 | |||||||
| rs1800588 | 15q22, LIPC | All women | 22,936 | 52.9 (14.6) | 54.9 (15.5) | 56.9 (15.5) | 2.0 (0.2) | <0.0001 |
| ≤8.8MET-hours/week | 11,445 | 51.3 (14.2) | 53.0 (15.1) | 54.4 (14.3) | 1.7 (0.2) | <0.0001 | ||
| >8.8MET-hours/week | 11,491 | 54.4 (14.9) | 56.8 (15.7) | 59.3 (16.3) | 2.3 (0.2) | <0.0001 | ||
| Test for interaction | 0.04 | |||||||
| rs1532624 | 16q13, CETP | All women | 22,195 | 51.3 (14.2) | 54.1 (15.0) | 57.5 (15.7) | 3.0 (0.1) | <0.0001 |
| ≤8.8MET-hours/week | 11,065 | 50.0 (14.1) | 52.2 (14.4) | 55.5 (15.2) | 2.7 (0.2) | <0.0001 | ||
| >8.8MET-hours/week | 11,130 | 52.6 (14.2) | 55.8 (15.4) | 59.4 (16.0) | 3.3 (0.2) | <0.0001 | ||
| Test for interaction | 0.02 | |||||||
| rs4939883 | 18q45, LIPG | All women | 22,763 | 54.2 (15.2) | 53.0 (14.7) | 51.6 (14.6) | −1.2 (0.17) | <0.0001 |
| ≤8.8MET-hours/week | 11,353 | 52.6 (14.8) | 51.0 (13.9) | 49.7 (14.0) | −1.4 (0.2) | <0.0001 | ||
| >8.8MET-hours/week | 11,410 | 55.7 (15.3) | 55.0 (15.1) | 53.7 (15.0) | −0.9 (0.2) | 0.0001 | ||
| Test for interaction | 0.14 | |||||||
Values for HDL (high-density cholesterol) levels are mean (±SD).
We observed similar associations between the above SNPs and ApoA1 levels (Table 3). As seen with HDL-C, the observed per-allele increase in ApoA1 for rs10096633 at LPL was greater among inactive women (interaction<0.001). Copies of the minor allele at rs1532624 (CETP) and rs1800588 (LIPC) were associated with a greater increases in ApoA1 levels amongst active women, compared to inactive women (P-interaction=0.02 and 0.09, respectively).
Table 3.
Mean ApoA1 (mg/dl) Levels per copy of the Minor Allele at Significant SNPs in the Entire Cohort and Across Median levels of Physical Activity
| Mean (SD) ApoA1 per allele, copy mg/dl | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome, Gene | N | 0 | 1 | 2 | β (SE) | P | |
| rs10096633 | 8p21, LPL | All women | 22833 | 150.6 (25.5) | 152.4 (25.8) | 153.2 (25.5) | 1.77 (0.31) | <0.0001 |
| ≤8.8MET-hours/week | 11390 | 148.1 (25.1) | 151.1 (25.0) | 152.4 (26.2) | 2.92 (0.43) | <0.0001 | ||
| >8.8MET-hours/week | 11443 | 153.0 (25.7) | 153.7 (26.6) | 154.0 (24.9) | 0.64 (0.45) | 0.15 | ||
| Test for interaction | 0.0003 | |||||||
| rs1800588 | 15q22, LIPC | All women | 22,831 | 149.3 (25.2) | 153.1 (26.0) | 158.2 (26.3) | 4.0 (0.3) | <0.0001 |
| ≤8.8MET-hours/week | 11,390 | 147.4 (24.6) | 150.6 (25.7) | 154.8 (25.9) | 3.6 (0.4) | <0.0001 | ||
| >8.8MET-hours/week | 11,441 | 151.2 (25.6) | 155.7 (26.0) | 161.6 (26.1) | 4.5 (0.4) | <0.0001 | ||
| Test for interaction | 0.09 | |||||||
| rs1532624 | 16q13, CETP | All women | 22,195 | 147.5 (25.0) | 151.6 (25.7) | 155.5 (25.9) | 4.0 (0.2) | <0.0001 |
| ≤8.8MET-hours/week | 11,065 | 145.9 (24.5) | 149.3 (25.1) | 152.9 (25.6) | 3.4 (0.3) | <0.0001 | ||
| >8.8MET-hours/week | 11,130 | 149.2 (25.3) | 153.8 (26.0) | 158.1 (25.9) | 4.4 (0.3) | <0.001 | ||
| Test for interaction | 0.02 | |||||||
| rs4939883 | 18q45, LIPG | All women | 22,809 | 151.8 (25.8) | 149.4 (25.0) | 146.6 (25.0) | −2.6 (0.3) | <0.0001 |
| ≤8.8MET-hours/week | 11,377 | 149.9 (25.4) | 146.8 (24.3) | 144.6 (24.1) | −2.7 (0.4) | <0.0001 | ||
| >8.8MET-hours/week | 11,432 | 153.9 (26.1) | 152.0 (25.5) | 148.8 (25.9) | −2.4 (0.4) | <0.0001 | ||
| Test for interaction | 0.59 | |||||||
Values for ApoA1(Apolipoprotein A1) levels are mean (±SD).
Higher HDL-C levels were observed among women with the beneficial as compared with deleterious genotype at rs10096633 in LPL (55.08 vs. 53.33mg/dL, P<0.0001), rs1800588 at LIPC (55.17 vs. 52.87mg/dL, P<0.0001), and rs1532624 at CETP (54.99 vs. 51.26 mg/dL, P<0.0001). However, only women who carried at least one copy of the minor allele at CETP had a reduced risk of MI (hazard ratio [HR] 0.72, 95% confidence interval [CI] 0.57–0.92; P=0.009) after adjusting for age, body mass index, diabetes, blood pressure, current smoking, and total cholesterol. Women with a beneficial genotype at rs10096633 (LPL) or rs1800588 (LIPC) were not at reduced risk of MI (LPL-HR 0.81, 95% CI 0.61–1.09, P=0.16; LIPC-HR 1.11, 95% CI 0.87–1.41, P=0.39).
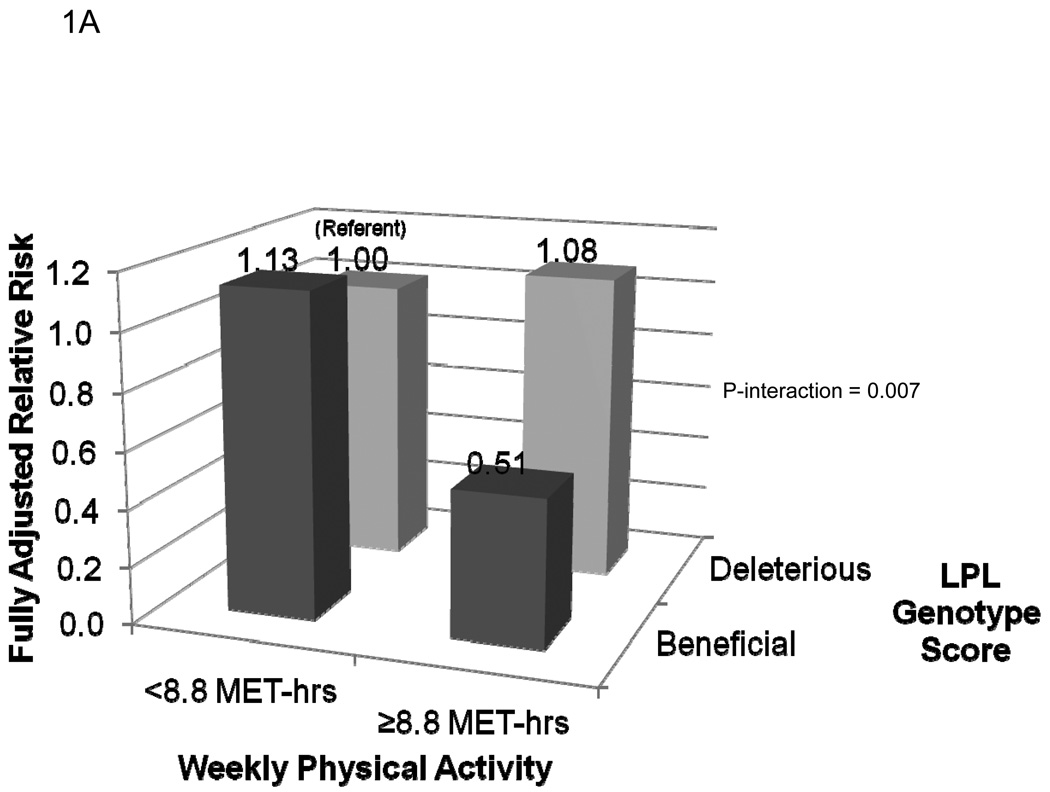
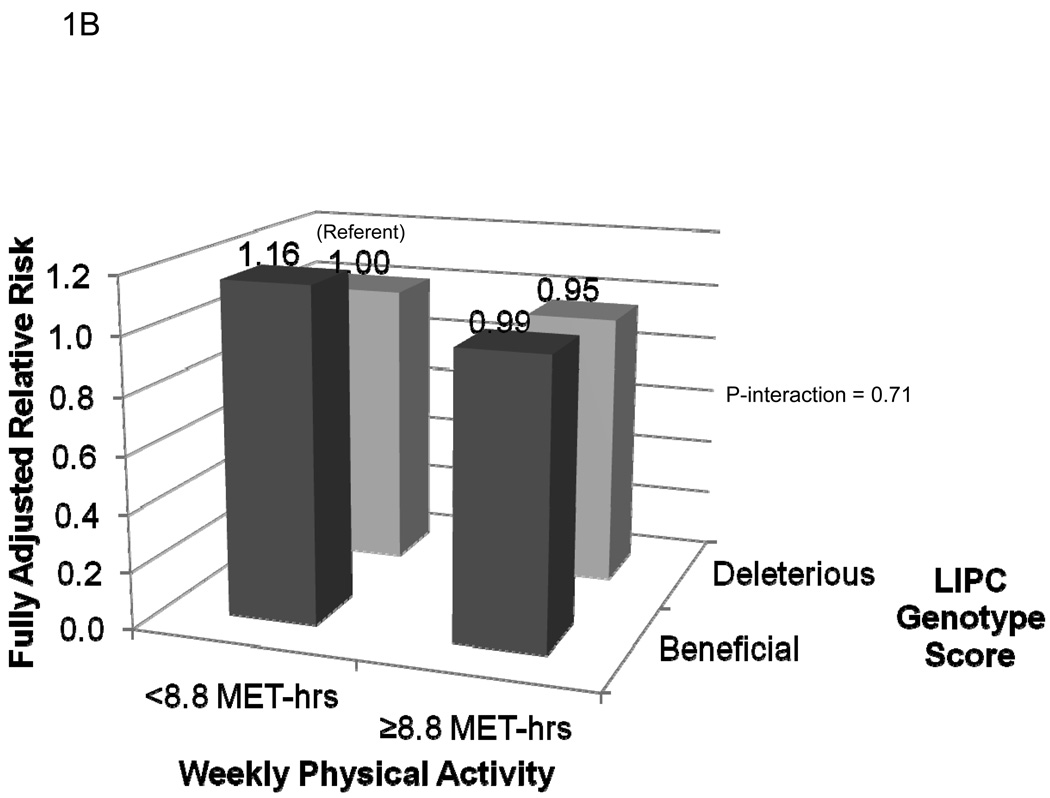
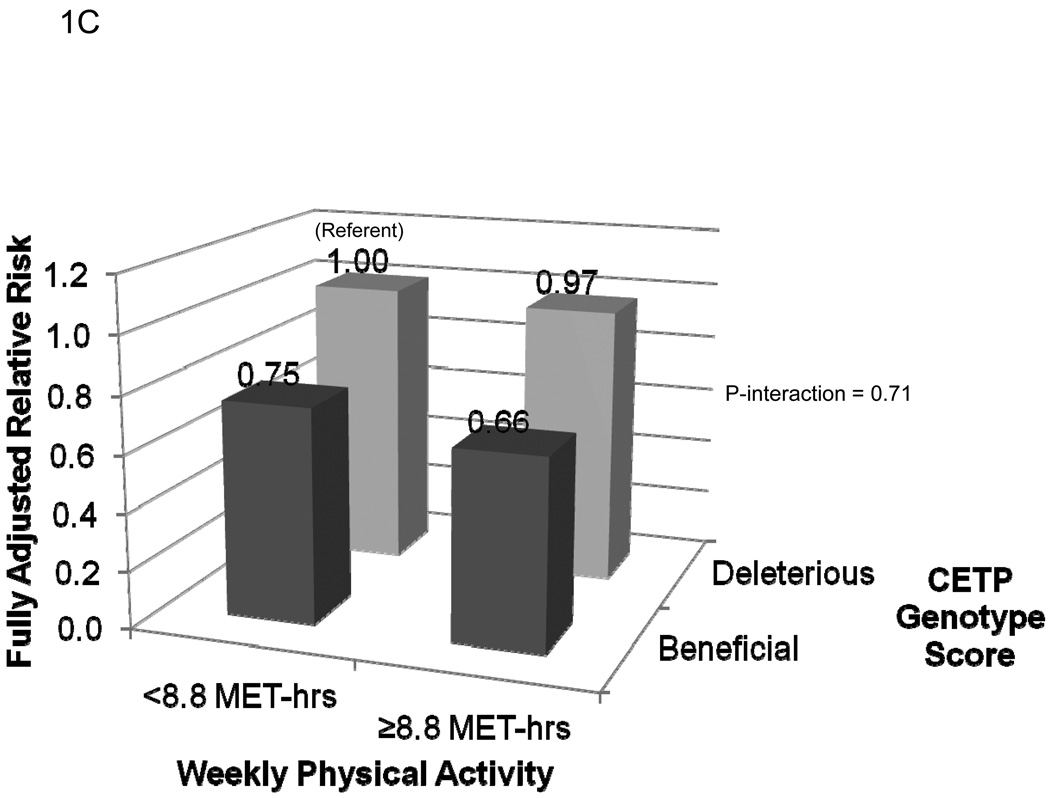
The fully adjusted risks of MI for each of the four categories of genotype score and activity level are presented in Figure 1. As shown in Figure 1A, compared to inactive women with a deleterious genotype at LPL, active carriers of at least one copy of the minor allele at rs10096633 were at reduced risk of MI (HR 0.51, 95% CI 0.30–0.86; P=0.01). By contrast, inactive women with the beneficial genotype at LPL were not at reduced risk of MI (HR 1.13, 95% CI 0.79–1.61; P=0.50; P-interaction = 0.007). The effect of physical activity on MI risk did not appear to be modified by genotype score at rs1800588 (LIPC) (P-interaction = 0.71, Figure 1B). Finally, the reduction in risk associated with a beneficial genotype at rs1532624 at CETP appeared to be similar in active and inactive women, regardless of physical activity level (P-interaction = 0.71, Figure 1C).
Figure 1.
Risk of incident myocardial infarction according to categories of leisure-time physical activity level and genotype score at the LPL (1A), LIPC (1B), and CETP (1C) loci. Women with at least one copy of the minor allele at each locus were defined as having a beneficial genotype at that locus, while those homozygous for the major allele were defined as having a deleterious genotype at that locus. Compared to those with a deleterious genotype score, high-density lipoprotein cholesterol levels among those with a beneficial genotype score at LPL (rs10096633) were 1.75 mg/dl higher, at LIPC (rs1800588) were 2.3 mg/dL higher, and at CETP (rs1800588) were 3.73 mg/dL higher (Each P<0.0001).
Discussion
In this study of 22,939 healthy U.S. Caucasian women, we report that the effects of SNPs at LPL, LIPC and CETP on HDL-C levels are modified by physical activity. In our study, the per-allele increase in HDL-C for rs1800588 at LIPC and rs1532624 at CETP was greater among active women, while the per-allele increase in HDL-C per copy of the minor allele of rs10096633 at LPL was larger among inactive women. Although minor allele carrier status at each of these loci was associated with higher levels of HDL-C, the associated reduction in risk of MI differed by SNP and by activity level. Minor allele carrier status at LPL was related to a 50% risk-reduction in MI only amongst active participants, and while carrier status at LIPC did not appear to afford protection from risk of MI, the carrier status at CETP was associated with approximately a 30% risk reduction in MI, regardless of activity levels.
We believe these data are of interest for several reasons. First, using a different study design and analytic approach than previous work in the field, we offer further evidence of the established role of LPL, LIPC, and CETP in modulating the response of HDL-C and ApoA1 to physical activity. Prior work has recognized that exercise-induced changes in lipid phenotypes, such as an increase in HDL-C, are related to decreases in plasma levels and activity of CETP,30, 31 increases in LPL activity in both plasma and skeletal muscle,20, 32, 33 and to decreases in post-heparin plasma LIPC activity.34 Several studies have shown that genetic variation within CETP can modulate alterations in HDL-C due to exercise, possibly via a differential response of CETP activity or mass to exercise.12, 18, 19, 35 Other studies have reported that physical activity modifies the effect of two common genetic polymorphisms in LPL on HDL-C.20, 36 Finally, while common variation in LIPC is known to affect enzyme activity,37 evidence that physical activity modifies the effects of genetic variation in LIPC on plasma LIPC activity and lipid phenotypes is inconsistent.21, 34, 38 While our findings also implicate LPL, LIPC, and CETP as important mediators of the response in HDL-C to physical activity, we extend those findings to women, who were absent from many the prior studies.20, 35, 38 We also measured leisure-time physical activity, which may be less likely to cause substantial charges in lipid metabolism and HDL-C levels than prescribed diet and exercise interventions, which were used in much of the previous work in this area.18, 19, 21, 34
Second, the magnitude of the per-allele effect of common polymorphisms on HDL-C levels varies according to level of physical activity. For example, the effect of variation at rs10096633 in LPL on HDL-C level is larger among sedentary than active women, while the effect of variation at rs1532624 in CETP on HDL-C levels is smaller among sedentary than active women. This differential modification provides a possible biological explanation for the wide variability in HDL-C response to exercise, and for the clinical observation that some individuals do not experience improvements in HDL-C level despite adopting an exercise regimen.17
Third, in contrast to rs1532624 at CETP, we report a lack of association between beneficial genotypes at rs10096633 in LPL and rs1800588 at LIPC and risk of incident MI. This is in spite of the fact that women with at least one copy of the minor allele at either locus had HDL-C levels that were, on average, 1.75–2.3 mg/dL higher than those with a deleterious genotype score at those loci. This lack of association, when viewed in the context of a strong association between CETP genotype and MI risk in our cohort7 and others12 highlights the understanding that increases in HDL-C levels due to variation in key lipid metabolism genes do not necessarily lead to the expected reduction in vascular risk.9, 10, 39, 40 Our observation that regular physical activity reduces MI risk among women with a beneficial LPL genotype, but not among those with a deleterious genotype, raises the additional possibility that the effects of lifestyle choices on cardiovascular events may vary by genotype. While one other study has reported evidence that physical activity alters the vascular risk conferred by the −480C>T mutation in LIPC, this report is the first of which we are aware that the effects of physical activity may differ according to variation within LPL.10 The biological mechanism by which physical activity might reduce the risk of MI among female carriers but not among non-carriers of the minor allele at rs10096633 is unclear. The identity and biological action of the true causal variant are unknown (it is unlikely to be rs10096633), as are the mechanisms by which it might alter LPL levels and/or function, HDL-C levels and/or function, and downstream vascular risk. While it is certainly possible that altered LPL catalyzes important functional changes in HDL-C that then affect MI risk, the design of the WHS prevents us from being able to tease out whether functional changes in HDL-C or another biologic process is the cause of the observed differences in MI risk.
Strengths and Limitations
Strengths of the present study include its large sample size and the careful collection of physical activity data at the time of blood sampling. However, a number of limitations need to be considered. First, our analytic approach selected only those SNPs related to HDL-C levels at a Bonferroni-corrected genome wide level of significance (P<5×10−8). This approach would not detect genes that had opposite per-allele effects in the two strata of physical activity, although the biological plausibility of such an interaction with a continuous exposure such as physical activity is uncertain. Second, while our sample size is large, our power to detect statistically significant interactions is dependent upon allele effect size and minor allele frequency. Third, physical activity, weight, height, diabetes and hypertension were assessed by self-report. Imprecise reporting of the variables may have occurred; however, any misclassification of this kind would be expected to bias the results towards the null since reports were prospective and predated the occurrence of MI. Lastly, our study only includes Caucasians and may not be generalizable to other groups.
We believe that the evidence of effect modification reported here is real, in part because the 3 reported loci have been implicated in prior reports of effect modification. In conjunction with findings from previously published work, these findings support the hypothesis that genetic determinants of key steps in HDL-C metabolism, involving the LPL, LIPC, and CETP genes, are differentially modified by physical activity. Finally, our data raise the possibility that the effects of common variants in these genes on MI risk may also be modified by activity level.
Conclusion
In summary, we report evidence that the effects of common polymorphisms in the LPL, LIPC, and CETP genes on HDL-C levels are modified by physical activity. We observed greater per-minor-allele increases in HDL-C for rs1800588 in LIPC and rs1532624 in CETP among active than inactive women, and smaller per-allele increases in HDL-C for rs10096633 at LPL among active than inactive women. Minor allele carrier status at LPL was related to risk-reduction in MI only amongst active participants, whereas carrier status at CETP was associated with reductions in risk of MI, regardless of activity status. Minor allele carrier status at LIPC did not appear to associate with MI risk. Common variation within these genes appears to influence the response of HDL-C to physical activity. The protection from MI afforded by higher plasma levels of HDL-C may depend both on absolute levels and variation in the genetic determinants of those levels.
Supplementary Material
Acknowledgements
Dr Ahmad and Dr. Everett had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding Sources
Dr Ahmad is supported by a T32 Training Grant from the NHLBI (T32 HL07575). Dr. Everett is supported by the American Heart Association (SDG 0835304N). The Women’s Health Study is supported by grants HL-43851 and CA-47988 from the National Heart, Lung, and Blood Institute and the National Cancer Institute, and by grants from the Donald W. Reynolds Foundation (Las Vegas, NV), the Leducq Foundation (Paris, France), and the Doris Duke Charitable Foundation (New York, NY). Genotyping was provided by Amgen, Inc (Cambridge, MA). The funding agencies played no role in the design, conduct, data management, analysis, or manuscript preparation related to this manuscript.
Footnotes
Journal Subject Codes: Acute myocardial infarction, Clinical genetics, Exercise/exercise testing/rehabilitation, Risk factors, Genetics of cardiovascular disease, Lipid and lipoprotein metabolism, Epidemiology.
Conflict of Interest Disclosures: Dr. Lee serves as a consultant to Virgin HealthMiles, and sits on its Scientific Advisory Board. No other author has any revelvant conflicts to disclose.
References
- 1.Ridker PM, Rifai N, Cook NR, Bradwin G, Buring JE. Non-HDL cholesterol, apolipoproteins A-I and B100, standard lipid measures, lipid ratios, and CRP as risk factors for cardiovascular disease in women. JAMA. 2005;294:326–333. doi: 10.1001/jama.294.3.326. [DOI] [PubMed] [Google Scholar]
- 2.Fenger M, Benyamin B, Schousboe K, Sorensen TI, Kyvik KO. Variance decomposition of apolipoproteins and lipids in Danish twins. Atherosclerosis. 2007;191:40–47. doi: 10.1016/j.atherosclerosis.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, Ordovas JM, Ripatti S, Parker AN, Miletich JP, Ridker PM. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chasman DI, Pare G, Zee RYL, Parker AN, Cook NR, Buring JE, Kwiatkowski DJ, Rose LM, Smith JD, Williams PT, Rieder MJ, Rotter JI, Nickerson DA, Krauss RM, Miletich JP, Ridker PM. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ Cardiovasc Genet. 2008;1:21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namboodiri KK, Kaplan EB, Heuch I, Elston RC, Green PP, Rao DC, Laskarzewski P, Glueck CJ, Rifkind BM. The Collaborative Lipid Research Clinics Family Study: biological and cultural determinants of familial resemblance for plasma lipids and lipoproteins. Genet Epidemiol. 1985;2:227–254. doi: 10.1002/gepi.1370020302. [DOI] [PubMed] [Google Scholar]
- 6.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Doring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridker PM, Pare G, Parker AN, Zee RYL, Miletich JP, Chasman DI. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 9.Andersen RV, Wittrup HH, Tybjaerg-Hansen A, Steffensen R, Schnohr P, Nordestgaard BG. Hepatic lipase mutations, elevated high-density lipoprotein cholesterol, and increased risk of ischemic heart disease: the Copenhagen City Heart Study. J Am Coll Cardiol. 2003;41:1972–1982. doi: 10.1016/s0735-1097(03)00407-8. [DOI] [PubMed] [Google Scholar]
- 10.Hokanson JE, Kamboh MI, Scarboro S, Eckel RH, Hamman RF. Effects of the hepatic lipase gene and physical activity on coronary heart disease risk. Am J Epidemiol. 2003;158:836–843. doi: 10.1093/aje/kwg230. [DOI] [PubMed] [Google Scholar]
- 11.Frikke-Schmidt R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis. 2010;208:305–316. doi: 10.1016/j.atherosclerosis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 13.Mora S, Cook N, Buring JE, Ridker PM, Lee IM. Physical activity and reduced risk of cardiovascular events: potential mediating mechanisms. Circulation. 2007;116:2110–2118. doi: 10.1161/CIRCULATIONAHA.107.729939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz RS. The independent effects of dietary weight loss and aerobic training on high density lipoproteins and apolipoprotein A-I concentrations in obese men. Metabolism. 1987;36:165–171. doi: 10.1016/0026-0495(87)90012-6. [DOI] [PubMed] [Google Scholar]
- 15.Kraus WE, Houmard JA, Duscha BD, Knetzger KJ, Wharton MB, McCartney JS, Bales CW, Henes S, Samsa GP, Otvos JD, Kulkarni KR, Slentz CA. Effects of the amount and intensity of exercise on plasma lipoproteins. N Engl J Med. 2002;347:1483–1492. doi: 10.1056/NEJMoa020194. [DOI] [PubMed] [Google Scholar]
- 16.Smith CE, Arnett DK, Tsai MY, Lai CQ, Parnell LD, Shen J, Laclaustra M, Junyent M, Ordovas JM. Physical inactivity interacts with an endothelial lipase polymorphism to modulate high density lipoprotein cholesterol in the GOLDN study. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leon AS, Gaskill SE, Rice T, Bergeron J, Gagnon J, Rao DC, Skinner JS, Wilmore JH, Bouchard C. Variability in the response of HDL cholesterol to exercise training in the HERITAGE Family Study. Int J Sports Med. 2002;23:1–9. doi: 10.1055/s-2002-19270. [DOI] [PubMed] [Google Scholar]
- 18.Wilund KR, Ferrell RE, Phares DA, Goldberg AP, Hagberg JM. Changes in high-density lipoprotein-cholesterol subfractions with exercise training may be dependent on cholesteryl ester transfer protein (CETP) genotype. Metabolism. 2002;51:774–778. doi: 10.1053/meta.2002.32730. [DOI] [PubMed] [Google Scholar]
- 19.Ayyobi AF, Hill JS, Molhuizen HO, Lear SA. Cholesterol ester transfer protein (CETP) Taq1B polymorphism influences the effect of a standardized cardiac rehabilitation program on lipid risk markers. Atherosclerosis. 2005;181:363–369. doi: 10.1016/j.atherosclerosis.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Senti M, Elosua R, Tomas M, Sala J, Masia R, Ordovas JM, Shen H, Marrugat J. Physical activity modulates the combined effect of a common variant of the lipoprotein lipase gene and smoking on serum triglyceride levels and high-density lipoprotein cholesterol in men. Hum Genet. 2001;109:385–392. doi: 10.1007/s004390100584. [DOI] [PubMed] [Google Scholar]
- 21.Grarup N, Andreasen CH, Andersen MK, Albrechtsen A, Sandbaek A, Lauritzen T, Borch-Johnsen K, Jorgensen T, Schmitz O, Hansen T, Pedersen O. The −250G>A promoter variant in hepatic lipase associates with elevated fasting serum high-density lipoprotein cholesterol modulated by interaction with physical activity in a study of 16,156 Danish subjects. J Clin Endocrinol Metab. 2008;93:2294–2299. doi: 10.1210/jc.2007-2815. [DOI] [PubMed] [Google Scholar]
- 22.Ruano G, Seip RL, Windemuth A, Zollner S, Tsongalis GJ, Ordovas J, Otvos J, Bilbie C, Miles M, Zoeller R, Visich P, Gordon P, Angelopoulos TJ, Pescatello L, Moyna N, Thompson PD. Apolipoprotein A1 genotype affects the change in high density lipoprotein cholesterol subfractions with exercise training. Atherosclerosis. 2006;185:65–69. doi: 10.1016/j.atherosclerosis.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein MS, Costanza MC, James RW, Morris MA, Cambien F, Raoux S, Morabia A. Physical activity may modulate effects of ApoE genotype on lipid profile. Arterioscler Thromb Vasc Biol. 2002;22:133–140. doi: 10.1161/hq0102.101819. [DOI] [PubMed] [Google Scholar]
- 24.Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 25.Wolf AM, Hunter DJ, Colditz GA, Manson JE, Stampfer MJ, Corsano KA, Rosner B, Kriska A, Willett WC. Reproducibility and validity of a self-administered physical activity questionnaire. Int J Epidemiol. 1994;23:991–999. doi: 10.1093/ije/23.5.991. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein AR, Sesso HD, Lee IM, Cook NR, Manson JE, Buring JE, Gaziano JM. Relationship of physical activity vs body mass index with type 2 diabetes in women. JAMA. 2004;292:1188–1194. doi: 10.1001/jama.292.10.1188. [DOI] [PubMed] [Google Scholar]
- 27.United States Department of Health and Human Services. Washington, D.C: U.S. Government; Physical Activity Guidelines for Americans. 2008
- 28.Ridker PM, Cook NR, Lee IM, Gordon D, Gaziano JM, Manson JE, Hennekens CH, Buring JE. A randomized trial of low-dose aspirin in the primary prevention of cardiovascular disease in women. N Engl J Med. 2005;352:1293–1304. doi: 10.1056/NEJMoa050613. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seip RL, Moulin P, Cocke T, Tall A, Kohrt WM, Mankowitz K, Semenkovich CF, Ostlund R, Schonfeld G. Exercise training decreases plasma cholesteryl ester transfer protein. Arterioscler Thromb. 1993;13:1359–1367. doi: 10.1161/01.atv.13.9.1359. [DOI] [PubMed] [Google Scholar]
- 31.Foger B, Wohlfarter T, Ritsch A, Lechleitner M, Miller CH, Dienstl A, Patsch JR. Kinetics of lipids, apolipoproteins, and cholesteryl ester transfer protein in plasma after a bicycle marathon. Metabolism. 1994;43:633–639. doi: 10.1016/0026-0495(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 32.Lithell H, Orlander J, Schele R, Sjodin B, Karlsson J. Changes in lipoprotein-lipase activity and lipid stores in human skeletal muscle with prolonged heavy exercise. Acta Physiol Scand. 1979;107:257–261. doi: 10.1111/j.1748-1716.1979.tb06471.x. [DOI] [PubMed] [Google Scholar]
- 33.Miyashita M, Eto M, Sasai H, Tsujimoto T, Nomata Y, Tanaka K. Twelve-week jogging training increases pre-heparin serum lipoprotein lipase concentrations in overweight/obese middle-aged men. J Atheroscler Thromb. 2010;17:21–29. doi: 10.5551/jat.2337. [DOI] [PubMed] [Google Scholar]
- 34.Teran-Garcia M, Santoro N, Rankinen T, Bergeron J, Rice T, Leon AS, Rao DC, Skinner JS, Bergman RN, Despres JP, Bouchard C. Hepatic lipase gene variant −514C>T is associated with lipoprotein and insulin sensitivity response to regular exercise: the HERITAGE Family Study. Diabetes. 2005;54:2251–2255. doi: 10.2337/diabetes.54.7.2251. [DOI] [PubMed] [Google Scholar]
- 35.Kuivenhoven JA, de Knijff P, Boer JMA, Smalheer HA, Botma G-J, Seidell JC, Kastelein JJP, Pritchard PH. Heterogeneity at the CETP gene locus : influence on plasma CETP concentrations and HDL cholesterol levels. Arterioscler Thromb Vasc Biol. 1997;17:560–568. doi: 10.1161/01.atv.17.3.560. [DOI] [PubMed] [Google Scholar]
- 36.Boer JM, Kuivenhoven JA, Feskens EJ, Schouten EG, Havekes LM, Seidell JC, Kastelein JJ, Kromhout D. Physical activity modulates the effect of a lipoprotein lipase mutation (D9N) on plasma lipids and lipoproteins. Clin Genet. 1999;56:158–163. doi: 10.1034/j.1399-0004.1999.560212.x. [DOI] [PubMed] [Google Scholar]
- 37.Cohen JC, Vega GL, Grundy SM. Hepatic lipase: new insights from genetic and metabolic studies. Curr Opin Lipidol. 1999;10:259–267. doi: 10.1097/00041433-199906000-00008. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Lopez-Ridaura R, Rimm EB, Rifai N, Hunter DJ, Hu FB. Interactions between the −514C->T polymorphism of the hepatic lipase gene and lifestyle factors in relation to HDL concentrations among US diabetic men. Am J Clin Nutr. 2005;81:1429–1435. doi: 10.1093/ajcn/81.6.1429. [DOI] [PubMed] [Google Scholar]
- 39.Wittrup HH, Tybjaerg-Hansen A, Nordestgaard BG. Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. A meta-analysis. Circulation. 1999;99:2901–2907. doi: 10.1161/01.cir.99.22.2901. [DOI] [PubMed] [Google Scholar]
- 40.Frikke-Schmidt R, Nordestgaard BG, Stene MC, Sethi AA, Remaley AT, Schnohr P, Grande P, Tybjaerg-Hansen A. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 2008;299:2524–2532. doi: 10.1001/jama.299.21.2524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.