Abstract
In this study we show the expression of flavin mononucleotide (FMN)-based fluorescent protein (FbFP) BS2 as a marker for gene expression in the opportunistic human anaerobic pathogen Bacteroides fragilis. B. fragilis 638R strain carrying osu::bs2 constructs showed inducible fluorescence following addition of maltose anaerobically compared to non-fluorescent cells under glucose repressed conditions. Bacteria carrying ahpC::bs2 or dps::bs2 constructs were fluorescent following induction by oxygen compared to non-fluorescent cells from the anaerobic control cultures. In addition, when these transcriptional fusion constructs were mobilized into B. fragilis IB263, a constitutive peroxide response strain, fluorescent BS2 was detected in both anaerobic and aerobic cultures confirming the unique properties of the FbFP BS2 to yield fluorescent signal in B. fragilis in the presence and absence of oxygen. Moreover, intracellular expression of BS2 was also detected when cell culture monolayers of J774.1 macrophages were incubated with B. fragilis ahpC::bs2 or dps::bs2 strains within an anaerobic chamber. This suggests that ahpC and dps are induced following internalization by macrophages. Thus, we show that BS2 is a suitable tool for the detection of gene expression in obligate anaerobic bacteria in in vivo studies.
Introduction
The use of fluorescent proteins in biomedical research started over ten years ago (Chalfie et al., 1994). Since then, fluorescent proteins proved to be extremely useful as reporter tools in several cellular processes such as tracking protein movements in the cell, monitoring mitochondrial redox potential and transcriptional reporters (Wachter, 2006). In bacteria, GFPs can be used to survey microorganisms in complex biological systems such as biofilms, soil and to visualize interactions of bacteria with plants or animal host tissues (Rosochacki & Matejczyk, 2002; Larrainzar et al., 2005; Hoppe et al., 2009; Chudakov et al., 2010). Furthermore, GFPs can be transcriptionally and translationally fused to bacterial genes and expressed in vivo as an alternative to immunofluorescence. It also can be used to examine the function and localization of the gene products (Margolin, 2000). Currently, GFPs are a cornerstone tool used in in vivo imaging, fluorescence resonance energy transfer (FRET) and quantitative transcriptional analysis. Several GFP-like derivatives have been engineered for better fluorescence and photostability, (Heim et al., 1995) as well as different color emissions (Shaner et al., 2007). However, in the catalytic formation of the chromophore, GFP requires the presence of molecular oxygen (Heim et al., 1994), thus rendering the protein colorless in anaerobic environments, making GFP unsuitable for use as a reporter gene in obligate anaerobic organisms.
Recent efforts to create a protein reporter for in vivo labeling and fluorescence either in the presence or absence of oxygen led to development of flavin mononucleotide (FMN)-based fluorescent proteins (FbFPs) (Drepper et al., 2007, Drepper et al., 2010). Commercial FbFPs are derived from the blue-light photoreceptors YtvA from Bacillus subtilis and SB2 from Pseudomonas putida that contain the light oxygen voltage (LOV) domains. The LOV domains were first identified in plant phototrophins (Huala et al., 1997) where they regulate several physiological processes such as phototropism, chloroplast relocation and stomatal opening (Briggs and Christie, 2002; Celaya and Liscum, 2005). LOV domain proteins were subsequently found in prokaryotes where they seem to participate in two-component systems for light sensing though the function of these light sensing phenotypes is not yet fully understood for most species (Losi and Gartner, 2008). LOV domains bind noncovalently to the oxidized FMN chromophore and when exposed to blue light (450 nm) undergo a reversible photocycle that leads to the formation of FMN-cysteine C(4a) thiol adduct which exhibits weak autofluorescence (Salomon et al., 2000). The photoactive cysteine residue in a truncated gene expressing only the LOV domain of YtvA protein (Cys53) from B. subtilis was substituted with an alanine by site-directed mutagenesis and adjusted for E. coli codon usage bias (Drepper et al., 2007). The modified protein, known as BS2, has a 25-fold increase in fluorescence intensity when compared to wild type YtvA and exhibits a maximal light absorption at 449nm and maximal emission at 495nm (Drepper et al., 2007).
An important characteristic of FbFPs, including BS2, is that its fluorescence signal is not affected by the lack of oxygen (Drepper et al., 2007). This property makes BS2 a useful tool to study gene expression in obligate anaerobes in different environmental conditions because of its ability to yield fluorescence in both anaerobic and aerobic condition. In this study we have used promoterless BS2 as a reporter gene to evaluate promoter activity in the anaerobe Bacteroides fragilis as a model organism. B. fragilis is an opportunistic human pathogen normally found as a component of microflora of the human lower intestinal tract (Smith et al., 2003). One characteristic of this species is its high aerotolerance which allows it to survive in aerobic environments for long period of time (Rocha & Smith, 1999) and to survive host cellular immune defense in extra-intestinal oxygenated tissues such as the intra-abdominal cavity (Rocha et al., 2007; Sund et al., 2008). Thus, in this study we have analyzed the promoter activities of two characterized essential oxidative stress response genes under the control of the transcriptional regulator OxyR, the alkyl hydroperoxide reductase (ahpCF) and the nonspecific DNA binding protein (dps) (Rocha et al., 2000) transcriptionally fused to promoterless BS2 fluorescent protein as reporter gene. In addition, we also demonstrate the anaerobic expression of fluorescent BS2 under control of the maltose/starch inducible promoter osu. We show in this work that the fluorescent peptide BS2 is a useful tool to evaluate expression B. fragilis genes in both anaerobic and aerobic conditions as well as in macrophage cell line assays.
Materials and methods
Strains and growth conditions
The B. fragilis strains 638R (Privitera et al., 1979) and IB263 (Rocha and Smith, 1998) used in this study were routinely grown on BHIS (brain heart infusion supplemented with L-cysteine, hemin, and NaHCO3) at 37°C under anaerobic conditions. Rifamycin (20 μg/ml), 100 μg/ml gentamycin, and 10 μg/ml erythromycin were added to the media when required. The E. coli DH10B strain was grown routinely in Luria-Bertani medium (L-broth) containing 100 μg/ml ampicillin or 50 μg/ml spectinomycin when appropriate. For oxygen induction, overnight cultures in BHIS were diluted 1:25 in fresh media and grown to mid-log phase at OD550nm of 0.5. The cultures were split and one half remained under anaerobic conditions. The other half was exposed to air in an orbital shaker incubator (250 rpm) at 37° C for 1h. Chloramphenicol at 100 μg/ml was added immediately prior to harvesting bacterial cells by centrifugation. Maltose at 0.5% was added into anaerobic cultures when required.
Construction of transcriptional fusions to promoterless bs2 gene
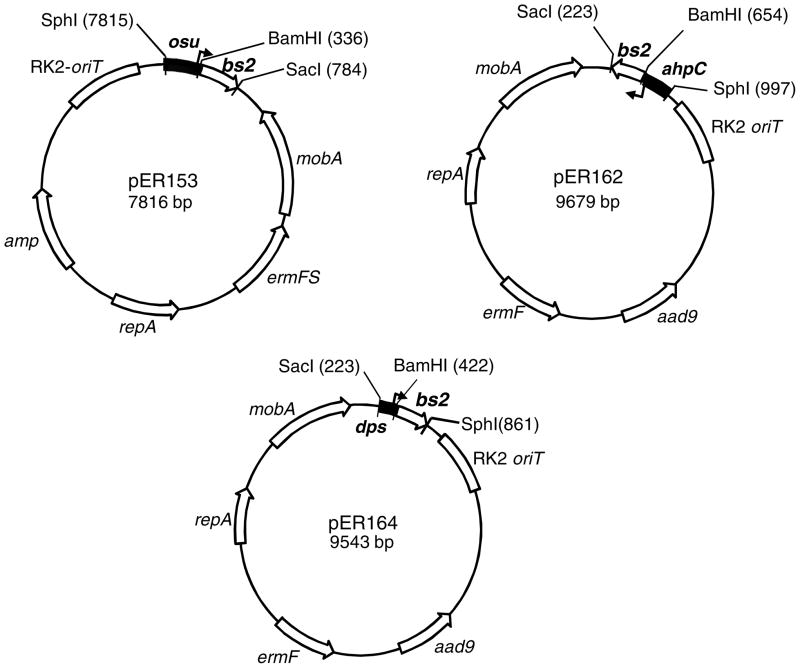
To clone the promoterless bs2 gene into a B. fragilis shuttle expression vector, the bs2 ORF (411 bp) from pGLOW-Bs2-stop (Evocatal GmbH, Dusseldorf, Germany) was PCR amplified using primers Bs2-BamHI-Forward (AAGAATGGATCCAAATAAGAAACAATTATG GCGTCGTTCCAGTCG) and Bs2-SstI-Reverse (GCCGAGCTCGCATGCCTGC). The oligo primer Bs2-BamHI-Forward was designed to place the ribosome binding site (RBS) of B. fragilis ahpC gene (Rocha and Smith, 1999) immediately upstream the bs2 ATG codon. This procedure was carried out to replace E. coli RBS region and insert a native RBS chromosomal region to optimize translation of bs2 gene in B. fragilis. The Bs2-SstI-Reverse primer was designed to contain the bs2 stop codon and restriction cloning sites from the original pGLOW-Bs2-stop plasmid except that HindIII site was replace with an SstI site. The PCR product containing a 462 bp DNA fragment was A-tailed and cloned into pGEM-T (Promega, Madisson, WI) according to manufacturer’s instructions to construct pER-151. pER-151 was digested with BamHI and SstI and the 447 bp DNA fragment containing the promoterless bs2 gene was cloned into the BamHI and SstI sites of pFD1045, a shuttle vector containing the maltose/starch and oxygen inducible promoter of the osu operon (Spence et al., 2006). This new construct, pER-153 (Fig 1), was conjugated into B. fragilis 638R by triparental mating according to standard protocols (Rocha et al., 1999). Transconjugants were selected on BHIS plates containing rifamycin (20 μg/ml), 100 μg/ml gentamycin, and 10 μg/ml erythromycin. The new strain, BER-85, was used for expression of BS2 under anaerobic conditions following addition of maltose. To construct the ahpC::bs2 transcriptional fusion, the 447 bp BamHI/SstI DNA fragment containing promoterless bs2 was cloned into the BamHI/SstI sites of pFD288 carrying 330 bp DNA fragment of the ahpC promoter region in the SphI/BamHI sites (Rocha and Smith, 1999). The new construct, pER162 (Fig 1) was conjugated into B, fragilis 638R and IB263 strains by triparental mating to construct BER-95 and BER-104 respectively. The dps::bs2 construct was obtained by cloning the 441 bp BamH/SphI promoterless bs2 gene into the BamHI/SphI sites of pUC19 containing 187 bp of the dps promoter region (Rocha et al., 2000). The new construct, pER 163, carrying the dps::bs2 transcriptional fusion was digested with SphI/SstI and cloned into the SphI/SstI sites of pFD288. The new construct, pER164 (Fig 1), was conjugated into B. fragilis 638R and IB263 strains by triparental mating to construct BER-96 and BER-105 respectively.
Figure 1.
Schematic representation of the shuttle vectors containing the transcription fusion constructs described in the materials and methods section. A partial restriction map relevant for the cloning strategy is shown in each plasmid. aad9: spectinomycin resistance in E. coli. ermF and ermFS: erythromycin resistance in Bacteroides. Broken arrows depict the transcription +1 nucleotide start site positioned at 54 bp, 39 bp and 62 bp upstream of the transcription fusion junction for osu, ahpC and dps promoter regions respectively.
Confocal Laser Microscopy
For the microscopy slides, 1 ml of bacterial cultures grown to mid-log phase in BHIS under conditions described above were centrifuged at 3000 × g for 3 min and washed once in 1 ml of phosphate buffer saline (PBS) [4.3 mM dibasic sodium phosphate, 1.47 mM monobasic potassium phosphate, 137 mM sodium chloride, 2.7 mM potassium chloride, pH 7.4]. Bacteria were suspended in 1 ml PBS and a drop of this suspension was added to each slide and allowed to air-dry. The coverslips were mounted with glycerol and the slides were analyzed with a Confocal Microscope Zeiss LSM 510 using an excitation of 450nm and an emission filter in the range of 475-525nm. For dual channel fluorescent color detection in slides stained with Alexafluor-546-phalloidin conjugate (Invitrogen, Carlsbad, CA) according to manufacturer’s instruction, an excitation at 556 nm and emission at 573 nm were also used.
Macrophage assay
The J774.1 macrophage cell lines was grown in Dulbecco’s modified Eagle’s medium [DMEM (Sigma-Aldrich, St. Louis, MO)] supplemented with 10% fetal bovine serum and 2 mM L-glutamine. The cells were grown over sterile coverslips placed inside of 6-well microplates at 37°C in a 5% CO2 humidified atmosphere. For all assays, 6-well plates were seeded with approximately 2 × 105 macrophages per mL and incubated until confluence was reached. The bacterial cell numbers were determined spectrophotometrically at 600 nm. The assay was carried out by inoculating B fragilis at approximate multiplicity of infection (MOI) of 100 into the 6-well plates under anaerobic conditions. Infected monolayers were incubated for 1h inside the anaerobic incubator to allow phagocytosis and internalization to occur. Then, the monolayers were washed three times with PBS without an antibiotic to remove unbound bacteria. The Cells were then fixed with 3.7% formaldehyde for 10 minutes and washed 3 times with PBS. Macrophages were stained with Alexafluor-546-phalloidin conjugate. The coverslips were removed from the wells and placed on top of glass slides for laser confocal microscopy analysis as previously described.
Results
In this study we show the use of the fluorescent protein BS2 as a reporter for in vitro and in vivo gene expression studies in the anaerobe B. fragilis. When the promoterless bs2 gene was cloned in fusion with the starch/maltose and oxygen inducible promoter osu (Spence et al., 2006), addition of maltose was able to induce expression of fluorescent BS2 under anaerobic conditions compared to uninduced culture controls. These results clearly demonstrate that expression of BS2 in cultures of B. fragilis BER-85 in the absence of oxygen yield intense fluorescence characteristic of BS2. In addition, exposure of BER-85 strain to atmospheric air also yielded intense fluorescence indicating that BS2 can be a useful fluorescent marker for gene expression in B. fragilis in both anaerobic and aerobic conditions (Fig. 2).
Figure 2.
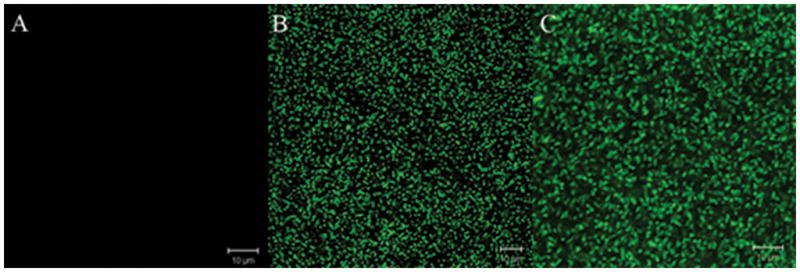
Confocal laser microscopy images of B. fragilis BER-85 (638R osu::bs2). (A) Anaerobic culture control. (B) Anaerobic culture after addition of Maltose for 1 h. (C) after exposure to oxygen for 1 h.
These findings prompted us to analyze the use of promoterless bs2 as a reporter gene by constructing bs2 transcriptional fusions to the oxygen and peroxide responsive promoters, ahpC and dps, which have been previously characterized in B. fragilis (Rocha et al., 2000). Both ahpC and dps expression are under control of the peroxide transcriptional regulator OxyR (Rocha et al., 2000). Thus, it seemed appropriate to investigate the expression of ahpC::bs2 and dps::bs2 constructs in response to oxygen and peroxide to further characterize expression of BS2 under both anaerobic and aerobic oxidative conditions. Fig. 3 shows that B. fragilis 638R carrying the ahpC::bs2 constructs (BER-95) were fluorescent compared to anaerobic culture control (Fig. 3A and Fig. 3B). When the constitutive peroxide response strain, IB263, was transformed with the ahpC::bs2 construct (BER-104), it produced fluorescence in both anaerobic and aerobic conditions (Fig. 3C and 3D). Similar findings were obtained when B. fragilis 638R carrying dps::bs2 (BER-96) was exposed to oxygen, it also showed increased fluorescence compared to the anaerobic culture control (Fig. 4A and 4B). In addition, the IB263 dps::bs2 strain (BER-105) also showed constitutive expression of BS2 independently of the presence or absence of oxygen confirming that the protein BS2 is a useful tool as a fluorescent image marker for gene expression in the anaerobe B. fragilis.
Figure 3.
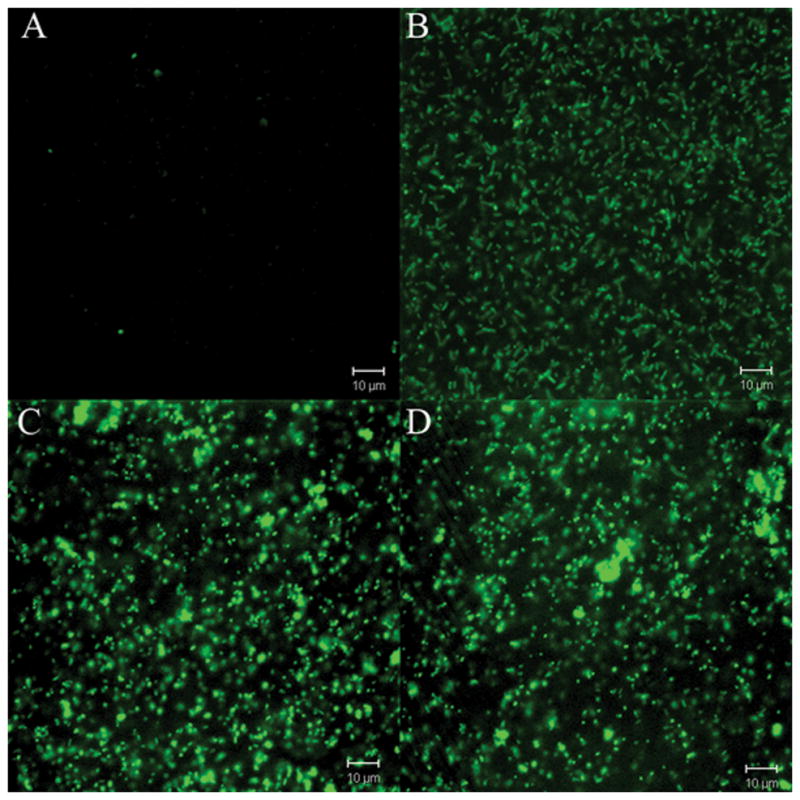
Confocal laser microscopy images of B. fragilis strains expressing FbFP BS2 fluorescent protein. Panels A and B) B. fragilis BER-95 (638R ahpC::bs2): (A) anaerobic culture control; (B) after exposure to oxygen for 1 h. Panels C and D) B. fragilis BER-104 (IB263 ahpC::bs2): (C) anaerobic culture control; (D) after exposure to oxygen for 1 h.
Figure 4.
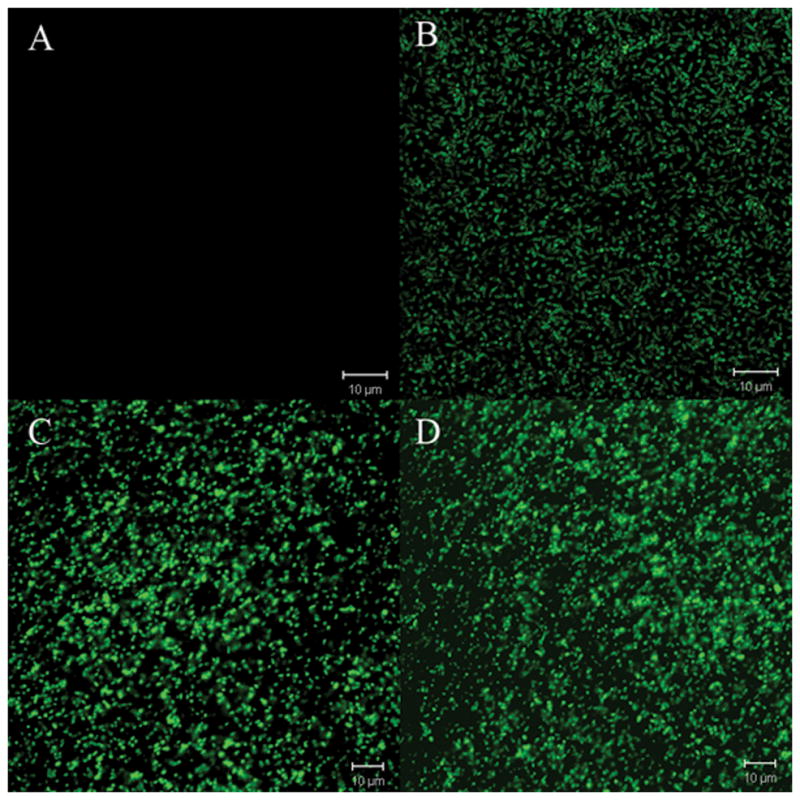
Confocal laser microscopy images of B. fragilis strains expressing FbFP BS2 fluorescent protein. Panels A and B) B. fragilis BER-96 (638R dps::bs2): (A) anaerobic culture control; (B) after exposure to oxygen for 1 h. Panels C and D): BER-105 (IB263 dps::bs2): (C) anaerobic culture control; (D) after exposure to oxygen for 1 h.
The oxidative stress response has been demonstrated to play an important role in the ability of the opportunistic intestinal colonizer B. fragilis to survive in intra-peritoneal experimental infections (Rocha et al., 2007: Sund et al., 2008). However, expression of oxidative response genes in vivo has not been extensively investigated in B. fragilis. Thus, to investigate whether, the ahpC and dps genes were induced following incubation with phagocytic cells, a J774.1 macrophage cell line assay in vitro was used to test if B. fragilis BER-95 and BER-96 express the peroxide response genes following cellular internalization by macrophages. In this study, we showed that expression of both ahpC (Fig. 5) and dps (Fig. 6) were visualized intracellularly as demonstrated by confocal laser microscopy showing the expression of BS2 fluorescent protein in internalized B. fragilis strains carrying ahpC::bs2 (BER-95) or dps::bs2 (BER-96) transcriptional fusion constructs. Using the Z-stack software function to analyze confocal laser microscopy image layers, we demonstrated that fluorescent B. fragilis cells were found to be in an intra-cellular compartment and not attached to the membrane surface of the macrophage cells. Thus, taken together these findings show for the first time that BS2 can be used as a fluorescent marker to analyze gene expression in obligate anaerobic bacteria in both in vitro and in vivo systems.
Figure 5.
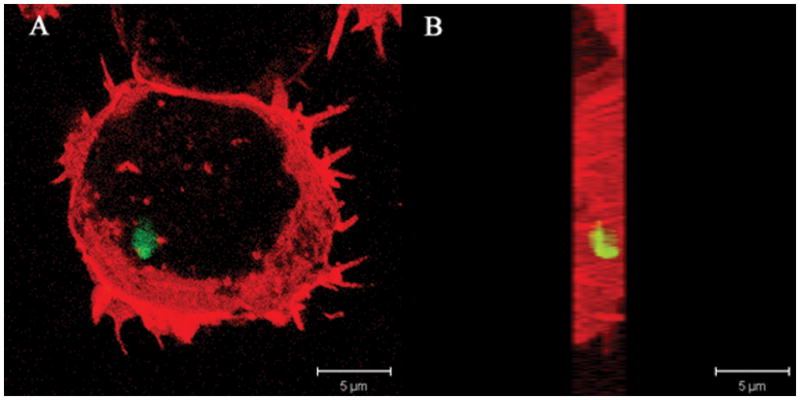
Confocal laser microscopy of intracellular expression of BS2 in B. fragilis 638R ahpC::bs2 (BER95) following internalization by murine macrophage cell line J774.1. A) Bacterial internalization was carried out for 1 h and visualized with laser confocal microscopy as describe in the material and methods section. (B) The Z-Stacks of the image layers obtained in panel A were piled up and rotated 90° angle.
Figure 6.
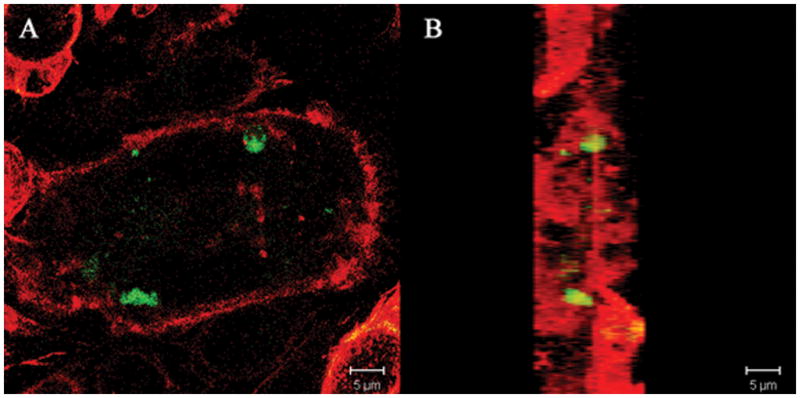
Confocal laser microscopy of intracellular expression of BS2 in B. fragilis 638R dps::bs2 (BER-96) following internalization by murine macrophage cell line J774.1. (A) Bacterial internalization was carried out for 1 h and visualized with laser confocal microscopy as describe in the material and methods section. (B) The Z-Stacks of the image layers obtained in panel A were piled up and rotated 90° angle.
Discussion
In addition to advances in fluorescent proteins derived from GFP, a new class of fluorescent proteins has recently been isolated and proven useful in environments deprived from oxygen. Drepper et al. (2007) demonstrated that FbFPs expressed in the facultative anaerobe Rhodobacter capsulatum in hypoxia was fully fluorescent. More recently, Drepper et al. (2010) have quantitatively monitored the fluorescent intensity of FpFP in vivo in E. coli under oxygen limitations by continuously measuring wavelength excitation at 460 nm and emission at 492 nm. A different study showed that FbFPs expressed in Candida albicans and Saccharomyces cerevisiae under anaerobic conditions render the cells fluorescent (Tielker et al. 2009). In this work we demonstrate that FbFPs expressed in the obligate anaerobe B. fragilis confer fluorescence to the cells when grown under anaerobic conditions. In the absence of oxygen, B. fragilis cells were remarkably fluorescent (Fig 2) presenting an emission in the range of 475-505nm when excited with light at 450nm in agreement with previous works using FbFPs as fluorescent marker (Drepper et al., 2007; Tielker et al., 2009).
Though GFP protein derivatives have been engineered to increase photostability, intensity, broad pH range tolerance, faster maturation rates and different colors which allow researchers to use multiple probes within the same image experimental set (Shaner et al., 2007), they are still dependent on the molecular oxygen to display their fluorescence. This requirement for oxygen for proper post-translational modification of the protein fluorophore, is a significant limitation to their use in anaerobic environments. Thus, our findings are important for the study of anaerobic bacteria as there is a lack of imaging tools to study molecular trafficking and gene expression in these organisms which require anaerobic conditions during their growth and metabolism during both in vitro and in vivo conditions. This is particular relevant with regard to B. fragilis because it will allow us to investigate this opportunistic anaerobic human pathogen under low or limited oxygen conditions similar to the ones that occurs during anaerobic infection in human tissues.
In this regard, as a first step to understand gene expression in B. fragilis during infection, we demonstrate in this study that the ahpC and dps genes are expressed following incubation with a cell line macrophage. These findings indicate that B. fragilis cells were internalized by macrophages and that its intra-cellular environment induced B. fragilis oxidative stress response as demonstrated by the up-regulation of the ahpC::bs2 and dps::bs2 transcription fusion. In animal models of intra-peritoneal infection, the B. fragilis oxidative stress response is required for survival (Sund et al., 2008). This is likely due to the fact that oxyR and dps mutant strains which are deficient in the regulation of the peroxide stress response and protection against oxidative DNA-damage are unable to survive in the peritoneal cavity. Our findings correlate with the fact that intra-cellular up-regulation of ahpC and dps expression may be an important defense for survival against phagocytic cells of the immune system. Though oxidative killing mechanisms of the phagocytic cells may contribute to B. fragilis oxidative stress response in vivo, the current study cannot rule out that other intra-cellular environmental conditions may also affect expression of ahpC and dps. One such factor is the depletion of iron availability in the phagolysosome compartment. Induction of ahpC and dps expression is also affected by iron limitation in vitro (data not shown) which could account for additional regulation of B. fragilis genes following internalization by phagocytic cells.
In conclusion, these results indicate that the FbFPs are suitable to be used as transcriptional fusion reporters in obligate anaerobic bacteria. There seems to be a great potential for FbFPs in the analysis of gene expression in vivo in anaerobic environments such as the human colon as well as in extra-intestinal infections when used in combination with modern in vivo imaging techniques.
Acknowledgments
This work was supported in part by NIH/NIAID research grants AI068659 and AI079183 to E.R.R. and grant AI40588 to C.J.S. Thanks to T. Whitehead for helpful discussions.
References
- Briggs WR, Christie JM. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sc. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- Celaya RB, Liscum E. Phototropins and associated signaling: providing the power of movement in higher plants. Photochem Photobiol. 2005;81:73–80. doi: 10.1562/2004-08-22-IR-282. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK, Wendorff M, Losi A, Gärtner W, Jaeger KE. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25:443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- Drepper T, Huber R, Heck A, Circolone F, Hillmer AK, Büchs J, Jaeger KE. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl Environ Microbiol. 2010;76 :5990–5994. doi: 10.1128/AEM.00701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hoppe AD, Seveau S, Swanson JA. Live cell fluorescence microscopy to study microbial pathogenesis. Cell Microbiol. 2009;11:540–550. doi: 10.1111/j.1462-5822.2009.01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Larrainzar E, O'Gara F, Morrissey JP. Applications of autofluorescent proteins for in situ studies in microbial ecology. Annu Rev Microbiol. 2005;59:257–277. doi: 10.1146/annurev.micro.59.030804.121350. [DOI] [PubMed] [Google Scholar]
- Losi A, Gärtner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- Margolin W. Green fluorescent protein as a reporter for macromolecular localization in bacterial cells. Methods. 2000;20:62–72. doi: 10.1006/meth.1999.0906. [DOI] [PubMed] [Google Scholar]
- Privitera G, Dublanchet A, Sebald M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis. 1979;139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Characterization of a peroxide-resistant mutant of the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1998;180:5906–5912. doi: 10.1128/jb.180.22.5906-5912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J Bacteriol. 1999;181:5701–5710. doi: 10.1128/jb.181.18.5701-5710.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Owens G, Jr, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/jb.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Tzianabos AO, Smith CJ. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J Bacteriol. 2007;189:8015–8023. doi: 10.1128/JB.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosochacki SJ, Matejczyk M. Green fluorescent protein as a molecular marker in microbiology. Acta Microbiol Pol. 2002;51:205–216. [PubMed] [Google Scholar]
- Salomon MJ, Christie M, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Rocha ER, Paster BJ. The medically important Bacteroides spp. in health and disease. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3. Springer-Verlag; New York: 2003. pp. 381–427. release 3.7. http://link.springer-ny.com/link/service/books/10125/.NY. [Google Scholar]
- Spence C, Wells WG, Smith CJ. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen. J Bacteriol. 2006;188:4663–4672. doi: 10.1128/JB.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, O'Rourke DP, Smith CJ. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–42. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Tielker D, Eichhof I, Jaeger KE, Ernst JF. Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryotic Cell. 2009;8:913–915. doi: 10.1128/EC.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter RM. The family of GFP-like proteins: structure, function, photophysics and biosensor applications. Introduction and perspective. Photochem Photobiol. 2006;82:339–344. doi: 10.1562/2005-10-02-IR-708. [DOI] [PubMed] [Google Scholar]