Abstract
We report that the dominant human missense mutations G303E and G296S in GATA4, a cardiac-specific transcription factor gene, cause atrioventricular septal defects and valve abnormalities by disrupting a signaling cascade involved in endocardial cushion development. These GATA4 missense mutations, but not a mutation causing secundum atrial septal defects (S52F), demonstrated impaired protein interactions with SMAD4, a transcription factor required for canonical bone morphogenetic protein/transforming growth factor-β (BMP/TGF-β) signaling. Gata4 and Smad4 genetically interact in vivo: atrioventricular septal defects result from endothelial-specific Gata4 and Smad4 compound haploinsufficiency. Endothelial-specific knockout of Smad4 caused an absence of valve-forming activity: Smad4-deficient endocardium was associated with acellular endocardial cushions, absent epithelial-to-mesenchymal transformation, reduced endocardial proliferation, and loss of Id2 expression in valve-forming regions. We show that Gata4 and Smad4 cooperatively activated the Id2 promoter, that human GATA4 mutations abrogated this activity, and that Id2 deficiency in mice could cause atrioventricular septal defects. We suggest that one determinant of the phenotypic spectrum caused by human GATA4 mutations is differential effects on GATA4/SMAD4 interactions required for endocardial cushion development.
The atrioventricular canal, the valve-forming region between the atria and ventricles, adopts a unique molecular and morphological program during cardiac development that requires coordinated activity of myocardial and endocardial lineages. The cardiac valve anlage is the endocardial cushion, a swelling composed of myocardial-derived extracellular matrix that becomes populated primarily by endocardial-derived mesenchymal cells (1). The development of a mature valve structure from the endocardial cushion requires multiple distinct steps (2), including activation and proliferation of endocardial cells, endothelial-to-mesenchymal transformation (EMT) of activated endocardial cells, and maturation of the cellularized cushions into functional valve leaflets.
Development of the atrioventricular endocardial cushions into the atrioventricular valves (tricuspid and mitral) and adjacent portions of the atrial and ventricular septae requires Tgf-β and Bmp signaling. At least eight Tgf-β or Bmp ligands are expressed during valve formation (3, 4), and gene ablation of individual ligands in model organisms produces phenotypes that range from pronounced valvuloseptal malformations to subtle valve maturation defects (5–18). Compound gene ablations of Bmp5/Bmp7 or Bmp6/Bmp7 produced more severe endocardial cushion defects than either single mutant did (16–18). These observations imply considerable redundancy of Tgf-β/Bmp signaling in the early functions of this pathway in endocardial cushion development.
Human atrioventricular septal defects (AVSDs) are defined by variable abnormalities of the mitral and/or tricuspid valves in conjunction with defects in the adjacent atrial or ventricular septae (19). The abnormalities produce substantial hemodynamic consequences that contribute to poor prognosis in affected patients and a virtually uniform requirement for surgical correction. Human genetic studies have demonstrated that mutations in the gene encoding cardiac transcription factor GATA4 cause familial atrial secundum defects and, less commonly, sporadic AVSDs (20–23).
We evaluated two families with dominant inheritance of AVSDs and identified two GATA4 missense mutations, G303E and G296S. We investigated the canonical TGF-β/BMP pathway effector SMAD4 and showed that G303E and G296S altered the transcriptional response to TGF-β/BMP activation. Deletion of Smad4 from the endocardium of mice caused severe maldevelopment of endocardial cushions and diminished expression of the Id2 gene. Together these studies define a transcriptional network directing endocardial cushion formation involving Gata4, Smad4, and Id2.
Results
Human Mutations in GATA4 Cause Endocardial Cushion Defects.
From a cohort of 103 probands with congenital heart defects, we sequenced all protein-encoding exons of candidate genes, including GATA4. Two GATA4 mutations were identified in probands from unrelated families (Fig. 1) with dominantly inherited AVSDs.
Fig. 1.
GATA4 mutations cause endocardial cushion defects. (A) Family A pedigree with cosegregation of GATA4 G303E mutation (+) with congenital heart disease (Left). Note that five mutation carriers had endocardial cushion defects (Right). (B) Family B pedigree with cosegregation of GATA4 G296S mutation (+) with congenital heart disease (Left). Note that six mutation carriers had endocardial cushion defects (Right). Electrophysiologic (EP) status and events of mutation carriers are indicated: AF, atrial fibrillation; EAR, ectopic atrial rhythm; LAD, left axis deviation, PM, pacemaker; SCD, sudden cardiac death; SR, sinus rhythm, SVT, supraventricular tachyarrhythmia.
Family A had six members with a spectrum of AVSDs (Fig. 1A). Sequence analyses of the proband revealed a heterozygous G-to-A substitution at nucleotide 4 in exon 4 of GATA4, which was also present in all affected Family A members (Fig. 1A), but absent from over 500 ethnically matched controls. This variant encodes replacement of a conserved glycine with glutamate at amino acid position 303 (denoted G303E), just distal to the carboxyl-terminal zinc finger of the transcription factor. We concluded that GATA4 G303E caused AVSDs in Family A.
Family B had five individuals with AVSDs and four individuals with electrophysiologic abnormalities (Fig. 1B). Sequence analyses of the proband revealed a heterozygous G-to-A transition at nucleotide 886 of GATA4, which was also present in all affected members of Family B (Fig. 1B), but absent from over 500 ethnically matched controls. This variant is predicted to substitute a conserved glycine with serine at amino acid position 296 (denoted G296S), located directly adjacent to the nuclear localization signal and carboxyl-terminal zinc finger. GATA4 G296S has been previously identified to cause atrial septal defects in three families (21, 24).
GATA4 Mutations Affect GATA4–SMAD4 Interactions.
Previous work demonstrated binding of the N-terminal portion of Smad4 to the second zinc-finger domain of Gata4 (25). This region of Gata4 (amino acids 248–335) encompasses residues 296 and 303, mutated in families A and B, respectively. Given the roles for Tgf-β/Bmp signaling in atrioventricular development, we asked whether these two GATA4 human mutations altered GATA4–SMAD4 interactions. Protein–protein interactions were analyzed by pulling down 35S-labeled GATA4 protein variants with wild-type SMAD4 (Fig. 2). SMAD4 efficiently precipitated wild-type GATA4. However, SMAD4 interactions with specific GATA4 mutants were greatly reduced. SMAD4 bound GATA4 G296S with 20% efficiency (P = 0.01) and GATA4 G303E with 30% efficiency compared with wild type (P = 0.002) (Fig. 2A). In contrast, SMAD4 bound GATA4 S52F, a previously reported GATA4 mutation that caused dominant secundum atrial septal defects but neither ASVDs nor valve malformations (21), with efficiency comparable to wild-type GATA4 (P = 0.15) (Fig. 2A). We concluded that the variable clinical consequences of GATA4 mutations reflected differences in GATA4–SMAD4 interactions.
Fig. 2.
GATA4 mutations cause disrupted GATA4–SMAD4 interactions. (A) Autoradiograms of GST-pulldown assays showed binding of wild-type and mutant GATA4 (G296S, G303E, and S52F) to SMAD4. Twenty percent of the in vitro-translated 35S methinonine-labeled mouse GATA4 protein used for binding assay (input) and the remaining GATA4 protein after precipitation by GST-SMAD4 are shown. (B) Quantification of the average of five independent GST pull-down assays. GATA4 protein pulled down was normalized to the relative input. GATA4 G296S and G303E diminished the GATA4–SMAD4 interaction (*P = 0.01; **P = 0.002), whereas the GATA4 mutation S52F did not (P = 0.15). (C–F) Endocardial haploinsufficiency of Gata4 and Smad4 causes atrioventricular canal defects. Transverse sections of Gata4fl/+; Smad4fl/+ (C), Tie2:Cre+; Gata4fl/+ (D), Tie2:Cre+; Smad4fl/+ (E), and Tie2:Cre+; Gata4fl/+; Smad4fl/+ (F) E14.5 hearts. (Magnification in C–F: 40×; G–J: 100×.) Atrioventricular septal defects [in F, boxed area (Upper) and asterisk (Lower)] can be seen in the double heterozygous Tie2:Cre+; Gata4fl/+; Smad4fl/+ heart. In contrast, Tie2:Cre+; Gata4fl/+ or Tie2:Cre+; Smad4fl/+ heterozygous animals show normal atrioventricular canal morphology (C–E).
Smad4 and Gata4 Genetically Interact in the Endocardium.
We tested whether Smad4 and Gata4 interact genetically in the context of endocardial cushion development in vivo. We used Tie2:Cre (26) to generate endocardial-specific knockouts using floxed conditional alleles of Smad4 (Smad4fl) (27) and Gata4 (Gata4fl) (28). Atrioventricular canal morphology of mice heterozygous for endocardial Gata4, Smad4, or both was analyzed at embryonic day 14.5 (E14.5). Animals heterozygous for either Gata4 (Tie2:Cre+; Gata4fl/+) (5/5) or Smad4 (Tie2:Cre+; Smad4fl/+) (5/5) in the endocardium were viable and displayed normal atrioventricular canal morphology in each case (Fig. 2 D and E). In contrast, double heterozygous Gata4; Smad4 embryos (Tie2:Cre+; Gata4fl/+; Smad4fl/+) displayed severe atrioventricular defects (5/5) and died by E12.5 (Fig. 2F). From these results we deduced that Gata4 and Smad4 genetically cooperate in the endocardium during atrioventricular valve formation.
Endocardial Smad4 Is Required for Endocardial Cushion Development.
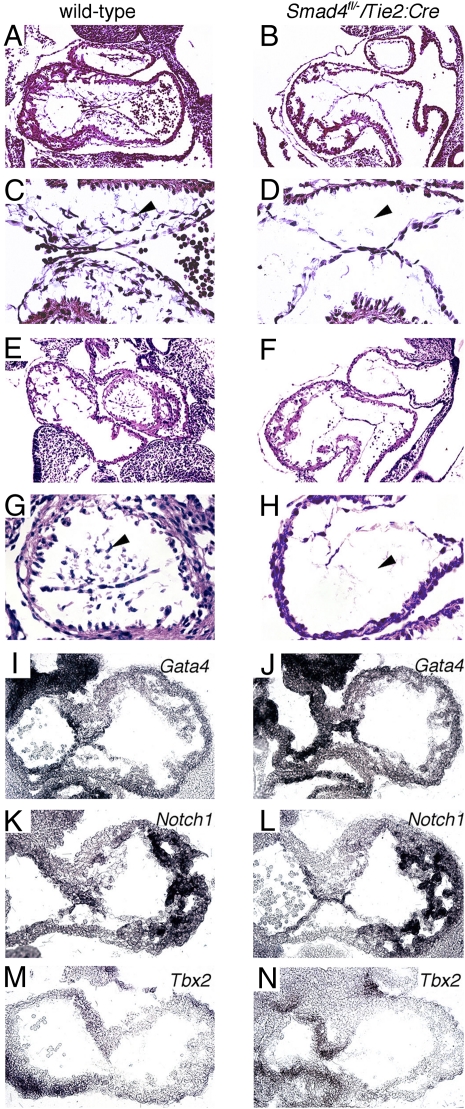
To further explore the consequences of endocardial Smad4 deficiency on atrioventricular valve formation, we generated endocardial Smad4 knockout embryos (Tie2:Cre+; Smad4fl/−). The developing atrioventricular endocardial cushions of Tie2:Cre+; Smad4fl/− mutant embryos were entirely acellular at E10.5 (Fig. 3 B and D). In contrast, the developing atrioventricular endocardial cushions of wild-type embryos were densely populated with mesenchymal cells (Fig. 3 A and C). The outflow tract cushions of Tie2:Cre+; Smad4fl/− embryos were also acellular (Fig. 3 F and H), in contrast to the well-populated outflow tract cushions of wild-type embryos at E10.5 (Fig. 3 E and G). The endocardial cushion structures of the atrioventricular canal and outflow tract were of normal size in both wild-type and Tie2:Cre+; Smad4fl/− embryos (Fig. 3 A–H). Furthermore, the myocardium of the atrioventricular canal and the cardiac chambers appeared morphologically normal in both wild-type and Tie2:Cre+; Smad4fl/− embryos (Fig. 3 A–H). We concluded that endocardial expression of Smad4 is required to promote normal cellularity in the developing endocardial cushion.
Fig. 3.
Endocardial loss of Smad4 causes acellular endocardial cushions. Parasagital heart sections showing the atrioventricular canal (A–D) and outflow tract (E–H) endocardial cushions from wild-type (A, C, E, and G) and Tie2:Cre+; Smad4fl/− (B, D, F, and H) embryos at E10.5. Tie2:Cre+; Smad4fl/− atrioventricular and outflow endocardial cushions are acellular (black arrowheads in D and H) compared with wild type (black arrowheads in C and G). In situ hybridization of sections from wild-type (A, C, E, G, I, and K) and Tie2:Cre+; Smad4fl/− (B, D, F, H, J, and L) embryos at E10.5. The patterns of expression for Gata4 (I and J), Notch1 (K and L), and Tbx2 (M and N) were comparable in wild-type and Tie2:Cre+; Smad4fl/− mutant embryos. (Magnification in A, B, E, F, I, J, K, L, M, and N: 40×; C, D, G, and H: 100×.)
We asked whether loss of Smad4 from the atrioventricular endocardium altered the expression of molecular regulators implicated in valve morphogenesis, which included Gata4, Notch1, and Tbx2 (34, 40, 43–45). In situ hybridization of atrioventricular canal sections from wild-type and Tie2:Cre+; Smad4fl/− embryos showed comparable pattern and intensity of expression for Gata4 (Fig. 3 I vs. J), Notch1 (Fig. 3 K vs. L), and Tbx2 (Fig. 3 M vs. N) at E10.5.
Smad4 Is Necessary for Endocardial Cushion EMT.
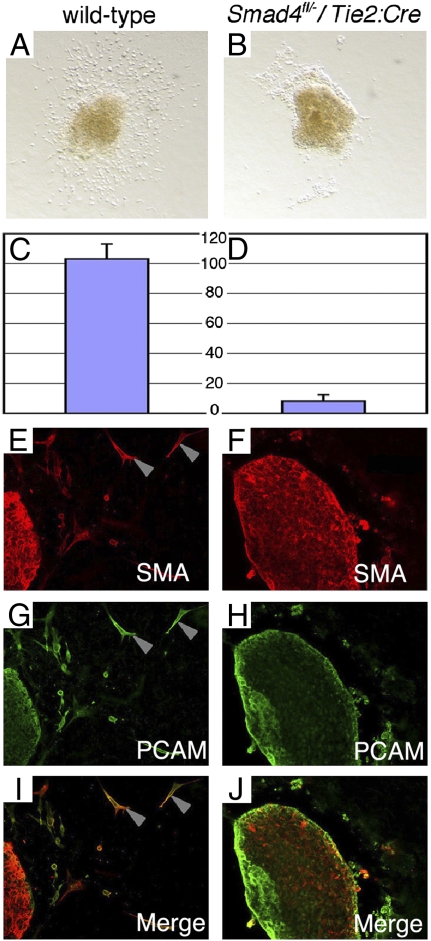
We next studied the transformation of endothelial cells to mesenchymal cells (EMT) in the endocardial cushions during valve morphogenesis using a collagen gel culture system (29). Wild-type endocardial cells from atrioventricular canal explants became motile and invaded the collagen matrix (Fig. 4A). Tie2:Cre+; Smad4fl/−endocardial cells from atrioventricular canal explants showed markedly reduced migration and invasion (Fig. 4B). The numbers of mutant cells entering the collagen matrix (8.2 ± 4.4) were markedly less than in wild-type atrioventricular explants (102.8 ± 10.2; P = 3.8 × 10−7) (Fig. 4 C vs. D).
Fig. 4.
Endocardial Smad4 is required for EMT in the endocardial cushion. (A–J) Atrioventricular cushion explants from wild-type (A, C, E, G, and I) and Tie2:Cre+; Smad4fl/− (B, D, F, H, and J) embryos. Mesenchymal cells were abundant in wild-type explants (A) but rare in mutant explants (B). Average numbers of invasive mesenchymal cells in wild-type explants (C: 102.8 ± 10.2 cells) was significantly greater (P = 3.8 × 10−7) than in mutant explants (D: 8.2 ± 4.4)(n = 5 each). Mesenchymal cells from control explants expressed Pecam (G and I: gray arrowheads), consistent with endocardial origin, and Sma (E and I; gray arrowheads), indicative of a mesenchymal fate. Cells from mutant explants did not express Sma (F and J).
We further characterized EMT in mutant and wild-type explants using immunohistochemistry. Endocardial cells express the surface cell adhesion protein Pecam, whereas mesenchymal cells express smooth muscle actin (Sma). Coexpression of both Pecam and Sma in endocardial cells is indicative of EMT. Cells surrounding the atrioventricular explants from both wild-type and Tie2:Cre+; Smad4fl/− embryos demonstrated Pecam expression, consistent with an intact endothelium in vivo (Fig. 4 G and H). Whereas abundant Sma-positive cells were found in wild-type atrioventricular canal explants (Fig. 4E), few if any Sma-positive cells were found in explants from Tie2:Cre+; Smad4fl/− embryos (Fig. 4F). Cells that coexpressed Sma and Pecam were abundant in the wild-type explants (Fig. 4I) but absent from Tie2:Cre+; Smad4fl/− explants (Fig. 4J). This result demonstrated failed EMT in Smad4-deficient endocardial cells.
Quiescent Endocardial Cushion Endocardium in Smad4 Mutant Embryos.
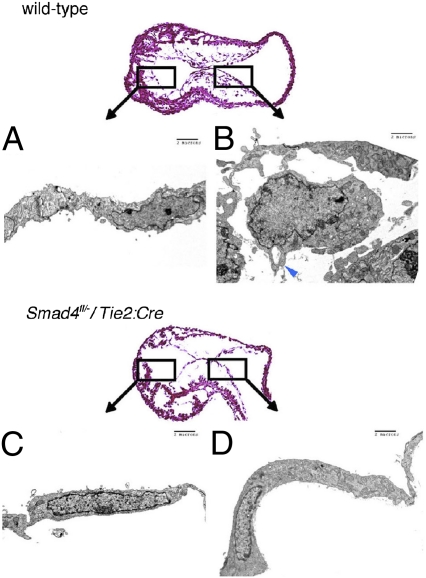
To assess for early morphological correlates of endocardial cell activation and EMT, endocardial Smad4-deficient and wild-type endocardial cells were evaluated by transmission electron microscopy. At E9.5, many wild-type endocardial cells were rounded, demonstrated filopodia and pseudopodia, and had lost cell–cell contacts (tight junctions) (Fig. 5B). In contrast, Tie2:Cre+; Smad4fl/− endocardial cells showed intact tight junctions without filopodia or pseudopodia (Fig. 5D). The morphology of Tie2:Cre+; Smad4fl/− endocardial cells was similar to the ventricular chamber endocardial cells from wild-type embryos (Fig. 5 D vs. A), which do not undergo EMT or participate in valve formation.
Fig. 5.
Quiescent endocardium in endocardial-deficient Smad4 embryos. Transmission electron microscopy of E9.5 wild-type (A and B) or Tie2:Cre+; Smad4fl/− (C and D) endocardium. Wild-type atrioventricular canal endocardial cells (B) were rounded with pseudopodia and filopodia (B: blue arrowhead). Tie2:Cre+; Smad4fl/− atrioventricular canal endocardial cells (D) demonstrated a thin cellular profile, maintenance of cellular junctions, and absence of pseudopodia and filipodia, similar to wild-type unactivated ventricular endocardium (A and C).
Smad4 Is Required for Proliferation of Endocardial Cushion Endocardium.
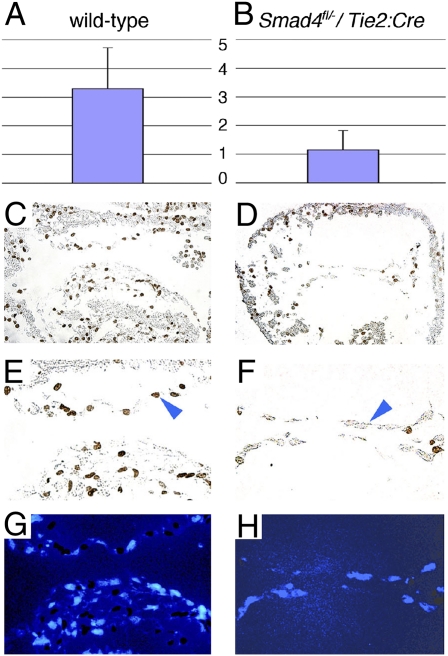
The quiescent cellular morphology of endocardial cells in Tie2:Cre+; Smad4fl/− embryos implied an early defect in the EMT sequence. To determine whether Smad4 was required for endocardial proliferation, we labeled embryos with BrdU at E9.5 and stained surface endocardium with anti-BrdU antibodies at E10.5 to identify proliferating cells. Actively proliferating cells at the surface of the endocardium were counted from four sections per embryo; proliferating cells undergoing EMT, present only in wild-type embryos, were specifically excluded. Surface atrioventricular canal endocardial cells of wild-type embryos had significantly more BrdU-labeled nuclei (3.24 ± 1.4; Fig. 6 A, C, and E) than those of Tie2:Cre+; Smad4fl/− embryos (1.04 ± 0.7, P = 1.8 × 10−5; Fig. 6 B, D, and F). We concluded that endocardial expression of Smad4 is required to promote proliferation during the activation process of the pre-EMT endocardial cell.
Fig. 6.
Endocardial proliferation defect in endocardial-deficient Smad4 embryos. (A and B) Significantly more surface endocardial cell proliferation in wild type (A: 3.24 ± 1.4) than in Tie2:Cre+; Smad4fl/− (B: 1.04 ± 0.7) embryos at E10.5 (P = 1.8 × 10−5). (C–H) Parasagital heart sections from wild-type (C, E, and G) and Tie2:Cre+; Smad4fl/− (D, F, and H) embryos treated with BrdU at E9.5 and stained for BrdU uptake (C–F) or DAPI (G and H) at E10.5.
Id2 Is Required for Atrioventricular Valve Morphogenesis.
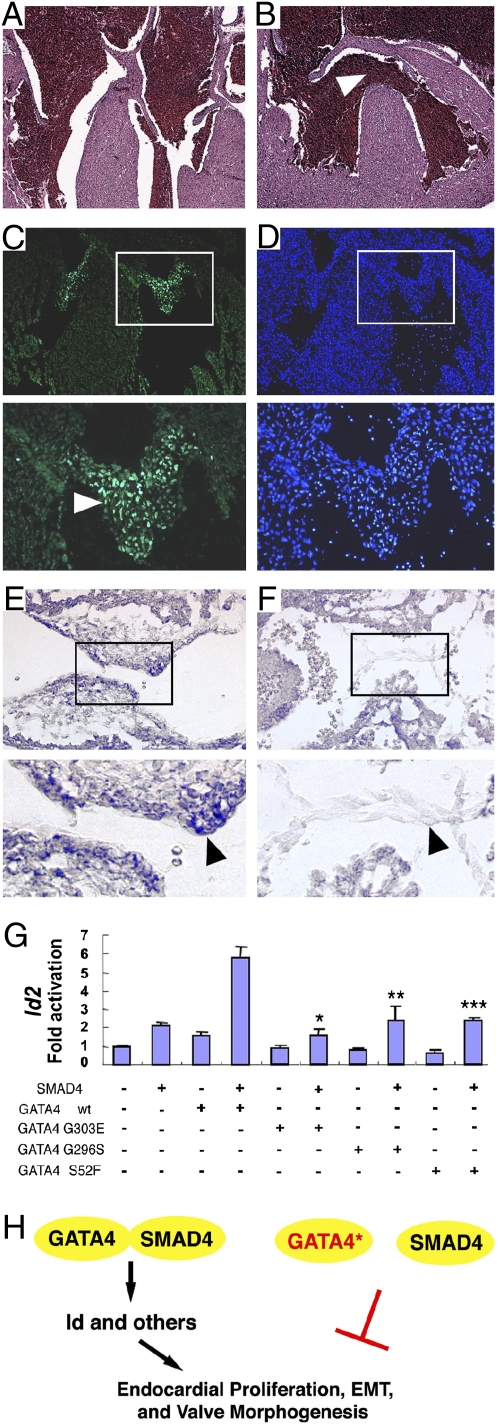
Id2, a member of a gene family encoding helix–loop–helix-containing transcriptional repressors (Id1–Id4), is a target of Tgf-β/Bmp signaling (30) and has been implicated in endocardial cushion development (31). Although no structural cardiovascular phenotype has been reported in Id2 knockout mice, we observed over 20% perinatal lethality of Id2−/− mutant newborn mice in the C57BL/6 genetic background (8 newborns with perinatal lethality among 36 sequential live-born Id2−/− pups). We found AVSDs and membranous ventricular septal defects in each Id2−/− perinatal lethal (n = 5) (Fig. 7B). In contrast, normal cardiac morphology with intact atrioventricular septation was observed in littermate-surviving Id2−/− mice (n = 8; P = 0.02; Fig. 7A) and in wild-type littermate controls (n = 12; P = 0.01). Immunohistochemistry at E14.5 demonstrated Id2 protein expression in the developing endocardial cushions, but not in the myocardium (Fig. 7C). Id2 expression was also detected by in situ hybridization in surface endocardium of the developing endocardial cushions at E10.5 (Fig. 7E). In contrast, expression of Id2 was substantially reduced in Tie2:Cre+; Smad4fl/− embryos on the surface endocardium of the atrioventricular canal (Fig. 7F).
Fig. 7.
Id2 is a target of Smad4 and Gata4 required for atrioventricular septation. (A and B) Id2−/− mice with perinatal lethality had atrioventricular septal defects (B: membranous ventricular septal defects) in contrast to surviving Id2−/− mice (A: normal atrioventricular septation). (Magnification: 40×.) (C and D) Id2 immunohistochemistry demonstrated expression in the endocardial cushion endocardium and mesenchyme (C: white arrowhead) at E14.5. (Right panels in C and D) DAPI staining. (Magnification in C and D: Upper panels—40×; Lower panels—100×.) (E and F) Id2 expression is decreased in Tie2:Cre+; Smad4fl/− (F: black arrowhead) compared with wild-type endocardium (E: black arrowhead) at E10.5. (Magnification in E and F: Upper panels—40×; Lower panels—100×. (G) Cooperative activation of a 1052-bp fragment of the Id2 promoter by Smad4 and Gata4. Luciferase constructs were transfected alone or with Smad4 or Gata4 expression vectors, singly and in combination. Mutant Gata4 constructs abrogated cooperative luciferase activity of Smad4 and Gata4. Compared with wild-type Smad4-Gata4: *Smad4-Gata4G303E, P = 0.001; **Smad4-Gata4G296S, P = 0.02; ***Smad4-Gata4S52F, P = 0.002. (H) GATA4–SMAD4 interaction is required for Id2 expression and cardiac valve development. Congenital heart disease-causing GATA4 mutations abrogate interaction with SMAD4, resulting in failed valve development.
Smad4 and Gata4 Cooperatively Activate the Id2 Promoter.
We evaluated transcriptional regulation of Id2 by Gata4 and Smad4. We assessed expression of the firefly luciferase reporter gene under control of a 1052-bp fragment from the Id2 promoter (from nucleotide +1584 to +532) containing three putative Smad4-binding sites (nucleotide positions 630–633, 977–980, and 1133–1136) and a putative Gata4-binding site (nucleotide positions 838–841). Compared with empty vector, transfection of CV-1 cells with either wild-type Smad4 or wild-type Gata4 expression constructs produced twofold activation of the Id2 promoter fragment (Fig. 7G). In contrast, cotransfection with both Smad4 and Gata4 produced sixfold cooperative transcriptional activation of the Id2 promoter fragment (Fig. 7G). Cotransfection with Smad4 and mutant Gata4 eliminated the cooperative activation (compared with wild-type Gata4-Smad4: Gata4G303E-Smad4, P = 0.001; Gata4G296S-Smad4, P = 0.02; Gata4S52F-Smad4, P = 0.002). These findings demonstrate that Gata4 mutations Gata4 G296S, Gata4 G303E, or GataS52F cause a loss of Smad4-dependent cooperative activation of the Id2 promoter.
Discussion
Role of Smad4 in Atrioventricular Canal Development.
Endocardial cushion development requires a series of events within endothelial cells to enable EMT, including proliferation and the acquisition of morphological and molecular features of mesenchymal cells (1). Removal of Smad4 from the endocardium resulted in defects in each aspect of the endocardial activation sequence required for EMT. Smad4-deficient endocardial cells failed to proliferate (Fig. 6) or undergo EMT (Fig. 4), retained morphological features of unactivated ventricular endocardium (Fig. 5), and showed decreased Id2 gene expression (Fig. 7). Furthermore, loss of Id2 alone was sufficient to cause AVSDs in some cases (Fig. 7). Gata4 and Smad4 cooperatively activated the Id2 promoter in vitro, and Gata4 mutations abrogated this cooperated activation (Fig. 7). Collectively, these data define a signaling network comprising endocardial Gata4, Smad4, and Id2 that regulates cardiac cushion development and is disrupted by human GATA4 mutations.
GATA4 and Human Congenital Heart Disease.
Gata4 associates with a variety of other transcriptional regulators, including Smad4, Nkx2-5, Tbx5, and Fog2 (21, 25, 32), implying that lineage-specific activity may be contextual and achieved through formation of specific combinatorial transcriptional complexes. Our data indicating that Smad4-dependent signaling in the endocardium was modulated by Gata4–Smad4 interactions supports this model. Smad4 may preferentially recruit Gata4 to endocardial enhancers, given its selective requirement in this lineage during atrioventricular canal development, and the differential clinical expression of GATA4 mutations provides a biological context for the importance of Gata4 and Smad4 as a transcriptional complex. GATA4 mutations produce an array of cardiovascular malformations that range from secundum ASDs (21, 33) to profound defects in endocardial cushion derivatives, including complete atrioventricular canal and electrophysiologic abnormalities (Fig. 1). Our data suggest that the lineage-specific impact of mutations on GATA4 function contributes to phenotype heterogeneity.
A role for Gata4 in transcriptional control of the conduction system is emerging (34). Given the electrophysiologic abnormalities observed in patients with GATA4 mutations G303E and G296S but not with other reported GATA4 mutations (24), we hypothesize that there are lineage-specific effects by Gata4 in conducting myocytes. Further studies will reveal whether Gata4 interacts with Tbx5 and Nkx2-5 (33) to regulate conduction system-specific transcription, including activation of Id2, participating in a described network that promotes normal structure and function of the cardiac conduction system (35). Because GATA4 mutations G303E and G296S but not S52F produced electrophysiologic abnormalities, we speculate that GATA4 may have SMAD-dependent functions in the cardiac conduction system as well as in the endocardial cushions.
The G303E and G296S mutations, in contrast to S52F, not only reduced GATA4 promoter activation but also attenuated SMAD4 interactions (Fig. 2A). Within the endocardial cushion, loss of GATA4–SMAD4 interactions due to specific mutations could reduce the responses to TGF-β/BMP or other developmental cues, altering differentiation and/or proliferation (36, 37). Given the requirement of endothelial-SMAD4 signaling for atrioventricular valve formation, we hypothesize that GATA4 mutations produced either a restricted constellation of congenital heart defects, predominantly secundum ASDs or more profound AVSDs with electrophysiologic abnormalities, depending on whether endocardial SMAD4-GATA4 signaling is preserved during heart development.
Materials and Methods
Human Subjects.
Studies were performed in accordance with institutional guidelines for human research and written informed consent was obtained from all participants.
Animal Handling.
Mice were analyzed as 129 SV inbred and mixed 129 SV/C57BL/6 genetic backgrounds. All protocols conformed to the Association for the Assessment and Accreditation of Laboratory Animal Care and the Harvard Medical School Animal Care and Use Committee.
Gene Expression Studies.
Mouse heart in situ hybridizations were performed as previously described (34). Id1, Id2, and Id3 probe templates were made from plasmid accession numbers BE945568, AI843393, and AI839283, respectively, from the Brain Molecular Anatomy Project library (38).
Plasmid Constructs.
The constructions of Nkx2-5 promoter-driven luciferase reporter and Id2 promoter-driven luciferase reporter, respectively, were described (35, 39, 40). Bacterial and mammalian SMAD4 expression vectors were detailed previously (25). All GATA mutants S52F, G296S, and G303E were generated by a two-step PCR mutagenesis protocol as described (40, 41).
GST-Pulldown Assays.
Cell Culture and Transfection.
Cultured CV-1 cells were maintained in DMEM plus 10% FBS. Transient transfections of plasmid-based expression vectors were performed using Lipofectamine 2000 based on the protocol provided by the manufacturer. Reporter assays were performed in 12-well per plate containers with reporter construct of 200 ng per well and expression vectors as noted and using empty expression vectors to maintain the total amount of DNA constant per well. Promoter activity was expressed as the ratio of luciferase activity induced by the presence of specific factor(s) in comparison with the control group usually in the presence of empty vector or as otherwise stated. Data are expressed as the mean ± SD from at least two independent assays performed in duplicate.
Acknowledgments
We thank Jose Rivera-Feliciano. This work was supported by grants from the Howard Hughes Medical Institute (to C.E.S.), the National Institutes of Health (to I.P.M., J.G.S., C.E.S.), the March of Dimes (to I.P.M.), GlaxoSmithKline (to I.P.M.), and the Schweppe Foundation (to I.P.M.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Armstrong EJ, Bischoff J. Heart valve development: Endothelial cell signaling and differentiation. Circ Res. 2004;95:459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- 3.Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Immunohistochemical localization of TGF beta 1, TGF beta 2, and TGF beta 3 in the mouse embryo: Expression patterns suggest multiple roles during embryonic development. J Cell Biol. 1991;115:1091–1105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somi S, Buffing AA, Moorman AF, Van Den Hoff MJ. Dynamic patterns of expression of BMP isoforms 2, 4, 5, 6, and 7 during chicken heart development. Anat Rec A Discov Mol Cell Evol Biol. 2004;279:636–651. doi: 10.1002/ar.a.20031. [DOI] [PubMed] [Google Scholar]
- 5.Brown CB, Boyer AS, Runyan RB, Barnett JV. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 6.Gaussin V, et al. Alk3/Bmpr1a receptor is required for development of the atrioventricular canal into valves and annulus fibrosus. Circ Res. 2005;97:219–226. doi: 10.1161/01.RES.0000177862.85474.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaussin V, et al. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao K, et al. Tgfbeta signaling is required for atrioventricular cushion mesenchyme remodeling during in vivo cardiac development. Development. 2006;133:4585–4593. doi: 10.1242/dev.02597. [DOI] [PubMed] [Google Scholar]
- 10.Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelial-mesenchymal transformation during cardiac valve formation. Dev Biol. 2007;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park C, et al. Bone morphogenetic protein receptor 1A signaling is dispensable for hematopoietic development but essential for vessel and atrioventricular endocardial cushion formation. Development. 2006;133:3473–3484. doi: 10.1242/dev.02499. [DOI] [PubMed] [Google Scholar]
- 12.Potts JD, Runyan RB. Epithelial-mesenchymal cell transformation in the embryonic heart can be mediated, in part, by transforming growth factor beta. Dev Biol. 1989;134:392–401. doi: 10.1016/0012-1606(89)90111-5. [DOI] [PubMed] [Google Scholar]
- 13.Song L, Fässler R, Mishina Y, Jiao K, Baldwin HS. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim RY, Robertson EJ, Solloway MJ. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev Biol. 2001;235:449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- 17.Solloway MJ, et al. Mice lacking Bmp6 function. Dev Genet. 1998;22:321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 19.Pierpont ME, Markwald RR, Lin AE. Genetic aspects of atrioventricular septal defects. Am J Med Genet. 2000;97:289–296. [PubMed] [Google Scholar]
- 20.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 21.Garg V, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 22.Li QY, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 23.Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 24.Sarkozy A, et al. CRELD1 and GATA4 gene analysis in patients with nonsyndromic atrioventricular canal defects. Am J Med Genet A. 2005;139:236–238. doi: 10.1002/ajmg.a.31018. [DOI] [PubMed] [Google Scholar]
- 25.Brown CO, III, et al. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 26.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 28.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Runyan RB, Markwald RR. Invasion of mesenchyme into three-dimensional collagen gels: A regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol. 1983;95:108–114. doi: 10.1016/0012-1606(83)90010-6. [DOI] [PubMed] [Google Scholar]
- 30.Kowanetz M, Valcourt U, Bergström R, Heldin CH, Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol. 2004;24:4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraidenraich D, et al. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science. 2004;306:247–252. doi: 10.1126/science.1102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crispino JD, et al. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001;15:839–844. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 34.Munshi NV, et al. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development. 2009;136:2665–2674. doi: 10.1242/dev.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moskowitz IP, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 36.Sirard C, et al. Targeted disruption in murine cells reveals variable require ment for Smad4 in transforming growth factor beta-related signaling. J Biol Chem. 2000;275:2063–2070. doi: 10.1074/jbc.275.3.2063. [DOI] [PubMed] [Google Scholar]
- 37.He W, et al. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 38.Bonaldo MF, et al. 1274 full-open reading frames of transcripts expressed in the developing mouse nervous system. Genome Res. 2004;14(10B):2053–2063. doi: 10.1101/gr.2601304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J Biol Chem. 2002;277:25775–25782. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, et al. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27:622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J Biol Chem. 2004;279:49091–49098. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]