Abstract
Microtubule plus-end-tracking proteins (+TIPs) localize to growing microtubule plus ends to regulate a multitude of essential microtubule functions. End-binding proteins (EBs) form the core of this network by recognizing a distinct structural feature transiently existing in an extended region at growing microtubule ends and by recruiting other +TIPs to this region. The nature of the conformational difference allowing EBs to discriminate between tubulins in this region and other potential tubulin binding sites farther away from the microtubule end is unknown. By combining in vitro reconstitution, multicolor total internal reflection fluorescence microscopy, and electron microscopy, we demonstrate here that a closed microtubule B lattice with incorporated GTPγS, a slowly hydrolyzable GTP analog, can mimic the natural EB protein binding site. Our findings indicate that the guanine nucleotide γ-phosphate binding site is crucial for determining the affinity of EBs for lattice-incorporated tubulin. This defines the molecular mechanism by which EBs recognize growing microtubule ends.
Keywords: Mal3, cytoskeleton, beryllium fluoride, EB1
Growing microtubule ends serve as a crucial binding platform for numerous regulators of microtubule dynamics as well as for proteins mediating interactions of microtubules with various subcellular structures (1). Most of these so-called plus-end-tracking proteins (+TIPs) target the microtubule end by interacting with proteins of the conserved end-binding family (2–4). EB proteins (EBs) have the unique property of autonomously binding to an extended region at the distal end of growing microtubules with high selectivity (2–7) and are considered the core of the dynamic +TIP network. Which distinct structural feature of tubulin is recognized by EBs in this end-associated region extending over several hundred nanometers (dozens of tubulin subunits) remains a central unresolved question.
A possibility is that EBs recognize the nucleotide state of microtubule lattice-incorporated tubulin (6). Microtubules polymerize by addition of GTP-loaded tubulin to growing microtubule ends followed by GTP hydrolysis at the exchangeable binding site and phosphate release (8, 9). These events are thought to result in a stabilizing cap of GTP or GDP/Pi tubulin at the microtubule end, whereas the microtubule lattice is composed of GDP-tubulin. Although the size of the GTP cap has been considered to be very small, because a minimal cap of one or two layers of GTP-tubulin protects the microtubule end from depolymerization (10, 11), recent evidence supports the notion of an extended GTP cap at growing microtubules (12). An alternative structural feature EBs could recognize is the several-hundred-nanometer-long, sheet-like extension that has been observed at growing microtubule ends in vitro (13, 14). However, it is unclear whether similarly long extensions are present in vivo. Finally, electron microscopic studies have visualized the fission yeast end-binding protein Mal3 on the seam of microtubules grown in vitro (15, 16). The seam is a lattice discontinuity, which is characterized by lateral α-to-β-tubulin contacts (A lattice) between protofilaments, whereas the majority of protofilaments form lateral α-to-α-tubulin and β-to-β-tubulin contacts (B lattice) both in vitro and in vivo (17, 18). It has remained unclear how seam binding is related to the property of EBs to accumulate at extended regions at growing microtubule ends. Because a change from A to B lattice would require a synchronous longitudinal shift of 4 nm between numerous tubulin subunits, it is difficult to envisage how the microtubule end could exist transiently in an A-lattice configuration. Thus, compelling experimental evidence providing an answer to the question of which tubulin conformation is recognized by EBs is still lacking.
Results
Mal3 and EB1 Bind Strongly to GTPγS Microtubules.
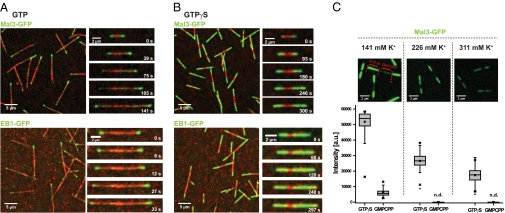
To test whether EBs recognize a nucleotide-dependent tubulin conformation, we studied microtubule plus-end tracking by EBs in vitro in the presence of GTP and GDP/Pi analogs. Microtubules were polymerized from surface-immobilized microtubule seeds stabilized by nonhydrolyzable guanosine-5-[(α,β)-methyleno]triphosphate (GMPCPP) in the presence of Alexa-568-labeled tubulin and different nucleotides. Using dual-color time-lapse total internal reflection fluorescence (TIRF) microscopy, we observed strong accumulation of GFP-labeled fission yeast Mal3 (Fig. 1A Upper) or Xenopus laevis end-binding protein 1 (EB1) (Fig. 1A Lower) at microtubule ends growing in the presence of 1 mM GTP. Conversely, only weak binding to GMPCPP seeds and to the GDP lattice of the growing segments could be observed, as shown previously (2, 4, 5, 7).
Fig. 1.
Mal3 and EB1 bind strongly to GTPγS microtubules. (A) Overview TIRF microscopy image (Left) and time series of images (Right) showing Mal3-GFP (green; Upper) or EB1-GFP (green; Lower) tracking the ends of growing Alexa-568-labeled microtubules (red) in the presence of GTP. Microtubules grow from surface-immobilized GMPCPP microtubule seeds (bright red). (B) TIRF microscopy images of Mal3-GFP (green; Upper) and EB1-GFP (green; Lower) strongly binding along the entire length of microtubules growing in the presence of GTPγS, but not to GMPCPP seeds (bright red). Mal3 was imaged in standard TIRF assay buffer, and EB1 in low-salt TIRF assay buffer with additional 4 mM MgCl2. (C) Effect of ionic strength on Mal3-GFP binding. The GFP channel of representative TIRF microscopy images (Upper) and box plots (Lower) of the measured intensities of 80 nM Mal3-GFP on GTPγS and GMPCPP microtubule segments at different salt concentrations. Buffer was standard TIRF assay buffer in the middle column, reduced by 85 mM KCl in the left column, and supplemented with 85 mM K-acetate in the right column. The fluorescence of Mal3-GFP on GMPCPP microtubule seeds was not detectable (n.d.) under the conditions used here in the middle and right columns. Protein concentrations were 80 nM Mal3-GFP, 600 nM EB1-GFP, and 25 μM Alexa-568-tubulin; nucleotides were used at 1 mM. Scale bars are as indicated.
Strikingly, in the presence of 1 mM GTPγS, a slowly hydrolyzable GTP analog with a modification at the γ-phosphate, both Mal3-GFP (Fig. 1B Upper) and EB1-GFP (Fig. 1B Lower) accumulated strongly along the entire microtubule lattice. Quantitative analysis of the fluorescence intensity revealed a large difference in Mal3-GFP binding to GTPγS versus GMPCPP microtubules (Fig. 1C). Although the fluorescence signal decreased with increasing ionic strength, as expected for an interaction with an electrostatic contribution, the large difference in EB binding to the two types of microtubule lattices remained (no fluorescence signal was detectable on GMPCPP seeds at elevated ionic strengths) (Fig. 1C). A similarly large difference in binding preference was found for EB1-GFP (Fig. S1 B and C). Recent studies reported weak preferential binding of human EB1 to GMPCPP microtubules in a low-ionic-strength buffer (3, 6). However, we found that this depends mostly on the presence of an artificial oligo-histidine sequence fused to EB1 (Fig. S1 A and C), shown recently to affect the EB1–microtubule interaction (19). Neither EB1-GFP (Fig. S1 B and C) nor Mal3-GFP (Fig. 1C) bind with strong preference to GMPCPP microtubules in the absence of an oligo-histidine tag. Thus, a GMPCPP-bound microtubule lattice does not mimic the EB protein binding site at growing microtubule ends.
The Guanine Nucleotide γ-Phosphate Binding Site in Tubulin Determines Mal3 Affinity.
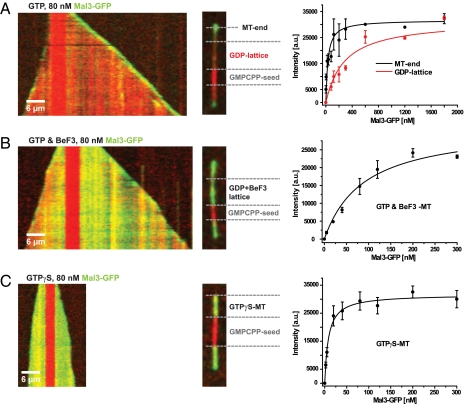
Focusing on Mal3 because of its higher solubility, we examined the importance of the γ-phosphate binding site in polymerized tubulin for EB binding. Kymograph analysis of growing microtubules revealed that in contrast to selective end accumulation in the presence of 1 mM GTP (Fig. 2A Left), the additional presence of 2 mM BeF3− led also to Mal3-GFP binding along the entire length of microtubules (Fig. 2B Left). BeF3− binds to the γ-phosphate binding site after GTP hydrolysis and phosphate release, mimicking an early hydrolysis transition state (20). The Mal3-GFP accumulation on these microtubules appeared similar to the situation in the presence of GTPγS (Figs. 1B Upper and 2C Left).
Fig. 2.
Binding characteristics of Mal3-GFP to the growing microtubule end, the GDP lattice, the GDP/BeF3− lattice, and GTPγS microtubules as measured using TIRF microscopy. (A) (Left) Kymograph of Alexa-568-labeled microtubules (red) grown from GMPCPP seeds (bright red) in the presence of 1 mM GTP and 80 nM Mal3-GFP (green) and an illustration of the different microtubule regions used for the intensity measurements. (Right) Binding curves of Mal3-GFP for microtubules grown in the presence of 1 mM GTP: averaged intensities of Mal3-GFP fluorescence at growing microtubule plus ends (black data points) and on the GDP lattice (red data points) as a function of the Mal3-GFP concentration and the corresponding hyperbolic fits (black and red lines) yielding dissociation constants of 31 ± 5.6 nM and 285 ± 43.9 nM, respectively. (B) Kymograph as in A, but in the presence of 1 mM GTP and additionally 2 mM BeF3−. (Right) Fluorescence intensity-based binding curves of Mal3-GFP binding to microtubules grown in the presence of 1 mM GTP and 2 mM BeF3−. The hyperbolic fit (line) to the data points yields a dissociation constant of 89 ± 13.9 nM. (C) Kymograph as in A, but in the presence of 1 mM GTPγS instead of GTP. (Right) Binding curve of Mal3-GFP to GTPγS microtubules. A hyperbolic fit (line) to the averaged intensity data yields a dissociation constant of 8 ± 1.7 nM.
We quantitatively characterized the similarity between GTPγS and GDP/BeF3− microtubules and the plus-end region of microtubules growing in the presence of GTP by determining the affinity of Mal3 for each of the different microtubule configurations. We measured the average steady-state fluorescence of microtubule-bound Mal3-GFP at the growing microtubule end or the microtubule lattice at various concentrations. In the presence of GTP, Mal3-GFP bound to both the growing microtubule plus-end region and to the GDP lattice in a concentration-dependent manner, albeit with different affinities (Fig. 2A Right). Assuming a simple binding equilibrium, we derived dissociation constants (Kd) of 31 ± 5.6 nM and 285 ± 43.9 nM for the growing microtubule end region and the GDP lattice, respectively, demonstrating an approximate 10-fold higher affinity of Mal3 for the growing end. On a GDP/BeF3− microtubule lattice, the affinity of Mal3-GFP was increased compared with a GDP microtubule lattice, yielding a Kd of 89 ± 13.9 nM (Fig. 2B). In the presence of GTPγS, Mal3-GFP bound even more strongly along the GTPγS microtubule segments, with a Kd of 8 ± 1.7 nM (Fig. 2C). This demonstrates that Mal3 binds with a similar high affinity to the GTPγS-bound microtubule lattice as to microtubule ends growing in the presence of GTP.
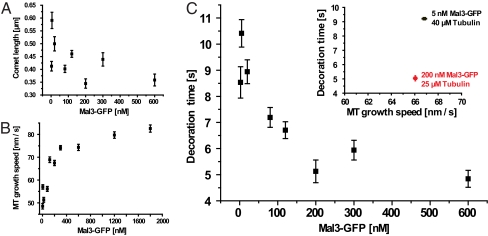
Because GTP γ-phosphate modifications (potentially inducing GTP hydrolysis intermediate-state conformations in polymerized tubulin) promoted Mal3 binding, we asked whether Mal3 binding affects the lifetime of the conformational state that it recognizes at growing microtubule ends in the presence of GTP. The lifetime of this conformational state can be determined from the length of the fluorescent Mal3-GFP comet tails (Fig. 3A and Fig. S2) and the growth velocity of the microtubules (Fig. 3B). Interestingly, Mal3 reduced this lifetime [previously called decoration time (5)] in a dose-dependent manner (Fig. 3C), indicating that it catalyzes the transition of tubulin from the recognized state at growing ends to the state tubulin adopts in the lattice distant from the ends. This is reminiscent of the proposal that EBs may promote the closure of extended sheets at growing microtubule ends into tubes (21).
Fig. 3.
Analysis of the lifetime of Mal3 binding sites in the plus-end region of growing microtubules in the presence of 1 mM GTP using TIRF microscopy. (A and B) Mean Mal3-GFP comet lengths (A) and mean speeds of microtubule plus-end growth (B) as a function of Mal3-GFP concentration. (C) Mal3-GFP decoration time of growing microtubule ends as obtained by dividing mean comet lengths by corresponding growth speeds. (Inset) Decoration times at different Mal3-GFP concentrations and similar microtubule growth speeds (adjusted by changing the tubulin concentration, as indicated). Unless stated otherwise, all experiments were performed with 25 μM Alexa-568-tubulin in standard TIRF assay buffer. All error bars are ±SEM.
GTPγS Microtubules Mimic the Growing Microtubule End Region.
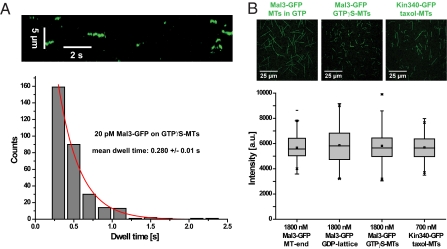
Imaging individual Mal3-GFP molecules (at reduced concentrations of 20 pM) revealed fast turnover of Mal3 on GTPγS microtubules (Fig. 4A). A monoexponential fit to the dwell time distribution of dimeric Mal3-GFP molecules (Fig. S3 A and B) showed that the mean dwell time of 0.28 ± 0.02 s (Fig. 4A) was similar to the mean dwell time of Mal3 at microtubule ends growing in GTP in the absence (5) and presence of additional unlabeled Mal3 (Fig. S4 Left). Similarity of the latter two values confirms that EBs do not show positive cooperativity of binding (2). Then, we determined the density of Mal3 binding sites on the lattice of GTPγS microtubules and on the plus ends as well as on the GDP lattice of microtubules growing in GTP. We measured the maximal average fluorescence intensity of microtubule-bound Mal3-GFP at saturating concentrations on each of these microtubule configurations (Fig. 4B) and compared these values to the maximal average fluorescence intensity of a GFP-labeled monomeric kinesin-1 variant known to bind in a 1:1 ratio to tubulin dimers on taxol-stabilized microtubules (22). This analysis demonstrated that the Mal3 dimer binds to each of these distinct microtubule configurations with one of its two calponin homology domains (23) per tubulin dimer at saturating conditions. Although monomeric Mal3 has a generally reduced affinity for microtubules (Fig. S5), similar to monomeric EB1 (24), we could show that Mal3 monomers also have a clear binding preference for GTPγS microtubules (Fig. 5), demonstrating that dimerization is not required for selective recognition of GTPγS microtubules. These experiments establish GTPγS microtubules as a static model that closely resembles the growing microtubule end with respect to Mal3 binding affinity, turnover, and density of binding sites.
Fig. 4.
Single-molecule binding/unbinding turnover and maximum number of binding sites on GTPγS microtubules. (A) TIRF microscopy imaging of single Mal3-GFP molecules at 20 pM: kymograph (time-space plot) showing single binding events of Mal3-GFP on a GTPγS microtubule (Upper) and a histogram showing the dwell time distribution (Lower). A monoexponential fit (red line) yields a mean dwell time of 280 ± 0.02 ms. (B) Maximum binding densities of Mal3-GFP on microtubules growing in the presence of 1 mM GTP or GTPγS in comparison with the density of monomeric kinesin-1 fused to GFP (Kin340-GFP) fully decorating a taxol-stabilized microtubule in the presence of the nonhydrolyzable ATP analog adenylyl-imidodiphosphate (AMPPNP). Example images (Upper) and box plots of averaged intensities (Lower) are shown. GFP fluorescence intensity values are corrected for differences between GFP variants (SI Materials and Methods). Concentrations were 25 μM Alexa-568-tubulin and 1 mM GTP or GTPγS in standard TIRF assay buffer supplemented with 5 mM AMPPNP and 4 mM MgCl2. Scale bars are as indicated.
Fig. 5.
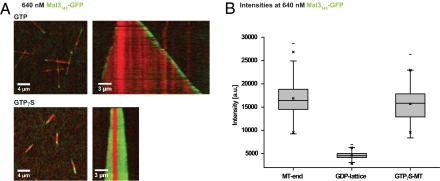
Microtubule binding selectivity of Mal3143 monomers. (A) Overview TIRF microscopy images (Left) and kymographs (Right) of 640 nM Mal3143-GFP (green) on Alexa-568-microtubules (red) grown in the presence of 1 mM GTP (Upper) or GTPγS (Lower). The kymograph extends over a time period of 360 s. Experiments were performed in standard TIRF assay buffer. (B) Box plot of intensity measurements of Mal3143-GFP at the microtubule end, GDP lattice, and GTPγS microtubules.
GTPγS Microtubules Are Closed B-Lattice Tubes.
To test whether this static model structure of a growing microtubule end shows characteristics of extended tubulin sheets or whether it consists mostly of lateral A-lattice contacts between protofilaments as previous observations might predict (15, 16), we adapted our microscopy assay for electron microscopy (EM). To distinguish GMPCPP microtubule seeds from GTPγS microtubule segments, we selectively labeled the biotinylated seeds with streptavidin-coated quantum dots, which are readily recognized both by negative-stain EM (Fig. 6A) as well as by cryo-EM (Fig. 6B). We demonstrated that GTPγS microtubule segments were closed tubes and not sheet-like structures, as revealed by the presence of moiré patterns (Fig. 6C) in tubes composed of fewer than 13 protofilaments and the distinct left–right asymmetry in 13-protofilament tubes. These patterns are seen in 2D projections due to the near- and far-side superposition of the densities of the protofilaments that make up the lattice of the 3D hollow tube (25). To determine the lattice type of the GTPγS tubes, we decorated them with a monomeric kinesin-1 construct which binds to microtubules in a rigor state (26). Diffraction analysis of these kinesin-decorated GTPγS microtubules revealed the presence of a predominantly B-lattice arrangement (Fig. 6C Lower Left), as indicated by the positions of the brightest diffraction peaks on the 1/8-nm layer line. In particular, the presence of a peak at Bessel order −1.5 strongly indicates a B lattice, because the lateral stagger of the protofilaments results in a left-handed 1.5-start helix of tubulin dimers (27, 28).
Fig. 6.
Electron microscopy of monomeric Mal3 on microtubules grown in the presence of different nucleotide analogs. (A) Selective structural tagging of GMPCPP seeds: negative-stain EM images showing a biotinylated GMPCPP microtubule seed labeled with streptavidin-coated quantum dots (Upper), and a quantum dot-labeled GMPCPP seed which has been elongated in the presence of 1 mM GTPγS (Lower). (B) Cryo-EM image of kinesin (rKinT93N340)-decorated microtubules polymerized in the presence of GTPγS from quantum dot-labeled GMPCPP seeds. (C) Lattice structure of an undecorated GTPγS microtubule (Upper Left), a Mal3143-decorated GTPγS microtubule (Upper Right), a kinesin-decorated GTPγS microtubule (Lower Left), and a kinesin-decorated GDP/BeF3− microtubule (Lower Right). Each of the four panels shows a cryo-EM image of a microtubule (Left), the Fourier-filtered representation of the respective image (Center), and the corresponding diffraction pattern (Right). The yellow lines in the diffraction patterns have been drawn through the positions of the dominant diffraction peaks. A strong peak at Bessel order −1.5 implies a B lattice, because the lateral stagger of the protofilaments in a B-lattice arrangement forms a left-handed, 1.5-start helix (of tubulin dimers). The fact that the dominant peaks are mirrored across the meridian (blue line) and the smeared-out nature of the diffraction spots in layer lines are further indications that GTPγS and GDP/BeF3− microtubules are tubes and not sheets, as there is contribution to the diffraction pattern from both the near and far sides of the microtubule.
To test which lattice structure GTPγS microtubules have when decorated with Mal3, we used monomeric Mal3, as it led to a more regular decoration than dimeric full-length protein. GTPγS microtubules retained the B-lattice structure when decorated with Mal3 (Fig. 6C Upper Right). As expected from fluorescence experiments (Fig. 4B), Mal3 decorated the GTPγS microtubules with a longitudinal spacing of 8 nm (Fig. 6C), indicating a binding ratio of one Mal3 monomer per tubulin dimer. Mal3-decorated GDP/BeF3− microtubules showed also largely B-lattice contacts (Fig. 6C Lower Right). This observation differs from the induction of mixed lattice contacts in GDP and GDP/taxol microtubules by Mal3 monomers (15). Thus, our results demonstrate that GTPγS microtubules have a closed B-lattice configuration and closely mimic the EB protein binding site at growing microtubule ends. This, together with the observation that microtubule protofilaments form mostly B-lattice contacts in vivo (17), leads us to conclude that growing end structures very likely adopt mostly B-lattice contacts in vivo when being decorated by EBs. The nucleotide state of lattice-incorporated tubulin is crucial for high-affinity EB binding.
Discussion
We demonstrate that EBs from two different organisms bind with high affinity to GTPγS microtubules, suggesting that these microtubules display the structural feature which is recognized by EBs at growing microtubule plus ends in living cells. Affinity, turnover, and stoichiometry of binding to GTPγS microtubules are similar to that at the region at microtubule ends growing in the presence of GTP. EBs can bind around the entire GDP lattice of microtubules in vitro (Fig. 4B and Fig. S5), but with about 10-fold lower affinity than to the end region (Fig. 2A), likely explaining their accumulation at growing microtubule ends in vivo. Because soluble tubulin was present in the higher micromolar range in our experiments, one can estimate that EBs discriminate even more strongly between soluble tubulin and lattice-incorporated tubulin, in agreement with the lack of a measurable interaction between EBs and soluble tubulin (2, 5).
The conformational feature responsible for high-affinity binding of EBs to microtubule end regions most crucially depends on conformational details of the guanine nucleotide γ-phosphate binding site of lattice-incorporated tubulin. Whereas GTPγS and to a lesser extent also GDP/BeF3− incorporation in a closed B lattice induce the conformation recognized by EBs, GMPCPP does not. How can this striking difference between GTP analogs be explained? GMPCPP is a strong microtubule nucleator, whereas, similar to GTP, GTPγS is not. GMPCPP-tubulin may allow for increased lateral tubulin interactions important for the nucleation process (29) due to the straight conformation of GMPCPP-tubulin (30, 31). Corresponding data for GTPγS-tubulin do not exist. However, it was recently shown that γ-tubulin, whose structure is very similar to that of α- and β-tubulin, has a rather curved structure with either GTPγS or GDP bound (29). In combination with structural data from bacterial tubulin orthologs, this has suggested that lattice incorporation, possibly accompanied by GTP hydrolysis and/or phosphate release, leads to straightening of otherwise curved GTP-tubulin (29, 32). In the context of this model, GTPγS might induce a slightly curved conformation of tubulin that is recognized by EBs when tubulin is incorporated into the microtubule lattice. In this view, EBs recognize a structural cap at growing ends of B-lattice microtubules which could be independent of the extent to which the growing end forms a 2D sheet (13, 14).
Interestingly, Mal3 monomers did not induce A-lattice contacts in GTPγS microtubules, in contrast to what was observed for GDP and GDP/taxol microtubules (15). This latter observation was interpreted as being in agreement with preferential binding of Mal3 dimers to the microtubule seam of GDP/taxol microtubules as observed by EM after freeze drying and metal shadowing (16). These experiments were, however, performed with Mal3 concentrations well above the Kd of Mal3 for the GDP lattice as measured in our TIRF microscopy experiments, suggesting that only part of the dynamically bound Mal3 was visible in these EM experiments, a part which possibly had a higher affinity for the seam (16). In our TIRF experiments here, we were unable to discriminate between seam and B-lattice binding on GDP microtubules because of technical limitations. Nevertheless, the similarity in affinity, stoichiometry of binding, and turnover rate of Mal3 on growing microtubule ends in the presence of GTP and on GTPγS microtubules strongly suggests that growing microtubule ends extend by forming B-lattice contacts between elongating protofilaments. In vivo, the Mal3 concentration (33) has been estimated to be in the low nanomolar range, which is in the range of the Kd which we have measured for Mal3 binding to the growing microtubule end (Fig. 2A). Thus, a substantial fraction of tubulin subunits at the growing microtubule end is expected to be occupied by Mal3 in vivo.
The end structure recognized by EBs is longer than the present consensus length of the minimal stabilizing GTP or GDP/Pi cap size of only a couple of tubulins along the protofilament axis (10, 11). Nevertheless, EBs recognize lattice-incorporated tubulin in a conformation which appears to be triggered by the γ-phosphate sensor at the exchangeable GTP binding site which lies between two tubulin heterodimers. The discrepancy between the length of the minimal GTP or GDP/Pi cap and the EB binding region could indicate that the nucleotide state of lattice-incorporated tubulin is not strictly coupled to the conformational state of tubulin, in agreement with the “structural plasticity” model of microtubule dynamics (9). Our observation that Mal3 reduces the lifetime of the recognized conformation at growing microtubule ends could therefore mean that EBs are able to exert a regulatory effect on the microtubule's structural plasticity.
In conclusion, GTPγS microtubules can be considered a static model for the growing microtubule end structure to which EBs bind. This system will therefore lend itself particularly well to structural cryo-EM studies and promises to lead to valuable insight also into the mechanism of recruitment of various other plus-end-binding proteins by EBs.
Materials and Methods
For protein biochemistry, microtubule copelleting assay, and negative-stain electron microscopy, see SI Materials and Methods.
TIRF Microscopy.
Dual-color TIRF microscopy imaging of fluorescently labeled microtubules in the presence of GFP-labeled EB1 or Mal3 was performed as described (5, 34). Dimly Alexa-568-labeled microtubules were polymerized from glass-immobilized, brightly labeled (33% Alexa-568-labeled tubulin) GMPCPP (Jena Bioscience)-stabilized microtubule seeds in the presence of EBs. Unless stated otherwise, the final solution was 25 μM tubulin (of which 10% is Alexa-568-labeled tubulin), 1 mM GTP (Sigma) or GTPγS (Roche), or 1 mM GTP + 2 mM BeF3− (Alfa Aesar) and varying concentrations of EBs in assay buffer. For experiments with EB1-GFP or oligo-His-EB1-GFP, low-salt TIRF assay buffer was used if not indicated otherwise: BRB80 (80 mM K-Pipes, pH 6.8, 2 mM MgCl2, 1 mM EGTA) with 10 mM 2-mercaptoethanol, 0.15% methylcellulose (4,000 centipoise; Sigma), and an oxygen scavenger system (35). For experiments with Mal3-GFP, standard TIRF assay buffer was used: low-salt TIRF assay buffer supplemented with additional 2 mM MgCl2 and 85 mM KCl. For experiments with excess unlabeled Mal3 or EB1, standard TIRF assay buffer was used. The longer mean dwell time found for EB1-GFP in these “spike experiments” (Fig. S4 Right) compared with previous measurements (2) is most likely a consequence of the lower salt concentration used here. For intensity comparisons between Mal3-GFP and Kin340-GFP on microtubules (Fig. 4B), standard TIRF assay buffer was supplemented with an additional 5 mM AMPPNP (Sigma) and 4 mM MgCl2. Mal3 or kinesin storage buffer was added in experiments where the respective protein was not present to compensate for storage buffer contributions to the reaction mixture. The temperature was kept at 30 °C. Time-lapse imaging was performed at 1 frame every 3 s, with 100-ms exposure time, using a magnification of 96×. Single-molecule data were obtained by recording streams of 60-s duration with an exposure time of 100 ms per frame and a magnification of 160× and elevated excitation intensity.
For binding curves, intensities from TIRF microscopy images were plotted as a function of the Mal3 concentration. To determine Kd, a one-site-binding model was fitted to the averaged binding curve I = Imax × c/(Kd + c) with the measured intensity I, the maximum intensity Imax at saturation, and the free Mal3-GFP concentration c. The total concentration of Mal3-GFP is here approximately equal to the free concentration c due to the small number of binding sites on microtubules compared with the total amount of Mal3 in the assay. For details, see SI Materials and Methods.
The determination of Mal3-GFP comet length, microtubule growth speed, and decoration time (Fig. 3 A–C and Fig. S2) was carried out as described (5, 34).
Quantum Dot Labeling of Biotinylated GMPCPP Seeds for Electron Microscopy.
Short biotinylated GMPCPP microtubules were generated from 18 μM tubulin and 11 μM biotin-labeled tubulin in BRB80 with 0.5 mM GMPCPP at 37 °C for 3 min. After 10-fold dilution with prewarmed BRB80, the seeds were centrifuged in a tabletop centrifuge at 15.300 × g for 8 min. Pelleted GMPCPP seeds were resuspended in 80 μL prewarmed BRB80. Forty microliters of the seed solution was mixed with Qdot 655 streptavidin conjugate (Invitrogen) at a final Qdot concentration of 250 μM. The mixture was incubated for 8 min at room temperature, diluted 10-fold, and pelleted as above. The labeled seeds were washed twice with prewarmed BRB80 to remove unbound Qdots and resuspended in 25 μL prewarmed BRB80. Labeled seeds were used immediately for EM sample preparation.
Cryoelectron Microscopy.
Holey-carbon C-flat grids (CF4-2-2; Protochips) were used for cryo-sample preparations. For the decoration of microtubules with monomeric Mal3143, the final reaction mixture contained 2% (vol/vol) of biotinylated GMPCPP seeds, 15 μM tubulin, 60 μM Mal3143, and 1 mM GTPγS in BRB80. A 4-μL sample was applied to the grid which was mounted in the Vitrobot Mark III (FEI). Before plunge freezing, the sample was incubated on the grid for 60 s at 37 °C and 100% ambient humidity. Samples were blotted for 3–4 s with blot offsets from −1 to 1 mm. For the analysis of the lattice structure of GTPγS microtubules, a monomeric rigor mutant of rat kinesin-1 rKinT93N340 (26) was used to decorate GTPγS microtubules. First, 15 μM tubulin was mixed with 5% Qdot-labeled GMPCPP seeds and 1 mM GTPγS in BRB80. The mixture was incubated for 10 min at 37 °C. Three microliters of the mixture was deposited onto a grid and adsorbed for 30 s at 37 °C. Subsequently, 3 μL of kinesin was added to a final concentration of 20 μM in a total volume of 6 μL on the grid. After a further 60 s of incubation at 37 °C, excess liquid was blotted away and the sample was plunge-frozen in liquid ethane using a homemade plunge-freezing device. Samples were transferred under liquid nitrogen to a Gatan 626 cryoholder. Cryoelectron microscopy data were collected on an FEI Tecnai F20 200 kV FEG transmission electron microscope (FEI) with a total dose of 25–35 electrons/Å2, a nominal magnification of 29,000×, and a defocus of −2.5 μm. The resulting pixel size corresponded to 7.6 Å on the specimen.
Supplementary Material
Acknowledgments
We thank Ivo Telley for maintaining the TIRF microscope setup and for help with statistical analysis, Rachel Santarella, Cindi Schwartz, James Riches, and John Briggs for technical advice with cryoelectron microscopy, Rob Cross for the monomeric kinesin head construct rKinT93N340, and Marie-France Carlier and Isabelle Arnal for stimulating discussions. We thank the European Molecular Biology Organization (Grant ALTF 1032-2009) and the Human Frontier Science Program Organization (Grant RGP0023/2008-C) for financial support for S.P.M. and T.S., respectively. A.H. and J.C. acknowledge financial support from the National Center for Research Resources (Grant P41-RR000592) and the National Institutes of Health (Grant T32 GM-065103), respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014758108/-/DCSupplemental.
References
- 1.Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Curr Biol. 2010;20:R528–R537. doi: 10.1016/j.cub.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 2.Bieling P, et al. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixit R, et al. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci USA. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 5.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 6.Zanic M, Stear JH, Hyman AA, Howard J. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One. 2009;4:e7585. doi: 10.1371/journal.pone.0007585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 9.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caplow M, Shanks J. Evidence that a single monolayer tubulin-GTP cap is both necessary and sufficient to stabilize microtubules. Mol Biol Cell. 1996;7:663–675. doi: 10.1091/mbc.7.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drechsel DN, Kirschner MW. The minimum GTP cap required to stabilize microtubules. Curr Biol. 1994;4:1053–1061. doi: 10.1016/s0960-9822(00)00243-8. [DOI] [PubMed] [Google Scholar]
- 12.Schek HT, III, Gardner MK, Cheng J, Odde DJ, Hunt AJ. Microtubule assembly dynamics at the nanoscale. Curr Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: Two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: A time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.des Georges A, et al. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat Struct Mol Biol. 2008;15:1102–1108. doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandblad L, et al. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 17.McIntosh JR, Morphew MK, Grissom PM, Gilbert SP, Hoenger A. Lattice structure of cytoplasmic microtubules in a cultured mammalian cell. J Mol Biol. 2009;394:177–182. doi: 10.1016/j.jmb.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song YH, Mandelkow E. Recombinant kinesin motor domain binds to β-tubulin and decorates microtubules with a B surface lattice. Proc Natl Acad Sci USA. 1993;90:1671–1675. doi: 10.1073/pnas.90.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu ZC, et al. Interactions between EB1 and microtubules: Dramatic effect of affinity tags and evidence for cooperative behavior. J Biol Chem. 2009;284:32651–32661. doi: 10.1074/jbc.M109.013466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlier MF, Didry D, Simon C, Pantaloni D. Mechanism of GTP hydrolysis in tubulin polymerization: Characterization of the kinetic intermediate microtubule-GDP-Pi using phosphate analogues. Biochemistry. 1989;28:1783–1791. doi: 10.1021/bi00430a054. [DOI] [PubMed] [Google Scholar]
- 21.Vitre B, et al. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 22.Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J Biol Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- 23.Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- 24.Komarova Y, et al. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chrétien D, Wade RH. New data on the microtubule surface lattice. Biol Cell. 1991;71:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 26.Crevel IM, et al. What kinesin does at roadblocks: The coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikkawa M. A new theory and algorithm for reconstructing helical structures with a seam. J Mol Biol. 2004;343:943–955. doi: 10.1016/j.jmb.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 28.Mandelkow EM, Schultheiss R, Rapp R, Müller M, Mandelkow E. On the surface lattice of microtubules: Helix starts, protofilament number, seam, and handedness. J Cell Biol. 1986;102:1067–1073. doi: 10.1083/jcb.102.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: Structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller-Reichert T, Chrétien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: Information from a slowly hydrolyzable analogue of GTP, guanylyl (α,β)methylenediphosphonate. Proc Natl Acad Sci USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buey RM, Díaz JF, Andreu JM. The nucleotide switch of tubulin and microtubule assembly: A polymerization-driven structural change. Biochemistry. 2006;45:5933–5938. doi: 10.1021/bi060334m. [DOI] [PubMed] [Google Scholar]
- 33.Katsuki M, Drummond DR, Osei M, Cross RA. Mal3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J Biol Chem. 2009;284:29246–29250. doi: 10.1074/jbc.C109.052159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bieling P, Telley IA, Hentrich C, Piehler J, Surrey T. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 2010;95:555–580. doi: 10.1016/S0091-679X(10)95028-0. [DOI] [PubMed] [Google Scholar]
- 35.Bieling P, Telley IA, Piehler J, Surrey T. Processive kinesins require loose mechanical coupling for efficient collective motility. EMBO Rep. 2008;9:1121–1127. doi: 10.1038/embor.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.