Abstract
Fetal exposure to environmental insults increases the susceptibility to late-onset neuropsychiatric disorders. Alcohol is listed as one of such prenatal environmental risk factors and known to exert devastating teratogenetic effects on the developing brain, leading to complex neurological and psychiatric symptoms observed in fetal alcohol spectrum disorder (FASD). Here, we performed a coordinated transcriptome analysis of human and mouse fetal cerebral cortices exposed to ethanol in vitro and in vivo, respectively. Up- and down-regulated genes conserved in the human and mouse models and the biological annotation of their expression profiles included many genes/terms related to neural development, such as cell proliferation, neuronal migration and differentiation, providing a reliable connection between the two species. Our data indicate that use of the combined rodent and human model systems provides an effective strategy to reveal and analyze gene expression changes inflicted by various physical and chemical environmental exposures during prenatal development. It also can potentially provide insight into the pathogenesis of environmentally caused brain disorders in humans.
Keywords: cross-species, human fetus, neuronal subtypes, microarray, Notch
Exposure of human fetuses to various physical and chemical environmental factors causes a wide variety of abnormalities with serious health consequences (1). The embryonic central nervous system is highly vulnerable to environmental exposures (EE), as it displays not only overt cytological malformations, but also a covert increase in the susceptibility to late-onset neuropsychiatric disorders (1–4). However, due to ethical, technical, and practical limitations, no pharmacological study has been performed on the developing human fetal cerebrum, which is particularly sensitive to physical and chemical agents due to a much more prolonged period of neurogenesis as well as longer distance neuronal migration and protracted differentiation (5). Magnetic resonance imaging (MRI) and postmortem examinations of the human brain reveal various consequences of fetal EE, but only long after initial exposure to harmful agents.
Most of our knowledge about the cellular and molecular mechanisms of brain damage caused by EE, therefore, has been gained from experimental studies in rodents. However, it is yet to be established to what extent the findings and insights obtained from rodent models apply to the larger, more complex and more slowly developing human brain. To overcome these limitations, we took advantage of recent progress in functional genomics, and performed coordinated cross-species transcriptome analysis with human in vitro (cortical slice culture) and mouse in vivo models of prenatal EE. There are two important expectations in this approach. First, by obtaining the gene expression changes caused by prenatal EE in both species (i.e., human in vitro and mouse in vivo models), we can enhance statistical power with a limited number of human experimental samples, while reducing the risk of picking up biologically irrelevant changes introduced by in vitro procedure in the human model. Second, subsequent wet analyses to further evaluate the biological meanings of obtained gene lists can take advantage of both models (e.g., direct relevance to human diseases in the human model and transgenic approaches in the mouse model).
We selected ethanol, the most common nongenetic contributor to the pathogenesis of various neurological dysfunctions, as a model prenatal stressor. Prenatal exposure to ethanol is known also as a risk factor for various cognitive dysfunctions (6–8), including a complex syndrome known as fetal alcohol spectrum disorder (FASD). In addition, ethanol has a technical advantage as a powerful pharmacological agent, which can mask subtle alterations of gene expression caused by heterogeneous genetic backgrounds among human samples. Above all, there is a comprehensive list of experimental literature on the effects of ethanol on the rodent brain (9–15).
The mouse cerebral cortex is highly vulnerable to ethanol exposure in the midgestational period, which is associated with extensive neurogenesis and neuronal migration to the cerebral cortex (9–15). A comparable stage in the human cerebral cortex is in the early midgestational period during which a large number of neurons migrate the longest distance to the superficial cortical layers (16, 17). This period in human is also the most vulnerable for the development of mental dysfunctions in FASD (18). Thus, our study has been focused on human cortical development between 15 and 18 gestational weeks (GW) and between mouse embryonic days (E) 14 and 16.
Results
Cross-Species Screening of Differentially Expressed Genes.
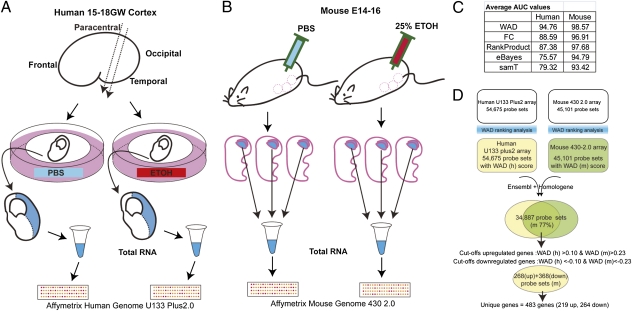
Total RNAs were isolated from human cortical slices cultured in a medium containing ethanol or PBS (control), and the cortical tissues of mouse embryos from dams injected with ethanol or PBS (control) (Fig. 1 A and B). Isolated RNAs were hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 and Mouse Genome 430 2.0, respectively. We first compared several ranking methods that are known for their superior performance with robust multichip average (RMA) preprocessed data (19). We obtained the area under the receiver operating characteristic (ROC) curve (AUC) (Fig.1C) for each ranking method with our midgestation fetal alcohol exposure (mFAE) vs. control data and found that the weighted average difference (WAD) method outperformed other methods for both human and mouse array data. Thus, in our study, we used the WAD for initial screening of differentially expressed genes.
Fig. 1.
Gene expression profiling using Human in vitro and mouse in vivo models. (A and B) Schematic illustration of the procedure for sample collection in human in vitro (A) and mouse in vivo (B) models of mFAE. (C) Comparison of the AUC values among different ranking methods, showing the best performance of the WAD for both human and mouse datasets. (D) Schematic diagram of the cross-species gene screening by the WAD. The CDEG of mFAE were selected as the up- and down-regulated genes with the human and mouse WAD statistics greater than 0.1 and 0.23 and less than −0.1 and −0.23, respectively. Cutoff was set empirically according to the qRT-PCR data (Table S1 and Fig. S1). Because of the lower AUC score with the human datasets than that with the mouse datasets, the cutoff setting was relaxed for the human datasets. The probe sets that showed expression changes in opposite directions between human and mouse were then eliminated from the list. Dataset S1 includes screened unique reference IDs, which were used for the matching of human and mouse probe sets and the corresponding WAD scores.
In the cross-species screening, we first matched human and mouse probe set IDs by using annotation tools (20). As shown in Fig.1D, 77% of the mouse probe sets matched the orthologous human probe sets. Subsequently, the cross-species conserved differentially expressed genes (CDEG), i.e., the genes with the expressions that significantly changed by mFAE in both human and mouse models, were selected as up-regulated (268 mouse probe sets) and down-regulated genes (368 mouse probe sets) (Dataset S1, see the setting of cutoff in Fig.1D and Fig. S1A). After putting together the probe sets of the same genes, the unique up-regulated and down-regulated genes finally ended up to be 219 and 264, respectively. Up- and down-regulation of the genes were validated by qRT-PCR for the genes selected randomly from around the top and bottom of up- and down-regulated gene lists (Fig. S1B). Table 1 shows the CDEG ranked in the top 15 of the up-regulated and down-regulated gene lists.
Table 1.
Genes affected by mFAE in the cerebral cortex
| Rank | Up-regulated |
Down-regulated |
||
| Gene symbol | WAD (m) | Gene symbol | WAD (m) | |
| 1 | ZIC1 | 0.92058 | SATB2 | −1.0327 |
| 2 | HSPA1A /// HSPA1B | 0.92018 | ZEB2 | −0.8069 |
| 3 | LOH11CR2A | 0.87649 | ZEB2 | −0.7446 |
| 4 | GNB1 | 0.86957 | BHLHB5 | −0.7002 |
| 5 | HSPA1A /// HSPA1B | 0.84755 | IMAGE: 5302168 | −0.6456 |
| 6 | LOC643167 /// RBM39 | 0.79163 | FAM49A | −0.6247 |
| 7 | HSPA1A /// HSPA1B | 0.75739 | JAKMIP1 | −0.6214 |
| 8 | ZIC1 | 0.71881 | DYNC1I1 | −0.6104 |
| 9 | PDLIM3 | 0.70814 | SFRP1 | −0.6098 |
| 10 | LOC643167 /// RBM39 | 0.70425 | ID2 /// ID2B | −0.608 |
| 11 | CALML4 | 0.64375 | IER5 | −0.5898 |
| 12 | DNAJC15 | 0.61996 | NR4A3 | −0.581 |
| 13 | HLA-B /// HLA-C | 0.58698 | GOLSYN | −0.5786 |
| 14 | HIST2H2BE | 0.56838 | FOXP1 | −0.5746 |
| 15 | HIST1H1C | 0.56202 | HIVEP2 | −0.5658 |
Top 15 CDEG (up- and down-regulated) according to the mouse WAD ranking. The list of CDEG was compared with the background list provided by Panther.
Biological Themes of the CDEG in Response to mFAE.
Biological annotation of the gene expression profiles changed by EE is expected to provide a prediction of potential biological events altered by EE. Using the DAVID database, we obtained the ranking of gene ontology (GO) terms enriched in the CDEG in response to mFAE (21) and visualized them as a hierarchical tree using GoSurfer software (22) (Fig. S2A). GO terms related to neural function and developmental events, such as cell migration and proliferation, as well as terms related to general cellular metabolism, were enriched. The list of GO generated by functional network analysis also included the terms related to nervous system development and function, cellular movement–cell interaction, and cell death–growth–proliferation as the most significantly altered biological events (Fig. S2B). We further sought the genes and signaling pathways that are potentially responsible for the cortical cellular phenotypes in response to mFAE. Among available analysis tools for the identification of enriched signaling pathways and biological themes, we used Panther, which encompasses the largest database of development-related signaling pathways and generates the maps of enriched genes in the signaling pathways in which they are involved (23) (Table 2 and Fig. S2 C and D). As shown in Table 2, a number of signaling pathways are listed as vulnerable to mFAE. By this analysis, we found that “TGF-β signaling pathway” is significantly altered (Table 2 and Fig. S2C). This finding is consistent with the previous observations in the developing rat cortex, which suggested that altered TGF-β signaling in response to mFAE may be involved in abnormal cell proliferation and neuronal migration (15, 24, 25). We also obtained a set of molecular clues to the potential defects in cortical development by mFAE, such as “PI3 kinase pathway” and “Notch signaling pathway” (Table 2). Many of these pathways, however, have pleiotropic functions, suggesting a complex nature of the impact of mFAE on cortical development. In the enriched gene lists (Dataset S1), we confirmed the reduction of DAB1, a critical regulator of neuronal migration (26–28) in both the human and mouse cortex by immunoblot analysis. Similarly, reduction of Neuropilin1, which is required for neuronal migration and axon guidance (29), was confirmed in the mouse cortex by immunohistochemistry (Fig. S3). Thus, it is likely that neuronal migration is one of the vulnerable processes to mFAE in cortical development.
Table 2.
Biological pathways enriched (uncorrected P < 0.05) in both human and mouse mFAE models were identified by Panther using human gene IDs
| Pathway | P value |
| PI3 kinase pathway | 0.00294 |
| Cortocotropin releasing factor receptor signaling pathway | 0.00383 |
| Wnt signaling pathway | 0.00408 |
| Apoptosis signaling pathway | 0.0047 |
| VEGF signaling pathway | 0.00528 |
| Angiogenesis | 0.00608 |
| EGF receptor signaling pathway | 0.0101 |
| Histamine H1 receptor-mediated signaling pathway | 0.0106 |
| Endothelin signaling pathway | 0.0134 |
| Glycolysis | 0.0164 |
| Notch signaling pathway | 0.0185 |
| Parkinson disease | 0.0189 |
| Histamine H2 receptor-mediated signaling pathway | 0.0198 |
| β3 adrenergic receptor signaling pathway | 0.0198 |
| Angiotensin II-stimulated signaling through G proteins and β-arrestin | 0.021 |
| T-cell activation | 0.023 |
| Fructose galactose metabolism | 0.0234 |
| B-cell activation | 0.028 |
| 5HT4 type receptor-mediated signaling pathway | 0.0297 |
| TSH-releasing hormone receptor signaling pathway | 0.0327 |
| TGF-β signaling pathway | 0.0332 |
| PDGF signaling pathway | 0.034 |
| Ras pathway | 0.0343 |
| Oxytocin receptor-mediated signaling pathway | 0.0344 |
| Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 0.0344 |
| Enkephalin release | 0.0367 |
| Interleukin signaling pathway | 0.0387 |
| Circadian clock system | 0.0396 |
| p53 pathway feedback loops 2 | 0.0416 |
| GABA-B_receptor_II_signaling | 0.0501 |
The enrichment score was generated using the binomial test. Analysis using mouse gene IDs also generated a list including the same pathways with slightly different ranking.
The affected genes/pathways also included those that are shared among a broad range of tissues and cell types. Of particular note, multiple probe sets of HSPA1A/1B (HSP70) and DNAJC15 (HSP40), members of heat shock proteins (HSPs), were ranked within the top 15 up-regulated genes (Table 1). Similarly, previous report that Heat Shock Factor1, a transcriptional regulator of HSPs, is activated in dissociated cortical neurons cultured with ethanol (30). HSPs are involved in a cellular defense pathway for promoting cell survival in response to various stress exposure, and our result suggests their involvement in cellular response to mFAE.
Increase of Notch and Wnt Signaling Activities by mFAE in the Cortex.
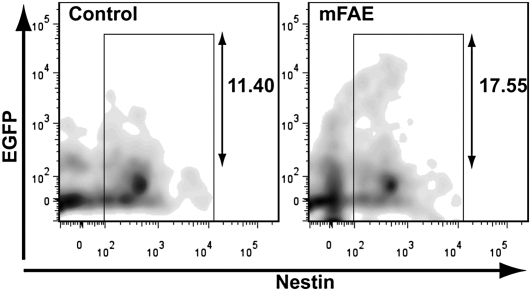
Wnt and Notch signaling pathways that play critical roles in regulating the proliferation and fate of neural stem cells (NSCs) (31, 32, 33) were also affected by mFAE (Table 2 and Fig. S2D). We further examined whether the terminal outputs of these signaling pathways are altered in vivo as a result of mFAE using Wnt (BAT-LacZ and Fos-LacZ) and Notch (Cp-GFP) reporter transgenic mouse lines. Flow cytometric analysis revealed that the percentages of reporter-expressing cells in Nestin+ NSCs were significantly increased in the cerebral cortex of both mouse lines at E16 (Fig. 2 and Fig. S4B), suggesting that the activation levels of canonical Wnt and Notch signaling in cortical NSCs are increased by mFAE.
Fig. 2.
Notch reporter analysis in the ethanol-exposed embryonic cortex. Flow cytometric analysis of the Notch signaling reporter activity in vivo. Cp-EGFP reporter transgenic mouse embryos were subjected to ethanol or PBS (control), and the numbers of EGFP+ and Nestin+ cortical cells were analyzed. The red numbers indicate the percentage of EGFP+ cells (in the range shown by two-headed arrows) in the total Nestin+ cells (within the boxed area). The percentage is increased by mFAE. Mean increase compared with the control is 30 ± 4.8% (SEM) (n = 16, P = 0.008 by t test).
Disturbance of Molecular Identities of Cortical Neuronal Subtypes by mFAE.
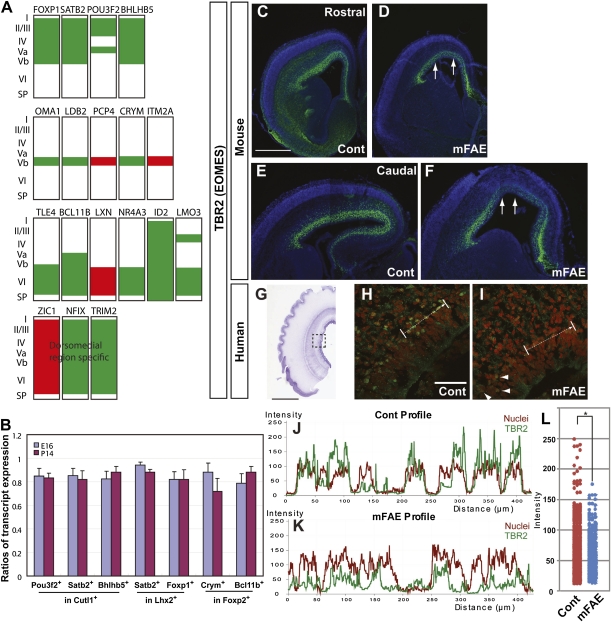
Among about 80 genes that are generally recognized to be expressed in layer- and/or area-specific neuronal subtypes in the developing neocortex (34–36), 18 genes (∼20%) were enriched in the up- or down-regulated gene lists. Strikingly, 14 of them (∼80%) including the 5 genes in the top 15 genes, SATB2, BHLHB5, ID2, NR4A3, and FOXP1, were all in the down-regulated gene list (Table 1 and Fig. 3A). Because many of these genes are known to play roles in the specification of cortical neuronal subtypes (37–40), our results suggest that reduction of their expression by mFAE might disturb the morphological and functional identities of specific neuronal subtypes. Immunoblot analysis showed less prominent but significant reduction of these gene products by mFAE in the entire cerebral cortex at E16. Immunohistochemistry, however, could not clearly discern the differences of their expression levels.
Fig. 3.
Molecular identities of neuronal subtypes were altered by mFAE. (A) Schematic diagrams showing up- (red) and down-regulation (green) of cortical layer and/or region-specific genes in the embryos exposed to ethanol. Expression patterns of the markers were depicted on the basis of previous reports (32–35). (B) The fractions of cells expressing the enriched (as altered expression by mFAE in microarray analysis) genes in cells expressing the reference nonenriched genes were calculated using the data obtained by flow cytometric analysis (Fig. S5C). The ratios of the corresponding fractions in mFAE vs. control-treated samples (set to 1.0) were presented for E16 and P14 (the mean ± SEM from the experiments using four dams per condition). P < 0.05 by Mann–Whitney u test. (C–I) Tbr2 immunohistochemistry (green) in the control (Cont) and ethanol-exposed E16 mouse (C–F) and GW17 human cortex (H and I). The nuclei were counterstained with DAPI (4',6-diamidino-2-phenylindole) (blue or red). Arrows in D and F indicate the regions that contain cells with weaker Tbr2 expression in rostral (C and D) and caudal (E and F) cortical domains. The dotted square in the thionin-stained human cortex (G) depicts the region presented at higher magnification in H and I. The wavy pial surface is an artifact introduced during the staining process. Arrows in H and I indicate the domain used to calculate the intensity of immunostaining shown in J and K. (J and K) Decreased intensity of TBR2 immunolabeling (green lines) concomitant with increased intensity of nuclear labeling (red lines) in neural precursors that show irregular distribution in the ethanol-exposed human cortex. (L) Dot plots of TBR2 labeling intensity in human samples. The labeling intensity was measured every 0.2 μm along the total length of 200 μm. P = 0.002 by Mann–Whitney u test. n = 8/8 mouse embryos from four dams, and 8/8 cultured slices from four human fetuses. Arrowheads in I indicate the cells showing relatively normal intensity of TBR2 immunolabeling in the region adjacent to the affected region. [Scale bars, 2 mm (C–F), 20 mm (G), and 0.2 mm (H and I).]
As an alternative method, we used flow cytometric analysis that enables unbiased counting of cells with combinatorial expression of these genes in the entire cortex. Satb2, Pou3f2, Bhlhb5, Foxp1, Bcl11b, and Crym were selected for the analysis from the enriched down-regulated gene list, on the basis of our immunolabeling results, which were consistent with the expression patterns available in public in situ hybridization databases (Fig. S5A, http://www.ncbi.nlm.nih.gov/projects/gensat/, http://www.brain-map.org/). After confirming the specificity of immunolabeling (Fig. S5B), we performed flow cytometric analysis of cortical cells dissociated from the control- and ethanol-exposed embryos at E16. As expected, the numbers of cells that express the enriched genes were all decreased by mFAE (Fig. S5C). We selected layer-specific neuronal markers, Cutl1 and Lhx2 (upper layers) and Foxp2 (lower layers) from the nonenriched gene list as the reference and analyzed the ratios of cells expressing the enriched genes to those expressing the reference genes. The ratios were significantly altered by mFAE. This analysis cannot distinguish whether the alteration is a result of altered number of cells or altered amount of transcript per cell. We also cannot completely exclude the possibility that the expression of reference gene is altered in inverse proportion to the possible alteration of the number of neurons within each layer, although it is not very likely. Nevertheless, the results suggest that the molecular identities of some neuronal subtypes in each cortical layer are disturbed by mFAE (Fig. 3B and Fig. S5C). Importantly, the analysis at P14 revealed that the discordance of molecular identities of neurons is not temporal, but persists long after mFAE (Fig. 3B and Fig. S5C). Another down-regulated gene in the enriched gene list was TBR2 (EOMES) (Dataset S1). The decrease of TBR2 expression by mFAE was consistently detectable by immunohistochemistry in both the human and mouse embryonic cortex (Fig. 3 C–L). As it is suggested that Tbr2 controls the production of upper cortical layer neurons (41, however, see ref. 42), its reduction by mFAE might contribute to the decrease of cells that express specific subtype markers in the upper layers.
Discussion
Before discussing the value of the combined rodent–human transcriptome analysis as a model system to study the response of the developing brain to EE, it is important to consider some constraints and technical issues involved in this approach.
One of the major problems in gene expression analysis is the discrepancy between mathematical and biological recognition of differential expression; i.e., mathematical tests permit the genes with arbitrarily small expression levels, regardless of biological meaningfulness (19, 43). The WAD metric considers both the average gene expression difference and relative average signal intensity, and gives higher ranks to the genes that are highly expressed on average over different conditions. Therefore, the WAD has a preference to provide biologically more significant sets of genes. Because potential biomarker genes generally have higher expression levels, this is useful for screening of biomarker-type genes expressed in tissues that contain cells with heterogeneous molecular properties, such as the cerebral cortex, as well as in tissues from heterogeneous genetic backgrounds, such as human specimens, because tiny expression changes due to the differences of cell contents and genetic backgrounds can be masked. The WAD does not take the variability of the datasets into account, unlike other traditional methods. Nevertheless, consistent with previous studies (19, 43), the WAD outperformed other methods with the greatest reproducibility and sensitivity as indicated by AUC in our study (Fig. 1), which had a limitation in the availability of pharmacologically treated human fetal tissues. Although cross-species transcriptome analysis using the WAD sacrifices the identification of genes with very low expression levels and genes that are secondarily affected by mFAE-induced damages in other tissues/organs (because of inclusion of an in vitro slice culture model), our study demonstrated that it serves as a powerful method with the use of human fetal tissues.
In the process of cross-species matching of probe sets, orthologous human probe sets were missing for 23% of mouse probe sets. Although this is partly because the orthologous genes do not exist in the human genome, the main reason for this is lack of corresponding probe sets of mouse genes on the human microarray. Nevertheless, the functional enrichment analysis using the Panther database allowed us to effectively identify the signaling pathways that are potential targets of mFAE. For example, although canonical target genes of Notch and Wnt signaling pathways, such as HES and AXIN, were not listed as CDEG partly because of the reason described above, we were able to identify these two signaling pathways as vulnerable targets of mFAE using Panther (Table 2).
Wnt signaling has been listed as an enriched pathway in the transcriptome analysis of the mouse headfold exposed to ethanol at earlier developmental stages, although whether its activity is increased or decreased is unknown (44). Our in vivo reporter assays revealed the overall increase of Wnt and Notch signaling activities in the cerebral cortex by mFAE (Fig. 2 and Fig. S4). This agrees with previous reports showing the effect of alcohol to maintain the self-renewal state of NSCs and increase gliogenesis concomitant with the decrease of neurogenesis (10, 31, 32, 45, 46), as all these effects are controlled by Wnt and/or Notch signaling activation (31, 32, 47). Reduced expression of TBR2, a marker for intermediate neuronal progenitors, by mFAE (Fig. 3), is also consistent with these results. A recent study using neurosphere cultures of the mouse fetal cortex reported an increase of Notch ligand Jagged-1 mRNA and the rate of NSC proliferation by the knockdown of specific micro-RNAs whose expression is suppressed by ethanol exposure (14). This mechanism might contribute to the increase of Notch signaling activity by mFAE. Regarding Wnt signaling, ZIC1 was listed as highly up-regulated CDEG (Table 1). Because Zic1 is a positive regulator of Wnt signaling and is suggested to function in the maintenance of neural precursor cells in an undifferentiated state (48), its up-regulation might underlie the increase of Wnt signaling activity by mFAE.
Our results revealed significant enrichment of the genes that are expressed in specific neuronal subtypes in the CDEG list. This indicates the disturbance of molecular identities of cortical neuronal subtypes by mFAE. Notably, the disturbed molecular identities were retained postnatally (Fig. 3B and Fig. S5C). Such persistent molecular disturbance can lead to morphological and functional defects of cortical neurons and might contribute to the neurological and psychiatric impairments in FASD patients, as well as people who were prenatally exposed to alcohol but were not diagnosed with FASD due to the lack of obvious anatomical abnormalities (8). Our study showed that most of these genes were down-regulated by mFAE (Fig. 3). Among them, SATB2 has a critical role in the specification of callosal projection neurons (39, 40) and ZEB2, when mutated, is known to cause Mowat-Wilson syndrome, which shows corpus callosum agenesis (37). Thus, the decrease of these genes might be responsible for the agenesis of corpus callosum in FASD subjects (49).
Several mechanisms are possibly involved in the alteration of molecular identities of neuronal subtypes by mFAE. Persistent transcriptional inhibition of specific genes via epigenetic genomic modification such as DNA methylation by alcohol exposure is one possible mechanism (50–52). Compromised production of some subtypes of neurons also might be involved. This is supported by finding of reduced TBR2 (Fig. 3), which presumably controls the production of upper layer neurons (41). The disruption of molecular identity in lower layers is unlikely due to altered neuronal production, because most lower layer neurons are produced before the embryos were exposed to ethanol at E14. Differential vulnerability of various neuronal subtypes to alcohol stress might be another mechanism (12, 53).
Collectively, the biological functions of identified genes/pathways/biological events as affected by mFAE well corresponded to a wide range of pathological features of FASD (54) as well as to the data obtained from animal models of ethanol's impact on brain development. Further analysis is required to reveal whether and how the altered gene regulation elicits phenotypes at the cellular level. A detailed future analysis at different time points and ethanol concentrations is also important for more faithful reproduction of ethanol's action in the pathogenesis of human FASD. Using ethanol as a model pharmacological agent, our study effectively identified significant initial pathological events that bridge the phenotypes in human diseases and the experimental data in animal models. The approach described here provides a framework for facilitating the findings of crucial biological pathways/signaling that are vulnerable to EE during human fetal development. Its broader application to chemical, biological, and physical stresses may help to decipher global molecular disturbances in environmentally induced human diseases.
Materials and Methods
Animal and Tissue Culture Models.
The embryonic human brain tissues were obtained from legal interruption of pregnancies. For the present study, we used six GW15-18 embryonic cortices that were obtained from the Human Fetal Tissue Repository at Albert Einstein College of Medicine. The research protocol was approved by the Yale Human Investigation Committee. All samples did not have medical histories of alcohol exposure during pregnancy. The tissues were stored in L-15 Leibovitz medium on ice and were cut at 300 μm using a tissue chopper within 3 h after the surgery. As a common problem in obtaining human fetal tissues, two brain samples out of six indicated the trace of damages as assessed by a large number of pyknotic cells and poor RNA integrity number (RIN) (Fig. S6). Because the medical histories displayed no linkage to abnormal and complicated phenotypic/functional defects of the fetuses, the damages were likely due to complications of the surgery. The remaining four samples that we used for the study all showed normal nuclear labeling as well as low level expressions of a cellular stress-inducible gene (HSP70) and an immediate early response gene (FOS) (Fig. S6). Each cortical slice was cultured on a membrane floating on the neurobasal medium containing 235 mg/dL ethanol or PBS (control) with supplements for 24 h in a 6-well tissue culture plate with a low evaporation lid (353046; Falcon). Sequential slices were put alternately into the plates containing exclusively either the ethanol-containing or control culture medium.
The pregnant mice were handled according to the protocols approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine. BAT-LacZ, FOS-LacZ, and Cp-GFP transgenic mice were purchased from The Jackson Laboratory (54–56) Wild-type CD-1 pregnant mice were purchased from Charles River. The pregnant mice were intraperitoneally injected with 25% ethanol (in PBS, final 2.0 g/kg weight) or PBS (control) once a day from E14 to E16. Three hours after the last injection, embryonic cortices were collected for analysis. No obvious effects of alcohol exposure were observed in gross morphology of the brains, except that very few embryos showed local cell migration defects in the cerebral cortex at E16 (3/28, control 0/33).
Cross-Species WAD Analysis and AUC Metric Analysis.
Microarray data were preprocessed using standard RMA algorithm in Partek Genomics Suite (Partek GS; Partek). Differential gene expression was analyzed using the WAD available as an R script (19) as well as other conventional methods for comparison. The CDEG across human and mouse were obtained using an R package, annotationTool (20) with HomoloGene (http://www.ncbi.nlm.nih.gov/homologene) and Ensembl (http://www.ensembl.org/index.html) databases. AUCs were obtained with the qRT-PCR data of 14 genes for each species (SI Materials and Methods) (18). All of the R scripts used in this work are available upon request.
CEL files used for this study are available at GEO (http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE23579).
Additional details are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. B. Poulos for human tissue acquisition, M. Torii and N. Sestan for their valuable comments, N. Iijima for technical advice, and A. Nakai for provision of antibodies. We also thank S. Ellis for technical assistance. This work was supported by the Kavli Institute for Neuroscience at Yale (K.H.-T. and P.R.), National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke NS038296-09 (to P.R.) and NIH/National Institute on Alcohol Abuse and Alcoholism K99AA018387 (to K.H.-T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE23579).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100903108/-/DCSupplemental.
References
- 1.Thompson BL, Levitt P, Stanwood GD. Prenatal exposure to drugs: Effects on brain development and implications for policy and education. Nat Rev Neurosci. 2009;10:303–312. doi: 10.1038/nrn2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakic P. Defects of neuronal migration and the pathogenesis of cortical malformations. Prog Brain Res. 1988;73:15–37. doi: 10.1016/s0079-6123(08)60494-x. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Ari Y. Neuro-archaeology: Pre-symptomatic architecture and signature of neurological disorders. Trends Neurosci. 2008;31:626–636. doi: 10.1016/j.tins.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35:528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakic P. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302:999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- 7.Riley EP, McGee CL. Fetal alcohol spectrum disorders: An overview with emphasis on changes in brain and behavior. Exp Biol Med (Maywood) 2005;230:357–365. doi: 10.1177/15353702-0323006-03. [DOI] [PubMed] [Google Scholar]
- 8.Levitt P. Developmental neurobiology and clinical disorders: Lost in translation? Neuron. 2005;46:407–412. doi: 10.1016/j.neuron.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Miller MW. Effects of alcohol on the generation and migration of cerebral cortical neurons. Science. 1986;233:1308–1311. doi: 10.1126/science.3749878. [DOI] [PubMed] [Google Scholar]
- 10.Miller MW, Nowakowski RS. Effect of prenatal exposure to ethanol on the cell cycle kinetics and growth fraction in the proliferative zones of fetal rat cerebral cortex. Alcohol Clin Exp Res. 1991;15:229–232. doi: 10.1111/j.1530-0277.1991.tb01861.x. [DOI] [PubMed] [Google Scholar]
- 11.Miller MW. Migration of cortical neurons is altered by gestational exposure to ethanol. Alcohol Clin Exp Res. 1993;17:304–314. doi: 10.1111/j.1530-0277.1993.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 12.Liesi P. Ethanol-exposed central neurons fail to migrate and undergo apoptosis. J Neurosci Res. 1997;48:439–448. doi: 10.1002/(sici)1097-4547(19970601)48:5<439::aid-jnr5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Mooney SM, Miller MW. Ethanol-induced neuronal death in organotypic cultures of rat cerebral cortex. Brain Res Dev Brain Res. 2003;147:135–141. doi: 10.1016/j.devbrainres.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: Evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powrozek TA, Miller MW. Ethanol affects transforming growth factor beta1-initiated signals: Cross-talking pathways in the developing rat cerebral wall. J Neurosci. 2009;29:9521–9533. doi: 10.1523/JNEUROSCI.2371-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: A review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- 17.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 18.McGee CL, Riley EP. Brain imaging and fetal alcohol spectrum disorders. Ann Ist Super Sanita. 2006;42:46–52. [PubMed] [Google Scholar]
- 19.Kadota K, Nakai Y, Shimizu K. A weighted average difference method for detecting differentially expressed genes from microarray data. Algorithms Mol Biol. 2008;3:8. doi: 10.1186/1748-7188-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn A, Luthi-Carter R, Delorenzi M. Cross-species and cross-platform gene expression studies with the Bioconductor-compliant R package ‘annotationTools’. BMC Bioinformatics. 2008;9:26. doi: 10.1186/1471-2105-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis G, Jr., et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 22.Zhong S, et al. GoSurfer: A graphical interactive tool for comparative analysis of large gene sets in Gene Ontology space. Appl Bioinformatics. 2004;3:261–264. doi: 10.2165/00822942-200403040-00009. [DOI] [PubMed] [Google Scholar]
- 23.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MW, Luo J. Effects of ethanol and transforming growth factor beta (TGF beta) on neuronal proliferation and nCAM expression. Alcohol Clin Exp Res. 2002;26:1281–1285. doi: 10.1097/01.ALC.0000026836.38681.58. [DOI] [PubMed] [Google Scholar]
- 25.Siegenthaler JA, Miller MW. Transforming growth factor beta1 modulates cell migration in rat cortex: Effects of ethanol. Cereb Cortex. 2004;14:791–802. doi: 10.1093/cercor/bhh039. [DOI] [PubMed] [Google Scholar]
- 26.Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 27.Sheldon M, et al. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 28.Olson EC, Kim S, Walsh CA. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J Neurosci. 2006;26:1767–1775. doi: 10.1523/JNEUROSCI.3000-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, et al. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11:36–44. doi: 10.1038/nn2018. [DOI] [PubMed] [Google Scholar]
- 30.Pignataro L, et al. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci. 2007;27:12957–12966. doi: 10.1523/JNEUROSCI.4142-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 32.Woodhead GJ, Mutch CA, Olson EC, Chenn A. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J Neurosci. 2006;26:12620–12630. doi: 10.1523/JNEUROSCI.3180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 34.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 35.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 36.Hevner RF. Layer-specific markers as probes for neuron type identity in human neocortex and malformations of cortical development. J Neuropathol Exp Neurol. 2007;66:101–109. doi: 10.1097/nen.0b013e3180301c06. [DOI] [PubMed] [Google Scholar]
- 37.Mowat DR, Wilson MJ, Goossens M. Mowat-Wilson syndrome. J Med Genet. 2003;40:305–310. doi: 10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi PS, et al. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 41.Arnold SJ, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kadota K, Nakai Y, Shimizu K. Ranking differentially expressed genes from Affymetrix gene expression data: Methods with reproducibility, sensitivity, and specificity. Algorithms Mol Biol. 2009;4:7. doi: 10.1186/1748-7188-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green ML, et al. Reprogramming of genetic networks during initiation of the fetal alcohol syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- 45.Miranda RC, Santillano DR, Camarillo C, Dohrman D. Modeling the impact of alcohol on cortical development in a dish: Strategies from mapping neural stem cell fate. Methods Mol Biol. 2008;447:151–168. doi: 10.1007/978-1-59745-242-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs JS, Miller MW. Proliferation and death of cultured fetal neocortical neurons: Effects of ethanol on the dynamics of cell growth. J Neurocytol. 2001;30:391–401. doi: 10.1023/a:1015013609424. [DOI] [PubMed] [Google Scholar]
- 47.Kalani MY, et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc Natl Acad Sci USA. 2008;105:16970–16975. doi: 10.1073/pnas.0808616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- 49.Riley EP, et al. Abnormalities of the corpus callosum in children prenatally exposed to alcohol. Alcohol Clin Exp Res. 1995;19:1198–1202. doi: 10.1111/j.1530-0277.1995.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 50.Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 51.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- 53.Miller MW, Hu H. Lability of neuronal lineage decisions is revealed by acute exposures to ethanol. Dev Neurosci. 2009;31:50–57. doi: 10.1159/000207493. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Guerri C, Bazinet A, Riley EP. Foetal Alcohol Spectrum Disorders and alterations in brain and behaviour. Alcohol Alcohol. 2009;44:108–114. doi: 10.1093/alcalc/agn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 56.Duncan AW, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.