Abstract
Ipl1/Aurora B is the catalytic subunit of a complex that is required for chromosome segregation and nuclear division. Before anaphase, Ipl1 localizes to kinetochores, where it is required to establish proper kinetochore–microtubule associations and regulate the spindle assembly checkpoint. The protein phosphatase Glc7/PP1 opposes Ipl1 for some of these activities. To more thoroughly characterize the Glc7 phosphatase that opposes Ipl1, we have identified mutations that suppress the thermosensitivity of an ipl1-2 mutant. In addition to mutations in genes previously associated with ipl1 suppression, we recovered a null mutant in TCO89, which encodes a subunit of the TOR complex 1 (TORC1), the conserved rapamycin-sensitive kinase activity that regulates cell growth in response to nutritional status. The temperature sensitivity of ipl1-2 can also be suppressed by null mutation of TOR1 or by administration of pharmacological TORC1 inhibitors, indicating that reduced TORC1 activity is responsible for the suppression. Suppression of the ipl1-2 growth defect is accompanied by increased fidelity of chromosome segregation and increased phosphorylation of the Ipl1 substrates histone H3 and Dam1. Nuclear Glc7 levels are reduced in a tco89 mutant, suggesting that TORC1 activity is required for the nuclear accumulation of Glc7. In addition, several mutant GLC7 alleles that suppress the temperature sensitivity of ipl1-2 exhibit negative synthetic genetic interactions with TORC1 mutants. Together, our results suggest that TORC1 positively regulates the Glc7 activity that opposes Ipl1 and provide a mechanism to tie nutritional status with mitotic regulation.
Ipl1/Aurora B, a conserved protein kinase whose activity is required for chromosome segregation and nuclear division, associates with three conserved proteins that constitute the chromosome passenger complex (CPC) (reviewed in ref. 1). During mitosis, this complex associates first with chromosomes and then with the inner centromeres between the two kinetochores before anaphase. At anaphase, the CPC migrates to the spindle midzone and to the cleavage furrow, where it has roles in spindle disassembly, chromosome decondensation, and cytokinesis (1). Before anaphase, Ipl1/Aurora B destabilizes microtubule–kinetochore interactions of mal-oriented chromosomes, chromosomes with an unoccupied kinetochore, or chromosomes on which both kinetochores associate with microtubules from the same pole. Chromosomes in ipl1 mutants fail to biorient on the mitotic spindle (2–4), and Aurora B is also required to correct mal-oriented chromosomes in vertebrate cells (5, 6). The signal for this correction mechanism appears to be the tension generated at bioriented kinetochores. Kineotochore proteins not under tension are phosphorylated by Ipl1/AuroraB, destablizing microtubule–kinetochore interactions and promoting microtubule turnover until proper biorientation is achieved (reviewed in ref. 7). This model is supported by recent in vitro studies showing that phosphorylation of a key kinetochore protein complex (Dam1) by Ipl1 reduces either its affinity for microtubules (8) or its association with the Ndc80 complex (9, 10), another key kinetochore complex.
Once under tension, Ipl1-dependent phosphorylation sites on Dam1 become dephosphorylated (11), indicating an active role for protein phosphatases. Experimental evidence indicates that the conserved type 1 protein phosphatase PP1 carries out this role. Mutations in the yeast PP1 gene, GLC7, suppress the temperature sensitivity of ipl1 mutants (12–14), activate the spindle assembly checkpoint (15, 16), and reduce the affinity of microtubules for kinetochore complexes (16). GLC7 mutations also enhance the phosphorylation state of the Ipl1 substrates, histone H3 (13), and Dam1 (14). PP1 also opposes Aurora B in metazoan systems (17).
PP1 is regulated by its interaction with a diverse group of PP1-binding proteins that target PP1 to specific subcellular locations, activate the catalytic subunit toward specific substrates, or inhibit its activity (18). Although two mammalian kinetochore proteins, KNL1 (19) and CENP-E (20), have recently been shown to have PP1-binding sites that are required for proper kinetochore function, the nature of the PP1/Glc7 activity that opposes Ipl1 in yeast is unclear. The Glc7-binding protein Fin1 helps target Glc7 to kinetochore proteins, and dysregulated Fin1 enhances the temperature sensitivity of an ipl1 mutant (21). However, a fin1 null mutant has no growth or chromosome segregation defect (22). Mutations in genes encoding the PP1-binding proteins Glc8/inhibitor 2 (23), Sds22 (24), Ypi1/Inh3 (25), and Shp1 (26) suppress the temperature sensitivity of ipl1 mutants, consistent with a model where they act with Glc7 to oppose Ipl1 phosphorylation. However, these proteins are not part of the kinetochore or spindle apparatus and may function, directly or indirectly, by regulating the nuclear accumulation of Glc7. Overexpression of two cytoplasmic Glc7-binding proteins, Gip3 and Gip4, suppresses the temperature sensitivity of an IPL1 mutant by titrating Glc7 from the nucleus (14).
To better understand the nature of the Glc7 activity that opposes Ipl1, we have carried out a genetic screen to isolate suppressors of ipl1-2, a well-characterized IPL1 mutant (27), and have uncovered a mutation in TCO89, which encodes a component of the TOR complex 1 (TORC1). We show here that reduced TORC1 activity suppresses defects in chromosome segregation in an ipl1-2 mutant, most likely by altering the activity of the opposing Glc7/PP1 phosphatase.
Results
Reduced TORC1 Activity Suppresses the Temperature Sensitivity of ipl1-2.
ipl1-2 strains grow at wild-type rates at 24 °C but fail to grow at 30 °C and above. To identify ipl1-2 suppressors, we subjected ipl1-2 strains to UV mutagenesis, identifying recessive revertants and performing tetrad analysis after crossing with an unmutagenized ipl1-2 strain, as described in SI Materials and Methods. In addition to predicted suppressor loci, including previously undescribed alleles of GLC7, SDS22, and YPI1, there was a recessive revertant (revertant 71) that showed a cold-sensitive phenotype, failing to grow at 14 °C. Subsequent tests revealed that revertant 71 is sensitive to 5 mM caffeine. The caffeine-sensitive suppressor of ipl1-2 in revertant 71 segregates as a single Mendelian allele (Fig. S1A), mapping ∼17 cM from IPL1 and very close to PPQ1 (Table S1), whose adjacent gene is TCO89, encoding a nonessential subunit of the TORC1 complex (28). TCO89 null mutants are reported to be hypersensitive to caffeine, suggesting that the cold-sensitive suppressor is an allele of TCO89. Sequence analysis of the TCO89 ORF from revertant 71 revealed multiple mutations that introduce four missense mutations into the N-terminal region of the protein and a stop codon at codon 645, deleting the last 156 amino acid residues from the Tco89 protein. Hereafter, we refer to this suppressor as tco89-71. Deletion of TCO89 confers an identical cold and caffeine-sensitive phenotype and suppresses the temperature sensitivity of ipl1-2 (Fig. S1B), indicating that loss of TCO89 function is responsible for the suppression phenotype.
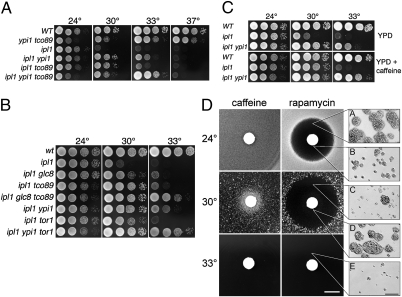
ipl1-2 mutant strains containing tco89-71 grow at 30 °C but not at higher temperatures (Fig. 1A). However, the ability of tco89-71 to suppress ipl1-2 is enhanced by additional weak suppressors of ipl1-2 in the Glc7 pathway. YPI1-GFP and glc8Δ each weakly suppress the temperature sensitivity of ipl1-2 (23, 25) but, together with tco89-71, allow ipl1-2 strains to grow well at 33 °C (Fig. 1A, row 6; Fig. 1B, row 5).
Fig. 1.
Reduction of TORC1 activity suppresses the temperature sensitivity of ipl1-2. (A) Cultures of WT (KT1113), YPI1-GFP tco89-71 (KT2998), ipl1-2 (KT1829), ipl1-2 YPI1-GFP (KT3007), ipl1-2 tco89-71 (KT3003), and ipl1-2 YPI1-GFP tco89-71 (KT3000) strains were serially diluted onto medium containing 1% yeast extract, 2% peptone, 2% glucose (YPD) and imaged after 40 h at the indicated temperatures. (B) Cultures of WT (KT1113), ipl1-2 (KT1829), ipl1-2 glc8Δ (KT3005), ipl1-2 tco89-71 (KT3003), ipl1-2 glc8Δ tco89-71 (KT3004), ipl1-2 YPI1-GFP tco89-71 (KT3007), ipl1-2 tor1Δ (KT3010), and ipl1-2 tor1Δ YPI1-GFP (KT3011) strains were treated as in A. (C) Cultures of WT (KT1113), ipl1-2 (KT1829), and ipl1-2 YPI1-GFP (KT3007) strains were serially diluted onto YPD medium and YPD containing 2.5 mM caffeine and treated as in A. (D) Diluted cultures of ipl1-2 (KT1829) were plated onto YPD medium, overlaid with a 7.5-mm paper disk containing 0.1 nM rapamycin or 2.5 μM caffeine, and incubated at the designated temperatures for 48 h (caffeine) or 72 h (rapamycin) before imaging. (Scale bar: 10 mm.) (Insets) Microscopic images were acquired from the designated locations on the plates after 72 h. (Scale bar: 200 μm.)
Tco89 is a nonessential component of the TORC1 complex (28). To determine if ipl1-2 suppression is due to a partial loss of TORC1 activity, we tested the ability of a tor1Δ mutation to suppress ipl1-2 thermosensitivity. tor1Δ mutants are viable because Tor1 and the related Tor2 have partially overlapping activities (29). As shown in Fig. 1B, tor1Δ ipl1-2 mutants grow at 30 °C, only slightly less well than tco89-71 ipl1-2 cells, and tor1Δ ipl1-2 YPI1-GFP cells grow at 33 °C. We also tested two pharmacological inhibitors of TORC1 for suppression: rapamycin, a specific inhibitor of TORC1 activity (30), and caffeine, whose predominant target in yeast also appears to be TORC1 (31, 32). As shown in Fig. 1C, caffeine suppresses the temperature sensitivity of ipl1-2 strains at 30 °C and enhances the suppression by YPI1-GFP at 33 °C. Rapamycin suppresses ipl1-2 but to a lesser extent than caffeine. The lower efficacy of rapamycin may be due to its extreme toxicity in our plate assay. To test this possibility, we placed filter discs impregnated with caffeine or rapamycin over a lawn of ipl1-2 cells and incubated the plates at permissive and restrictive temperatures. In the case of caffeine at 30 °C (Fig. 1D, Left panels), a halo of growing cells was observed near the caffeine source, as expected from the serial dilution growth tests in Fig. 1C. With rapamycin (Fig. 1D, Right panels), a large zone of inhibition was observed around the disk at 24 °C and 30 °C. Cells within the halo from the 24 °C plate grew into microcolonies (Fig. 1D, Insets A and B). Most cells within the halo from the 30 °C plate, but far from the disk, divided only a few times (Fig. 1D, Inset C), similar to cells grown at 33 °C (Fig. 1D, Inset E). In contrast, cells within the halo close to the disk (within 5 mm) grew into microcolonies (Fig. 1D, Inset D). Thus, rapamycin at high concentration suppresses the rapid cell death caused by the ipl1-2 mutation. However, as for tor1Δ and tco89-71, no suppression was observed at 33 °C. Together, these results indicate that a reduction in TORC1 activity suppresses the temperature-sensitive growth of ipl1-2.
TORC1 activity is required for many aspects of cell growth, including protein synthesis, ribosome biogenesis, and nutrient uptake. One simple explanation for the ipl1-2 suppression at 30 °C is that the slower growth rate conferred by tco89-71 lengthens the cell cycle and allows proper bipolar chromosome attachment on the mitotic spindle. Therefore, we tested whether ipl1-2 mutants can grow at nonpermissive temperatures on media that confer slower growth. However, we found no evidence for suppression of thermosensitive growth on rich media (1% yeast extract, 2% peptone) containing 0.1% glucose, 2% galactose, 2% ethanol, and 3% glycerol or on synthetic medium containing 2% glucose. Thus, the effect of TORC1 reduction on ipl1-2 mutants is unlikely to be caused simply by slower growth due to reduced TORC1 activity.
TCO89 Null Mutations Suppress Chromosome Loss in ipl1-2 Mutant Cells.
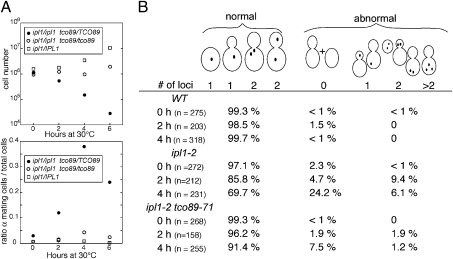
A key role of Ipl1 is to phosphorylate kinetochore proteins to ensure proper bipolar attachment of chromosomes to the mitotic spindle. Loss of Ipl1 activity results in improper chromosome attachments and increased rates of chromosome loss. To determine if a TORC1 deficiency directly suppresses chromosome loss in the ipl1-2 mutant, we first monitored rates of chromosome III loss in diploid strains homozygous for both ipl1-2 and tco89-71. Diploid yeast strains monosomic for chromosome III are able to mate due to the loss of heterozygosity of the MAT locus on chromosome III. Therefore, chromosome III loss can be assessed by the acquisition of mating ability. Diploid yeast strains were grown at 24 °C to midlog phase and shifted to 30 °C. Cell viability and mating ability were assayed after the temperature shift. In the ipl1-2 strain with wild-type TCO89, loss of cell viability and increase in mating ability were evident after 2 h at 30 °C. After 4 h at 30 °C, cell viability dropped 10-fold, and cells with mating ability increased 10-fold (Fig. 2A). In contrast, the viability and mating ability of the homozygous tco89-71 ipl1-2 strain did not change following the temperature shift.
Fig. 2.
The tco89-71 mutation suppresses the chromosome loss defect due to ipl1-2. (A) WT (KT113 × KT1963), ipl1-2 (KT1963 × KT2941), and ipl1-2 tco89-71 (KT2941 × KT3002) diploid strains were grown to log phase in YPD medium at 24 °C, shifted to 30 °C for the designated times, and assayed for cell viability (Upper) and mating ability (Lower) at each time point. (B) Distribution of GFP-tagged chromosome IV in wild-type (KT2088), ipl1-2 (KT3031), and ipl1-2 tco89-71 (KT3030) cells after incubation for 2 and 4 h at 30 °C. Cells were grouped into those with a normal fluorescent pattern and those with an abnormal pattern representing loss or missegregation of chromosome IV or chromosome XV, which contains the GFP-tagged lacI gene.
We also assayed for loss of chromosome IV that was tagged with a lacO array and visualized with GFP-LacI (33). To quantify chromosome loss, we distinguished between cells with a normal fluorescent pattern and those with an abnormal pattern or no fluorescence. Loss of fluorescence is due to loss of the fluorescently tagged chromosome IV or loss of chromosome XV, which encodes the GFP-tagged lac repressor. Wild-type, ipl1-2, and ipl1-2 tco89-71 strains were grown to midlog phase at 24 °C and then shifted to 30 °C for 2 or 4 h. After 4 h at 30 °C, over 30% of the ipl1-2 cells contained either no fluorescent chromosomes or an abnormal arrangement of chromosomes (Fig. 2B). In contrast, less than 9% of ipl1-2 tco89-71 cells had an abnormal fluorescent pattern. Together, these results indicate that ipl1-2 strains lose chromosomes rapidly at 30 °C and that loss of TORC1 activity suppresses this defect.
TCO89 Null Mutations Partially Restore Ipl1-Dependent Phosphorylation in ipl-2 Mutant Cells.
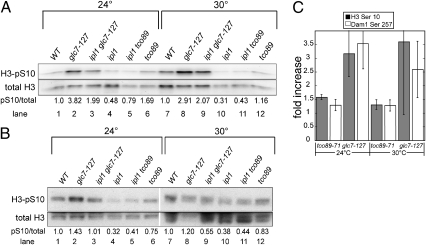
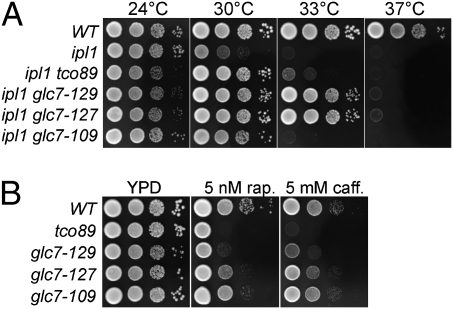
The suppression of ipl1-2 mutations by TORC1 deficiency could be due to a partial restoration of Ipl1 substrate phosphorylation, either by stimulation of Ipl1 activity or inhibition of Glc7 phosphatase activity. Alternatively, TORC1 could act on another component of the kinetochore or spindle to suppress ipl1-2. For example, mutations in the Set1 methyltransferase also suppress the temperature sensitivity of ipl1-2, most likely by reducing Dam1 methylation (34). To distinguish between these alternatives, we assayed phosphorylation levels on histone H3 Ser10 and Dam1 Ser257, known Ipl1 substrates (11, 13). Immunoblot analysis on whole-cell extracts using phosphospecific antibodies to H3 Ser10 and Dam1 Ser257 was used to assay phosphorylation levels in ipl1-2 cells (Fig. 3 A and B). As a control, we assayed phospho H3 Ser10 levels in a glc7-127 mutant, which was previously shown to restore phospho H3 Ser10 levels in an ipl1-2 mutant background (13). Although we observed a high level of variability between experiments in every case (three separate experiments for H3 Ser10 and four for Dam1Ser257), the tco89-71 mutation raised the phosphorylation levels of H3 Ser10 and Dam1 Ser257 at both 24 °C and 30 °C, relative to the ipl1-2 strain (Fig. 3C). The glc7-127 mutation is more effective at increasing phosphorylation of both substrates. This is in line with the observation that glc7-127 is a much better suppressor than tco89-71, restoring growth of an ipl1-2 strain to 33 °C (Fig. 4A) (13). These results indicate that reduced TORC1 levels suppress ipl1-2 by partially restoring Ipl1 substrate phosphorylation and suggest that TORC1 is acting directly or indirectly on Ipl1 or Glc7.
Fig. 3.
The tco89-71 mutation partially restores phosphorylation of Ipl1 substrates. (A) Immunoblot analysis of WT (KT1113), glc7-127 (KT1969), glc7-127 ipl1-2 (KT1968), ipl1-2 (KT1829), tco89-71 ipl1-2 (KT3003), and tco89-71 (KT2959) extracts with phospho-specific antibody to histone H3 serine 10 and with antibody to total histone H3. (B) Dam1 phosphorylation on Ser257 of WT (KT1113), glc7-127 (KT1969), glc7-127 ipl1-2 (KT1968), ipl1-2 (KT1829), tco89-71 ipl1-2 (KT3003), and tco89-71 (KT2959) extracts was examined by immunoblotting with anti-S257 P and the overall level of Dam1 was examined using anti-Dam1. The ratio of the signal from the anti-S257 P antibody to the anti-Dam1 antibody is presented below each lane in A and B. (C) The fold increase in phosphorylation of H3 Ser10 and Dam1 Ser257 in extracts from ipl1-2 tco89-71 and ipl1-2 glc7-127 strains over extracts from ipl1-2 strains. The results represent the average of three separate experiments for histone H3 and four separate experiments for Dam1. Error bars, SD.
Fig. 4.
Correlation between suppression of ipl-2 and sensitivity to TORC1 depletion. (A) Cultures of WT (KT1112), ipl1-2 (KT1963), ipl1-2 tco89-71 (KT3002), ipl1-2 glc7-129, (KT3076), ipl1-2 glc7-127 (KT1965), and ipl1-2 glc7-109 (KT1964) strains were serially diluted onto YPD medium and imaged after 40 h at the indicated temperatures. (B) Cultures of WT (KT1112), tco89-71 (KT2958), glc7-129 (KT1774), glc7-127 (KT1967), and glc7-109 (KT1966) strains were serially diluted onto YPD medium containing 5 nM rapamycin or 5 mM caffeine and imaged as in A.
Loss of TORC1 Activity Reduces Nuclear Accumulation of Glc7.
Mutations in GLC7 that confer mitotic defects and suppress the temperature sensitivity of ipl1-2 show strong genetic interactions with mutations in other components of the Glc7 pathway (25). If TORC1 regulates the activity of Glc7, then we would expect to observe similar genetic interactions between such GLC7 alleles and mutations in components of TORC1. To test this, we crossed tco89-71 and glc7 mutants and characterized the double mutants following tetrad analysis. We found that glc7-127 and glc7-129, which suppress the temperature sensitivity of ipl1-2 at 33 °C (13), genetically interact with tco89-71; that glc7-127 tco89-71 mutants are slower growing than either single mutant; and that glc7-129 tco89-71 mutants are inviable, germinating but failing to grow into macrocolonies. In contrast, the glc7-109 mutation, which suppresses ipl1-2 weakly (Fig. 4A), exhibits no obvious genetic interaction with tco89-71. A similar relationship was observed for the growth rate of GLC7 mutants treated with rapamycin and caffeine (Fig. 4B). The glc7-129 mutant is the most sensitive to caffeine and rapamycin, glc7-127 confers intermediate sensitivity, and glc7-109 cells show the least sensitivity. We also note that YPI1-GFP tco89-71 mutants grow more slowly than either single mutant (Fig. 1A).
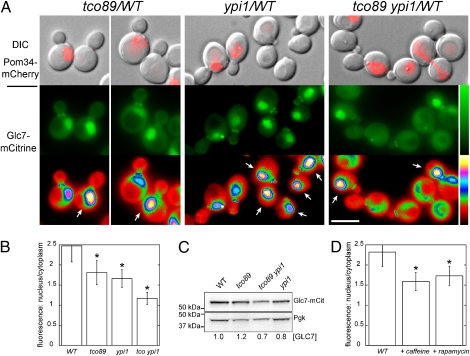
Mutations in the GLC7 pathway that suppress the temperature sensitivity of ipl1 mutants do so in many cases by reducing nuclear levels of Glc7 (14, 24–26). We therefore monitored the subcellular localization of functional, chromosomally encoded Glc7-mCitrine in tco89-71 and YPI1-GFP mutants (Fig. 5A). Glc7 is predominantly located in the nucleus, at the spindle pole bodies during anaphase, and at the bud neck (35). The tco89-71 mutation results in a significant reduction in the levels of nuclear Glc7-mCitrine fluorescence (Fig. 5A). As reported (25), the YPI1-GFP allele also causes a reduction in nuclear Glc7-mCitrine fluorescence and results in bright fluorescent puncta in many cells. In tco89-71 YPI1-GFP cells, nuclear Glc7-mCitrine levels are reduced further, to the point where it was difficult to distinguish the nuclear compartment by mCitrine fluorescence. Quantitation of Glc7-mCitrine fluorescence in the nucleus and the cytoplasm revealed a significant reduction (P < 10−21) in the ratio of nuclear-to-cytoplasmic Glc7-mCitrine in the mutants (Fig. 5B). Ypi1-GFP fluorescence does not contribute to the Glc7-mCitrine signal because Ypi1-GFP is expressed at low levels (25) and GFP fluorescence from YPI1-GFP cells is at the level of background autofluorescence (Fig. S2). Total Glc7-mCitrine levels in whole-cell extracts from the four strains were similar, as assayed by immunoblot (Fig. 5C), indicating that tco89-71 does not greatly influence total levels of Glc7-mCitrine.
Fig. 5.
Tco89 is required for the normal nuclear localization of Glc7. (A) Subcellular distribution of Glc7-mCitrine in WT (KT3252), tco89-71 (KT3245), YPI1-GFP (KT3247), and tco89-71 YPI1-GFP (KT3250) strains at log phase in YPD medium grown at 30 °C. WT and mutant cells were imaged in the same field and distinguished by Pom34-mCherry fluorescence in the mutant cells. (Top panels) Fluorescence images of Pom34-mCherry overlaid on differential interference contrast (DIC) images. (Bottom and Middle panels) Fluorescence images of Glc7-mCitrine. The images in the lowest panel are displayed using a six-shade look-up table to better distinguish fluorescence levels between WT and mutant cells. (Scale bar: 5 μm.) (B) Quantitative analysis of nuclear and cytoplasmic Glc7-mCitrine fluorescence of cells (n ≥ 67 cells) imaged in A. The ratio of nuclear-to-cytoplasmic fluorescence is presented for each panel in A. Error bars, SD. Significantly different levels from WT, *P < 10−20, according to the unpaired Student's t test. (C) Immunoblot analysis with anti-GFP antibody of extracts from strains imaged in A. Glc7-mCitrine levels are relative to the loading control (Pgk1) and normalized to the WT. (D) Quantitative analysis of nuclear and cytoplasmic Glc7-mCitrine fluorescence in WT cells (KT3242) after treatment with 10 mM caffeine or 10 nM rapamycin. n ≥ 50 cells. Error bars, SD. Significantly different levels from YPD-grown control, *P < 10−19, according to the unpaired Student's t test.
We also quantified nuclear and cytoplasmic Glc7-mCitrine fluorescence after a 2-h treatment of cells with 10 mM caffeine or 10 nM rapamycin. The ratio of nuclear-to-cytoplasmic fluorescence was reduced by these treatments (Fig. 5D), supporting the hypothesis that TORC1 is a positive regulator of Glc7 and is required for maintaining the proper levels of Glc7 in the nucleus.
Gip4 and Gip3 are cytosolic Glc7-binding proteins whose overexpression suppresses the temperature sensitivity of an IPL1 mutant by redistributing Glc7 from the nucleus to the cytoplasm (14). Gip4 is also hyperphosphorylated in cells treated with cycloheximide (36), a treatment that hyperactivates TORC1 (37). To determine if Gip4 mediates our TORC1-dependent response, we assayed the ability of the tco89-71 mutation to suppress ipl1-2 temperature sensitivity in a gip4Δ mutant background. However, we observed no influence of the gip4Δ mutation (or gip3Δ gip4Δ) on suppression of ipl1-2 by tco89-71 (Fig. S3).
Discussion
Reduction in TORC1 activity caused by loss of either nonessential subunit Tco89 or Tor1, or by the pharmacological inhibitors caffeine and rapamycin, suppresses the temperature sensitivity of ipl1-2. Partial restoration of histone H3 Ser10 and Dam1 Ser257 phosphorylation in ipl-2 mutant cells by the tco89 mutation suggests that TORC1 acts as either an inhibitor of Ipl1 activity or an activator of Glc7. Our data showing that nuclear Glc7 levels are reduced under conditions of lower TORC1 activity and the genetic interactions between the Glc7 and TORC1 pathways support the latter hypothesis. The phosphoprotein phosphatases PP2A and Sit4 are major downstream targets of TORC1 (38). Our work now provides evidence that PP1/Glc7 is also regulated by TORC1.
TORC1 regulates many aspects of cell growth, including protein synthesis, nutrient uptake, and autophagy in response to nutritional stress. It is generally believed that nutritional signals such as amino acid availability regulate TORC1 activity. Downstream targets include RNA polymerase subunits and transcription factors regulating nutrient uptake, ribosome biogenesis, and autophagy (reviewed in ref. 29). In addition to Gip4, which does not appear to be the relevant target here, we note two Glc7 regulators (Shp1 and Reg1) among the TORC1 targets identified by screening the rapamycin-sensitive phosphoproteome by mass spectrometry (36, 39). Shp1 is hyperphosphorylated at Ser106 and Ser108 following rapamycin treatment (36). Shp1, a cofactor for the AAA-ATPase Cdc48, was originally identified by a mutation that suppresses the lethality caused by overexpression of Glc7 (40). Cdc48, like its metazoan ortholog p97, acts in diverse physiological pathways, including membrane fusion and protein degradation (reviewed in ref. 41). Recently, Cdc48 and Shp1 were shown to be necessary for the nuclear accumulation of Glc7. More importantly, Shp1 depletion suppresses the temperature sensitivity of ipl1-321 (26). Interestingly, CDC48/p97 has also been found to inactivate Aurora B in vertebrate cells by extracting Aurora B from chromosomes at the end of anaphase (42). Together, these results suggest that Cdc48 and its cofactor Shp1 may regulate Ipl1 substrate phosphorylation by regulating either Glc7 or Ipl1. SHP1 is an essential gene in our background, precluding a simple genetic assay to determine if Shp1 and Cdc48 act downstream of TORC1 to regulate Glc7 activity, but it will be an important target for future studies.
Reg1 phosphorylation has been reported to be reduced at Ser572 and Ser576 (36) or S570 (39) upon rapamycin treatment. Reg1 is thought to negatively regulate Snf1, the yeast ortholog of AMP-activated protein kinase, by targeting Glc7-dependent dephosphorylation of Thr210 (43). Snf1 is required for growth on nonpreferred carbon sources and for response to other forms of cellular stress (reviewed in ref. 44). It remains to be determined if phosphorylation of Reg1 influences Ipl1 substrate phosphorylation.
In conclusion, we show here that down-regulation of TORC1 can suppress the temperature sensitivity of an IPL1 mutant by restoring Ipl1 substrate phosphorylation, most likely by inhibition of the opposing Glc7/PP1 activity. Because many of the components of these regulatory pathways are evolutionarily conserved, it will be important to determine if this regulation occurs in metazoans. Song et al. (45) reported that Aurora B and mammalian TOR together regulate the G1-S checkpoint in T lymphocytes. The authors observed the association of the two in immune complexes and found that rapamycin inhibits Aurora B kinase activity. However, this connection is outside the established role for Aurora B in regulating mitosis. The proposed mechanism is also the opposite from what we observe: that reduced TORC1 activity suppresses defects caused by mutations in IPL1. Generally, TORC1 has been shown to be a key regulator of cell growth, regulating protein synthesis, ribosome biogenesis, and autophagy in response to growth factors, nutritional cues, and other cell stresses. Our work now ties TORC1 to the regulation of chromosome segregation and provides a mechanism to integrate nutritional signals with chromosome segregation and mitosis. Perhaps during nutritional deprivation, when TORC1 activity is reduced, Ipl1/AuroraB activity can be effectively increased by decreasing nuclear Glc7 phosphatase activity. Our findings provide an effective means of tying cell growth control to regulatory aspects of chromosome segregation.
Materials and Methods
The yeast strains used in this work are listed in Table S2 and are congenic to strain JC482 (46). Microscopy data in Fig. 5 were acquired as described in SI Materials and Methods. For quantitative analysis of nuclear and cytoplasmic fluorescence in Fig. 4 B and D, images in five focal planes, 0.5 μm apart, were flattened into 2D projections and nine adjacent pixels were quantitated as described in SI Materials and Methods.
Yeast media, mutagenesis, genetics, immunoblot analysis, and microscopy are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Shile Huang for reagents and reading the manuscript, Doug Kellogg for advice on protein purification, Jennifer Larson for reading the manuscript, and Sue Biggins for helpful discussion. This work was supported by National Science Foundation Grant MCB-0517204 (to L.C.R.) and by Biotechnology and Biological Sciences Research Council Grant BB/G003440/1 (to M.J.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014406108/-/DCSupplemental.
References
- 1.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: Conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 2.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 3.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 5.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 6.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka TU, Stark MJ, Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat Rev Mol Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- 8.Gestaut DR, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by Aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci. 2009;122:4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francisco L, Wang W, Chan CSM. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu J-Y, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 14.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloecher A, Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sassoon I, et al. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emanuele MJ, et al. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009;183:1591–1595. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- 23.Tung HY, Wang W, Chan CS. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol Cell Biol. 1995;15:6064–6074. doi: 10.1128/mcb.15.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peggie MW, et al. Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J Cell Sci. 2002;115:195–206. doi: 10.1242/jcs.115.1.195. [DOI] [PubMed] [Google Scholar]
- 25.Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1032–1045. doi: 10.1091/mbc.E07-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng YL, Chen RH. The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J Cell Sci. 2010;123:2025–2034. doi: 10.1242/jcs.066043. [DOI] [PubMed] [Google Scholar]
- 27.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinke A, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 29.Soulard A, Cohen A, Hall MN. TOR signaling in invertebrates. Curr Opin Cell Biol. 2009;21:825–836. doi: 10.1016/j.ceb.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 31.Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 2006;281:31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- 32.Wanke V, et al. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- 33.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: Anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloecher A, Tatchell K. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Biol. 2000;149:125–140. doi: 10.1083/jcb.149.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber A, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 38.De Virgilio C, Loewith R. Cell growth control: Little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 39.Soulard A, et al. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Guha S, Volkert FC. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol Cell Biol. 1995;15:2037–2050. doi: 10.1128/mcb.15.4.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jentsch S, Rumpf S. Cdc48 (p97): A “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Ramadan K, et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 43.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 44.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song J, Salek-Ardakani S, So T, Croft M. The kinases aurora B and mTOR regulate the G1-S cell cycle progression of T lymphocytes. Nat Immunol. 2007;8:64–73. doi: 10.1038/ni1413. [DOI] [PubMed] [Google Scholar]
- 46.Cannon JF, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.