Abstract
Microtubules are integral to neuronal development and function. They endow cells with polarity, shape, and structure, and their extensive surface area provides substrates for intracellular trafficking and scaffolds for signaling molecules. Consequently, microtubule polymerization dynamics affect not only structural features of the cell but also the subcellular localization of proteins that can trigger intracellular signaling events. In the nematode Caenorhabditis elegans, the processes of touch receptor neurons are filled with a bundle of specialized large-diameter microtubules. We find that conditions that disrupt these microtubules (loss of either the MEC-7 β-tubulin or MEC-12 α-tubulin or growth in 1 mM colchicine) cause a general reduction in touch receptor neuron (TRN) protein levels. This reduction requires a p38 MAPK pathway (DLK-1, MKK-4, and PMK-3) and the transcription factor CEBP-1. Cells may use this feedback pathway that couples microtubule state and MAPK activation to regulate cellular functions.
Keywords: kinase, gene regulation
The state of microtubule polymerization varies from cell to cell, within cells, and at various times throughout the cell cycle (1). Localized microtubule depolymerization, for example, is associated with neurite outgrowth and synapse formation (2), and differences in cell shape correspond to differences in the amount and structure of microtubules (1). Given the many proteins that are associated with these cytoskeletal elements (3), their effective concentrations in the cytoplasm should also change with these conditions, potentially having a great impact on multiple cellular functions.
The touch receptor neurons (TRNs) of the nematode Caenorhabditis elegans provide a striking model for studying the contribution of microtubules to these functions. Only the six TRNs (ALML/ALMR, AVM, PLML/PLMR, and PVM), which enable the worm to sense touch to the body (4), have 15-protofilament microtubules. These microtubules associate in bundles and fill the neurite processes (5, 6). These large-diameter microtubules can be eliminated by loss-of-function mutations in the β-tubulin gene mec-7 (4, 7) or α-tubulin gene mec-12 (8, 9). The resulting cells have the 11-protofilament microtubules seen in other C. elegans neurons (6, 8) but cannot sense touch and have additional defects in axonal transport and synapse formation (10, 11). All stable microtubules can be eliminated using mec-7 or mec-12 gain-of-function mutations, which presumably interfere with general microtubule polymerization and result in greater transport defects (10). The more severe effect on microtubules can also be mimicked by growth of the animals on 1 mM colchicine, which selectively disrupts all of the microtubules in these cells after the animals have hatched (6, 10).
In this study, we used genetic and pharmacological approaches to disrupt TRN microtubules and found that these manipulations decrease general protein production in these neurons. This reduction can be suppressed by mutations affecting the PMK-3 p38 MAPK (mitogen-activated protein kinase) pathway. This pathway regulates synapse formation (12), axon termination (13), and neurite regeneration (14) in C. elegans. Because microtubule disruption can affect all three processes, microtubule-dependent signal transduction of gene activity through this MAPK kinase pathway may regulate these activities.
Results
Loss of Microtubules Results in a Reduction of TRN Gene Expression.
TRN differentiation, including the expression of the MEC-7 and MEC-12 tubulins, requires the LIM homeodomain transcription factor MEC-3 (15–17). While studying expression of GFP from the mec-3 promoter, we discovered the unexpected result that several mutations in mec-7 and mec-12 decreased GFP fluorescence (for a summary of genes and mutant alleles used in this paper, see Table 1). This decrease occurred in a time-dependent manner and was apparent 24 h after hatching, i.e., during the third of the four larval stages (Fig. 1A and Fig. S1A). Although the reduction of GFP fluorescence varied among the TRNs (and from animal to animal), ALM neurons demonstrated the most consistent decrease. In addition, animals with mec-7 and mec-12 gain-of-function mutations and wild-type animals grown on 1 mM colchicine generally suffered a greater reduction in GFP fluorescence than animals with putative loss-of-function mutations.
Table 1.
Genes and mutant alleles used in this study
| Gene | Description | Mutant alleles |
| mec-3 | Transcription factor required for TRN specification | e1338 |
| Tubulin genes | ||
| mec-7 | β-tubulin | Loss-of-function: u142, u440, u443; |
| Gain-of-function: u18, u283 | ||
| mec-12 | α-tubulin | Loss-of-function: e1607 (putative) |
| Gain-of-function: u63†, u241 | ||
| Partial loss-of-function: e1605* | ||
| TRN marker genes | ||
| mec-2 | Auxillary channel protein | NA |
| mec-17 | TRN-specific enzyme (acetytransferase) | NA |
| mec-18 | TRN-specific enzyme (luciferase) | NA |
| unc-119 | Pan-neural protein | NA |
| Cre genes | ||
| dlk-1 | Dual leucine zipper kinase/MAP kinase kinase kinase | km12, u815-u818, u820, u821 |
| mkk-4 | MAP kinase kinase | ju91, ok1545 |
| pmk-3 | p38 MAP kinase | ok169 |
| cebp-1 | Basic region leucine zipper transcription factor | tm2087, u819 |
*Recessive mutation that does not eliminate 15-protofilament microtubules.
†Recessive gain-of-function mutation.
Fig. 1.
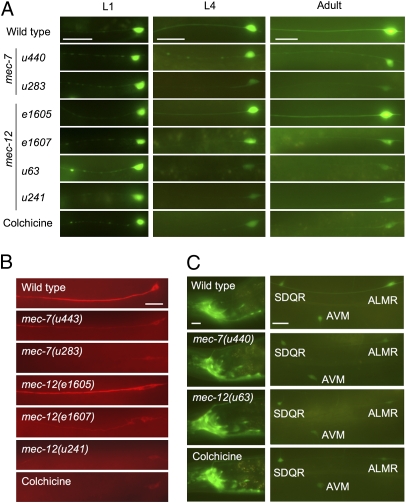
Protein levels are reduced in TRNs when microtubules are disrupted. (A) Fluorescence in Pmec-3gfp-expressing ALM neurons in L1 larvae, L4 larvae, and adults 48 h after hatching is reduced by mutation of mec-7 or mec-12 or growth in 1 mM colchicine. The mec-12(e1605) is an exception. (B) MEC-18 immunoreactivity is similarly reduced in adult ALM neurons. (C) Similar conditions reduce fluorescence from Punc-119gfp in the ALM and AVM TRNs but not in other cells (e.g., SDQ and nerve ring neurons). Nerve ring (Left); anterior midbody (Right). (Scale bars, 20 μm.)
Loss of function mutations in other mechanosensory genes expressed in these cells (mec-1, mec-4, mec-5, mec-6, mec-8, and mec-9) did not cause a reduction of this fluorescence (Fig. S2). Furthermore, animals with the mec-12(e1605) missense mutation, which are touch insensitive but retain 15-protofilament microtubules in the TRNs (10), also retained normal GFP levels (Fig. 1A and Fig. S1A). These results indicate microtubule disruption and not touch insensitivity alone causes the decrease of expression and suggest that the microtubules have a specific role in maintaining protein concentrations.
Microtubule disruption reduced the steady-state levels of many TRN proteins, including the mec-3–dependent expression (16, 17) of the endogenous proteins MEC-18 (Fig. 1B) and MEC-2 (Fig. S3) as well as a MEC-17::GFP fusion (16) (Fig. S4). MEC-18 and MEC-17 are diffusely expressed at high levels throughout the TRN process (16, 18, 19). MEC-2 is a component of the mechanoreceptor channel complex that is localized in regular puncta throughout the TRN process (20, 21). Transport of MEC-2 from the cell body to these puncta is disrupted in mec-7 and mec-12 mutants and animals treated with colchicine (10, 20), but levels of MEC-2 in the cell body of these animals did not significantly increase (mec-7 mutants are shown in Fig. S3). These results suggest that disrupting microtubules affects both the distribution and the expression of the MEC-2 protein.
The decrease of protein levels was not limited to mec-3–dependent or even TRN-specific genes. The unc-119 promoter is expressed in all nerve cells (22); its expression in the TRNs is mec-3 independent (Fig. S5). Nonetheless, GFP expressed from the unc-119 promoter was also reduced in the TRNs in mec-7 and mec-12 mutants and animals grown on colchicine plates (Fig. 1C and Fig. S1B). The level of GFP in other neurons was not affected.
This reduction of protein expression is due, at least in part, to a reduction in steady-state levels of mRNA. mec-3 transcript levels were significantly reduced in mec-7 and mec-12 mutants as well as colchicine-treated animals (Fig. S6A) as were the steady-state mRNA levels for mec-2, mec-17, and mec-18 (Fig. S6B). Because MEC-3 protein regulates it own transcription as well as that of the other genes (15, 16), we cannot rule out the possibility that all of the effects involve the regulation of protein levels. Unfortunately, we cannot test whether other genes are regulated at the level of transcription, because no gene is known that is both mec-3 independent and selectively expressed in the TRNs. However, because mec-17 mRNA reduction is minimal in colchicine-treated animals, but MEC-17::GFP protein levels were significantly reduced (Fig. S4), posttranscriptional effects of microtubule disruption definitely occur.
The level of reduction of mRNA transcripts varied for different genes. Differential reduction might reflect different affinities of MEC-3 binding at specific promoters; the reduction would be less for promoters with greater affinity for MEC-3. In addition, the transcript levels of some genes (mec-2 and mec-17) were not diminished as significantly or consistently in colchicine-treated animals as in mec-7 and mec-12 mutants (Fig. S6B). These differences may be due to a later effect of colchicine treatment on gene expression. Unlike tubulin mutations, which would affect embryonic neurons, colchicine may affect TRN microtubules only after hatching.
Protein Losses Caused by Microtubule Disruption Are Suppressed by Mutations in dlk-1 and cepb-1.
To understand how disruption of microtubules reduced protein levels in the TRNs, we used a short-lived GFP to identify mutants that failed to show this microtubule-dependent reduction. Normally, GFP is a very stable protein (23), and this stability may account for the finding that colchicine reduces, but does not abolish, GFP fluorescence in the TRNs of adults. GFP, however, can be made unstable by fusing a RING-finger domain from an E3 ubiquitin ligase to it (24). Adopting this strategy, we fused GFP and the RING-finger domain from the mouse E3 ubiquitin ligase PJA1/C-1 (25, 26). Animals expressing Praja::GFP from the TRN-specific mec-18 promoter (producing the integrated arrays uIs43 and uIs44) generated sufficient fusion protein to produce detectable fluorescence throughout their lifespan. When these animals were grown in the presence of colchicine, however, GFP fluorescence was undetectable 48 h after hatching (Figs. 2A and 3A). To identify genes needed for the colchicine-dependent reduction in expression, we mutated these animals with EMS, grew their F2 progeny in the presence of colchicine, and screened for adults with fluorescent TRNs.
Fig. 2.
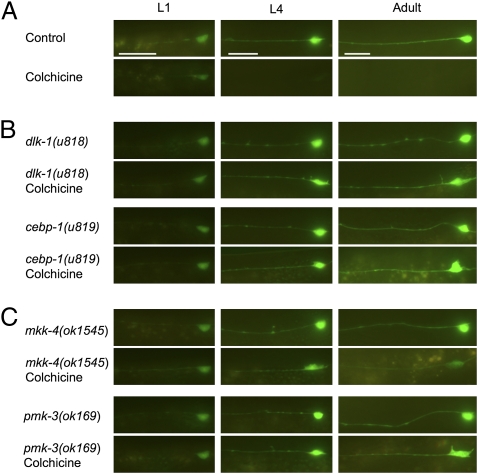
Mutations in several genes result in the colchicine-resistant expression (Cre) of short-lived PRAJA::GFP. (A) Colchicine abolishes TRN fluorescence in wild-type animals expressing uIs44 (Pmec-18praja::gfp) in a time-dependent manner. (B) Mutants defective in dlk-1 and cebp-1 retain TRN fluorescence when grown on colchicine. (C) Mutants defective in mkk-4 and pmk-3 also display Cre phenotypes, although it is incomplete in mkk-4 animals. An ALM neuron is shown in all panels; similar changes were seen for the other TRNs. (Scale bars, 20 μm.)
Fig. 3.
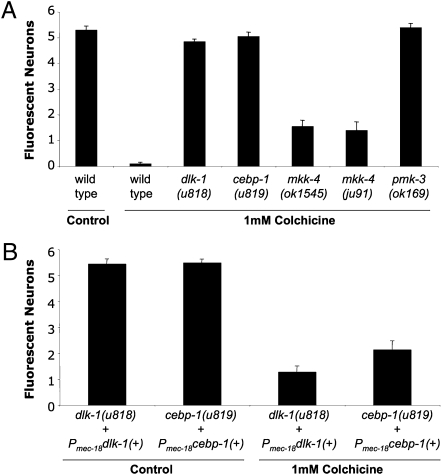
Extent of the Cre phenotype in dlk-1, cebp-1, mkk-4, and pmk-3 adults. The Cre phenotype was measured as the number of fluorescing TRNs (maximum six) seen in uIs44-expressing adults that had been grown on 1 mM colchicine. (A) Although completely penetrant in all four mutant backgrounds, expressivity is incomplete in mkk-4 mutants. (B) dlk-1(u818) and ceb-1(u819) Cre phenotypes can be rescued by expressing a wild-type copy of the gene under the TRN-specific mec-18 promoter (Pmec-18). The mean ± SEM is indicated for n ≥ 20 animals.
We obtained seven suppressor strains with recessive mutations from animals representing 13,640 haploid genomes (Figs. 2B and 3A). This phenotype is designated Cre for colchicine-resistant expression. Six Cre mutations were alleles of the dlk-1 gene (SI Materials and Methods). All six noncomplementing alleles had mutations in the coding sequence of dlk-1, including three missense mutations (u815, u816, and u820), two nonsense mutations (u817 and u818), and one frameshift mutation (u821). Previously known dlk-1 mutant alleles—ju476, a missense mutation (12), and km12, a deletion of the gene—failed to complement u818 for the Cre phenotype. The dlk-1 gene encodes a dual leucine zipper-bearing MAPKKK (mitogen-activated protein kinase kinase kinase) of 928 amino acids that is expressed in neurons and the pharynx (12). Expression of dual leucine zipper-bearing kinase (DLK)-1 under the mec-18 promoter rescued the dlk-1(u818) Cre phenotype (Fig. 4B), verifying the identity of the gene and demonstrating that DLK-1 acts in the TRNs in a cell-autonomous manner.
Fig. 4.
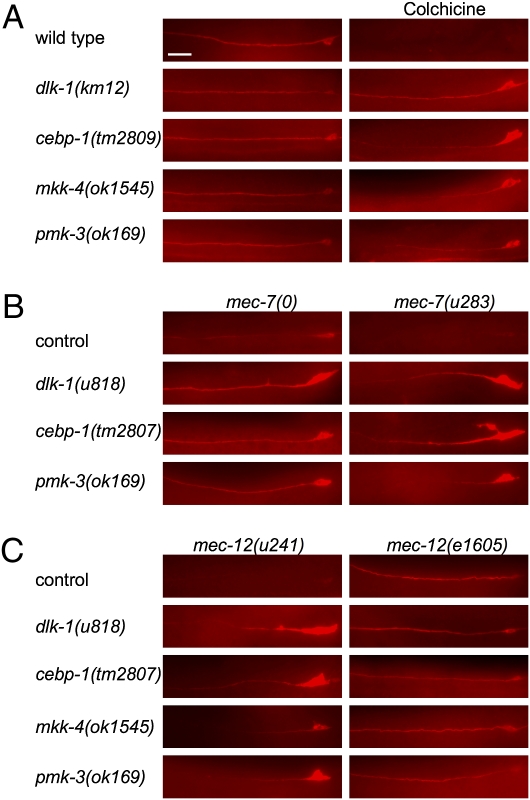
Mutations in genes for p38 MAPK pathway proteins and cebp-1 suppress the reduction of endogenous MEC-18 protein caused by microtubule defects. MEC-18 immunoreactivity in ALM neurons is shown for (A) colchicine-treated animals, (B) mec-7 mutants, and (C) mec-12 mutants. mec-7(0) denotes null allele; dlk-1 double mutants were generated with mec-7(u440); cebp-1 and pmk-3 were generated with mec-7(u443). (Scale bar, 20 μm.)
The remaining Cre mutation (u819) is an allele of cebp-1. cebp-1(tm2087), a deletion of the gene, failed to complement the u819 Cre phenotype. cebp-1 encodes a putative basic region leucine zipper transcription factor (27); the u819 mutation changes a conserved alanine to a valine in its DNA-binding domain. The u819 Cre phenotype was rescued by expression of wild-type cebp-1 from the mec-18 promoter (Fig. 4B), confirming the identity of the gene and demonstrating its cell-autonomous function.
Mutations in Other Members of a p38 MAP Kinase Pathway Suppress Protein Production Defects.
Mammalian DLK regulates several neuronal functions, including differentiation (28), migration (29), morphogenesis (30), axonal growth (31), apoptosis (32), and gene expression (33). C. elegans DLK-1 also functions in multiple ways in neurons, including presynaptic differentiation, axon termination, and axonal regeneration (12–14). However, unlike its mammalian counterpart, which works predominately through the c-jun NH2-terminal kinase (JNK) MAP kinase pathway (34), C. elegans DLK-1 acts through a p38 MAP kinase pathway that acts upstream of cebp-1 (35). DLK-1 is believed to phosphorylate the MAPKK MKK-4, which in turn phosphorylates the p38 MAPK PMK-3 (12). To determine whether the Cre phenotype produced by the dlk-1 mutations used the same pathway, we crossed a Pmec-18praja::gfp transgene (uIs44) into mkk-4(ok1545, ju91) and pmk-3(ok169) backgrounds and tested GFP fluorescence in TRNs in mutants grown on 1 mM colchicine.
mkk-4 animals had an incomplete Cre phenotype compared with dlk-1 animals. Although GFP fluorescence was seen in adult cells, often the fluorescence was fainter and restricted to only a few of the six TRNs (Figs. 2C and 3A). pmk-3 animals, however, were completely Cre; all six cells in adults were fluorescent in the presence of colchicine (Figs. 2C and 3A). These results suggest that the p38 MAP kinase pathway is needed for colchicine-induced gene reduction, but mkk-4 may be partially redundant in the pathway. To test this hypothesis, we generated uIs44 doubles with all available MAPKK mutants including sek-1, ZC449.3, VZC374L.1, jkk-1, and mek-1, and tested for a Cre phenotype. None of these mutants retained TRN fluorescence in any of the six TRNs. Because mkk-4 and these other MAPKK genes are clustered together on the X chromosome, we were unable to examine double mutants to test for redundancy, and RNAi experiments using dsRNA in a sensitized lin-35(n745) background (36) failed to produce Cre phenotypes.
Cre Mutants Suppress Reduction of Endogenous Gene Expression but Not Other Consequences of Microtubule Disruption.
In addition to suppressing reduction of the Pmec-18praja::gfp transgenes, Cre mutations prevented other TRN expression defects caused by the disruption of microtubules. All four Cre mutations suppressed reduction of endogenous MEC-18 (Fig. 4) and MEC-2 protein levels (pmk-3 is shown in Fig. S3) caused by colchicine and tubulin mutations. In addition, mec-18 message levels were restored in cebp-1 mutants grown on colchicine plates (Fig. S7). In contrast, these mutations did not prevent the touch insensitivity or the defective distribution of MEC-2 puncta produced by microtubule defects (11) (Fig. S3). Furthermore, we found that TRN cell bodies were misshaped and larger in animals with these mutations and disrupted microtubules (11) (Figs. 2 and 4). Interestingly, Cre mutants in a mec-12(e1605) background failed to show an increase of MEC-18 expression or the aberrant soma morphology, suggesting that all of these phenotypes require loss of the 15-protofilament microtubules. Taken together, these results suggest that the four Cre genes are needed only for a subset of functions related to microtubule disruption.
Colchicine Eliminates Microtubules in the Cre Mutant TRNs.
Several studies suggest that MAP kinase pathways can modulate the state of microtubules. The MAP kinase JNK1 is required for maintenance of neuronal microtubules in mice (37), and DLK activation of JNK1 results in the inactivation of microtubule-associated proteins that promote stability (31). Furthermore, inhibition of p38 MAP kinase activity increases stability of microtubules in growth cones of cultured dorsal root ganglion (DRG) mouse neurons (38). If loss-of-function mutations in Cre genes were similarly promoting microtubule stability, they might confer resistance to depolymerization by colchicine; the lingering presence of these colchicine-resistant microtubules in the TRNs could then account for the Cre phenotype.
To test whether TRN microtubules are more stable in pmk-3 mutants, we looked for microtubules in electron micrographs of TRN somas in wild-type and pmk-3 animals grown on control and colchicine plates. pmk-3 mutants on average appeared to have slightly more microtubules when grown on control plates (Table 2 and Fig. S8 A and B), but this increase was not statistically significant. In both groups of animals, colchicine eliminated all microtubules in the soma, whereas other characteristic TRN physical attributes, such as the extracellular mantle (4), were maintained (Table 2 and Fig. S8 C and D). These results suggest that an increase in microtubule stability does not account for the Cre phenotype in pmk-3 mutants.
Table 2.
Quantification of TRN microtubules in ALM somas of control and colchicine-treated wild-type and pmk-3 animals
| No. of microtubules in ALM soma |
||
| Animal | Control | Colchicine-treated |
| Wild type | 35 ± 8 | 0 ± 0 |
| pmk-3(ok169) | 46 ± 2 | 0 ± 0 |
Animals were grown at 20 °C for at least three generations on either control or 1 mM colchicine plates. Values are mean ± SE; n = 5 cells per condition.
Discussion
These experiments demonstrate a role for microtubules in regulating TRN protein levels. In the absence of large-diameter microtubules, protein levels of all tested markers were significantly reduced in the TRNs, and conditions that interfere with general microtubule polymerization produced an even greater reduction. The decreases in protein production caused by tubulin mutations and colchicine treatment was suppressed by mutations in a p38 MAP kinase pathway and the CEBP-1 transcription factor. Although the mechanism underlying this resulting Cre phenotype is unclear, electron micrographs did not show a stabilization of microtubules in Cre mutants treated with colchicine.
One mechanism whereby microtubule state could affect p38 MAPK signaling is by microtubules serving as docking sites and reservoirs for one or more of the signaling molecules. Microtubules are docking sites for a wide range of proteins including MAPKs, PI3Ks, cAMP-dependent kinases, phospholipases, GTPases, and transcription factors (3). Depolymerization of microtubules can release these diffusible factors, which can directly or indirectly affect changes in gene expression (3). For example, in mammalian cells, release of the Smad2 protein from microtubules results in its phosphorylation and nuclear translocation where it can oligomerize with other Smad proteins to induce transcriptional changes in the cell (39).
In C. elegans TRNs, DLK-1 (or another signaling molecule) may be similarly scaffolded onto the microtubules. Depolymerization of the microtubules would release DLK-1 to activate p38 MAPK signaling (Fig. 5). This model is consistent with the findings that mammalian DLK-1 protein (DLK/MUK/ZPK) associates with microtubules in cultured mouse cortical neurons (29). In C. elegans the p38 MAPK pathway has been implicated in neuronal process growth, the formation of synapses, and axonal regeneration (12–14), processes that are accompanied by localized microtubule instability (40–42). An intriguing possibility is that microtubule state regulates these processes by sequestering or releasing factors that modulate the p38 signaling pathway.
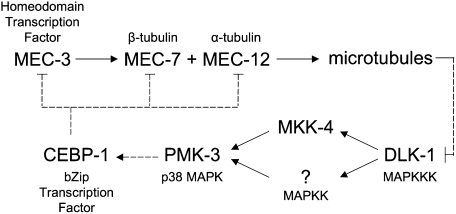
Fig. 5.
Model for microtubule-based regulation of gene expression. Interactions are derived from data in this paper and others (4, 8, 12, 16, 35). Direct positive (→) and negative (⊣) regulation is indicated by solid lines. Dashed lines indicate parts of the pathway that may be indirect. The MEC-3 transcription factor is also required for maintenance of its own expression (15). The question mark denotes an unidentified MAPKK believed to be partially redundant with MKK-4.
When activated by stress, the p38 signaling pathway can affect transcription (43). The CEBP-1 transcription factor may mediate this effect, because it acts downstream of the MAPK pathway in C. elegans (35) and is required for the gene expression changes described here. In mice, the C/EBP family of transcription factors regulates cell proliferation and differentiation, metabolism, and the response to inflammation and other stresses (27). In cultured rat neurons and injured Aplysia neurons, phosphorylation of C/EBPβ results in its nuclear translocation (44, 45). In our model, MAPK activation might similarly promote nuclear localization of CEBP-1 in response to microtubule disruption. Although the effects of phosphorylation on C. elegans CEBP-1 localization has not been investigated, the protein is found in both the soma and the synapses of motor neurons, and the stability of its mRNA is dependent upon DLK-1 and the p38 MAP kinase pathway (35).
The exact relationship between microtubules and the MAPK signaling appears to be complex. MAP kinases are potent regulators of microtubule dynamics. In mice, active DLK can reduce microtubule stability through both JNK (31) and p38 MAPK signaling pathways (38). However, the state of microtubules also regulates MAPK signaling, as depolymerization of microtubules activates these signaling pathways in cultured mammalian cells (46–49). These dual processes suggest a feedback mechanism with DLK acting as a monitor of microtubule state: the activation of DLK by microtubule depolymerization could further reduce microtubule stability.
Localized disruption of microtubules and the subsequent activation of MAP kinase signaling could be important for several neuronal functions. Bray found that localized application of colchicine to cultured sensory neurons resulted in their branching, as depolymerization of microtubules generated preterminal growth cone-like structures (50). This result demonstrated that the ability to generate growth cones was repressed by neuronal microtubules. Microtubules are absent at the tips of growth cones (40), suggesting that signaling molecules normally bound to microtubules are free to pursue their regulatory functions at growing axon terminals. When microtubules reach this space, they may sequester these molecules to prevent further kinase signaling; indeed, failure to control p38 MAPK signaling results in overextended axons and poor synapse formation (12). The presence of microtubules may provide an additional means of negatively regulating MAP kinase function. Moreover, controlling microtubule state may regulate a variety of cellular activities through analogous sequestration mechanisms.
Materials and Methods
Generation, Growth, and Maintenance of Nematode Strains.
C. elegans strains were cultured at 20 °C as previously described (51). Isolation and initial characterization of mec-7 and mec-12 mutants have been described previously (4, 7–10, 52). [All of the protein-coding sequences involved in this work were verified by PCR-based sequencing (20) at GeneWiz.]
Gene expression experiments were performed using strains TU2562 expressing uIs22 (Pmec-3gfp), TU2769 expressing uIs31 (mec-17::gfp; ref. 18), and TU3266 containing an integrated transgene, uIs57, expressing Punc-119gfp (and several nongfp-containing DNAs that do not effect GFP expression; ref. 53).
Previously existing dlk-1(km12, ju476), mkk-4(ju91, ok1545), pmk-3(ok169), and other MAPKK mutant strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. cebp-1(tm2807) and ZC449.3(tm1344) knockout strains were obtained from the National Bioresource Project in Japan.
Colchicine Treatment.
The effects of colchicine on C. elegans touch sensitivity were tested by growing animals for multiple generations on standard NGM agar plates (51) containing 1 mM colchicine (6).
Immunochemistry.
Whole-mount immunochemistry was carried out as described previously (52). Primary rabbit polyclonal MEC-18 antibody (54) and T6739 monoclonal anti-acetylated tubulin antibody (Sigma) were used at a 1:1,000 dilution; rabbit polyclonal MEC-2 antibody (21) was used at a 1:200 dilution. Secondary antibodies Cyanine Cy3 goat anti-rabbit IgG (Jackson ImmunoResearch), Rhodamine TRITC goat anti-mouse IgG (Jackson ImmunoResearch), and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) were all used at a 1:1,000 dilution.
Light Microscopy.
For fluorescence microscopy, animals were anesthetized using 0.3 M 2–3 butanedione monoxime in 10 mM Hepes and observed using a Zeiss Axiophot microscope. Images for all figures except Figs. S3 and S5 were taken with Plan-APOCHROMAT 63× objective; Fig. S3 images were taken using a Plan NEOFUAR 40× objective, and Fig. S5 was taken with a Plan NEOFUAR 25× objective. All images were taken with a Diagnostic Instruments Spot 2 camera at the same settings. To make images clearer for publication, all images in a set were treated equally to enhance contrast and brightness. Images for reported observations were made on at least 20 animals per mutation or condition.
Isolation of Cre Mutants.
We constructed a plasmid vector (TU819) containing Pmec-18praja::gfp by first isolating the Praja domain from a mouse cDNA library by PCR using the Praja primers from ref. 25. Nested PCR was then used to generate a DNA fragment fusing the Praja domain to the 5′ end of gfp; the PCR also introduced an ATG 5′ to the praja::gfp fusion and KpnI and ApaI cloning sites flanking the fragment. [The sequence for GFP was taken from the pPD95.75 expression vector (a gift from Andy Fire, Stanford University, Stanford, CA; Fire laboratory vector kit; http://www.addgene.org/pgvec1?f=c&cmd=showcol&colid=1)]. KpnI and ApaI cloning sites were then used to replace gfp with praja::gfp in the pPD95.75 vector. A 0.4-kb PCR fragment containing the mec-18 promoter was then inserted into the HindIII–BamHI sites before praja::gfp. The Pmec-18praja::gfp expressing strains TU3066 and TU3067, containing the integrated arrays uIs43 and uIs44, respectively, were generated by germline transformation of lin-15(n765ts) animals with plasmid TU819 and lin-35(+) DNA and exposure to γ-radiation (55).
TU3066 animals were mutagenized with EMS as described (51) and then placed onto 1 mM colchicine NGM plates. Parents were allowed to lay 30–50 eggs overnight at 25 °C, and F2 progeny were screened for Cre phenotypes. Mapping and characterization of Cre mutants are described in SI Materials and Methods.
Electron Microscopy.
Wild-type and pmk-3 animals were cultured for several generations on standard NGM or plates supplemented with colchicine and prepared for transmission electron microscopy using standard methods (56). Adults were fixed with 3.5% glutaraldehyde and 1% paraformaldehyde in 0.12 M sodium cacodylate. Eighty-nanometer transverse sections were cut and poststained with uranium acetate and lead citrate. For each genotype and culture condition, five ALM cell bodies were examined in the plane of the nucleus with a Philips CM10 electron microscope using a Morada digital camera (Olympus). Microtubules were counted in at least five sections per cell, covering a distance of at least 5.6 μm.
Touch Sensitivity.
Touch sensitivity of worms was tested by stroking the animals with an eyebrow hair attached to a toothpick (4). Wild-type animals respond to touches to the anterior body by moving backward and to posterior touches by accelerating forward. Each animal was tested 10 times by alternately touching the anterior and posterior.
Methods used to produce Figs. S1, S3B, S6, and S7 are available in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Shifang Zhang for the MEC-18 antibody, Dong Yan and Yishi Jin for suggestions and information about cebp-1, and Dave Hall for assistance with electron microscopy. This research was supported by National Institutes of Health Grant GM30997 (to M.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101360108/-/DCSupplemental.
References
- 1.Alberts B, et al. Molecular Biology of the Cell. 4th Ed. New York: Garland; 2002. [Google Scholar]
- 2.Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 3.Janmey PA. The cytoskeleton and cell signaling: Component localization and mechanical coupling. Physiol Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- 4.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Thomson JN. Organization of neuronal microtubules in the nematode Caenorhabditis elegans. J Cell Biol. 1979;82:278–289. doi: 10.1083/jcb.82.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalfie M, Thomson JN. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J Cell Biol. 1982;93:15–23. doi: 10.1083/jcb.93.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage C, et al. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3:870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- 9.Fukushige T, et al. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J Cell Sci. 1999;112:395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- 10.Bounoutas A, O'Hagan R, Chalfie M. Touch sensitivity in Caenorhabditis elegans. Curr Biol. 2009;19:1362–1367. doi: 10.1016/j.cub.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bounoutas A, Zheng Q, Nonet ML, Chalfie M. The multipurpose 15-protofilament microtubules in C. elegans have specific roles in mechanosensation. Genetics. 2009;183:607–617. doi: 10.1016/j.cub.2009.06.036. 601SI–604SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakata K, et al. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Grill B, et al. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989;3(12A):1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 17.Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- 18.O'Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 19.Gu G. A Molecular Model of Mechanosensory Transduction in Caenorhabditis elegans in Department of Biological Sciences. New York: Columbia Univ Press; 1998. [Google Scholar]
- 20.Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, et al. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Altun-Gultekin Z, et al. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- 23.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 24.Poyurovsky MV, et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell. 2003;12:875–887. doi: 10.1016/s1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 25.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 26.Mishra L, et al. Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene. 1997;15:2361–2368. doi: 10.1038/sj.onc.1201405. [DOI] [PubMed] [Google Scholar]
- 27.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robitaille H, Proulx R, Robitaille K, Blouin R, Germain L. The mitogen-activated protein kinase kinase kinase dual leucine zipper-bearing kinase (DLK) acts as a key regulator of keratinocyte terminal differentiation. J Biol Chem. 2005;280:12732–12741. doi: 10.1074/jbc.M411619200. [DOI] [PubMed] [Google Scholar]
- 29.Hirai S, et al. MAPK-upstream protein kinase (MUK) regulates the radial migration of immature neurons in telencephalon of mouse embryo. Development. 2002;129:4483–4495. doi: 10.1242/dev.129.19.4483. [DOI] [PubMed] [Google Scholar]
- 30.Hirai S, et al. The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J Neurosci. 2006;26:11992–12002. doi: 10.1523/JNEUROSCI.2272-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eto K, Kawauchi T, Osawa M, Tabata H, Nakajima K. Role of dual leucine zipper-bearing kinase (DLK/MUK/ZPK) in axonal growth. Neurosci Res. 2010;66:37–45. doi: 10.1016/j.neures.2009.09.1708. [DOI] [PubMed] [Google Scholar]
- 32.Hébert SS, et al. The mixed lineage kinase DLK is oligomerized by tissue transglutaminase during apoptosis. J Biol Chem. 2000;275:32482–32490. doi: 10.1074/jbc.M006528200. [DOI] [PubMed] [Google Scholar]
- 33.Oetjen E, et al. Inhibition of membrane depolarisation-induced transcriptional activity of cyclic AMP response element binding protein (CREB) by the dual-leucine-zipper-bearing kinase in a pancreatic islet beta cell line. Diabetologia. 2006;49:332–342. doi: 10.1007/s00125-005-0087-1. [DOI] [PubMed] [Google Scholar]
- 34.Hirai S, Izawa M, Osada S, Spyrou G, Ohno S. Activation of the JNK pathway by distantly related protein kinases, MEKK and MUK. Oncogene. 1996;12:641–650. [PubMed] [Google Scholar]
- 35.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehner B, et al. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang L, Jones Y, Ellisman MH, Goldstein LS, Karin M. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev Cell. 2003;4:521–533. doi: 10.1016/s1534-5807(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 38.Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Dong C, Li Z, Alvarez R, Jr, Feng XH, Goldschmidt-Clermont PJ. Microtubule binding to Smads may regulate TGF beta activity. Mol Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 40.Gordon-Weeks PR. Microtubules and growth cone function. J Neurobiol. 2004;58:70–83. doi: 10.1002/neu.10266. [DOI] [PubMed] [Google Scholar]
- 41.Roos J, Hummel T, Ng N, Klämbt C, Davis GW. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 42.Erez H, et al. Formation of microtubule-based traps controls the sorting and concentration of vesicles to restricted sites of regenerating neurons after axotomy. J Cell Biol. 2007;176:497–507. doi: 10.1083/jcb.200607098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitmarsh AJ. A central role for p38 MAPK in the early transcriptional response to stress. BMC Biol. 2010;8:47. doi: 10.1186/1741-7007-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metz R, Ziff E. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 1991;5:1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- 45.Sung YJ, Povelones M, Ambron RT. RISK-1: A novel MAPK homologue in axoplasm that is activated and retrogradely transported after nerve injury. J Neurobiol. 2001;47:67–79. doi: 10.1002/neu.1016. [DOI] [PubMed] [Google Scholar]
- 46.Wang TH, et al. Microtubule-associated protein tau in development, degeneration and protection of neurons. J Biol Chem. 1998;273:4928–4936. doi: 10.1016/j.pneurobio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Stone AA, Chambers TC. Microtubule inhibitors elicit differential effects on MAP kinase (JNK, ERK, and p38) signaling pathways in human KB-3 carcinoma cells. Exp Cell Res. 2000;254:110–119. doi: 10.1006/excr.1999.4731. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Zhu X, Chen Y, Wang X, Chen R. p38 and JNK MAPK, but not ERK1/2 MAPK, play important role in colchicine-induced cortical neurons apoptosis. Eur J Pharmacol. 2007;576:26–33. doi: 10.1016/j.ejphar.2007.07.067. [DOI] [PubMed] [Google Scholar]
- 49.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 50.Bray D, Thomas C, Shaw G. Growth cone formation in cultures of sensory neurons. Proc Natl Acad Sci USA. 1978;75:5226–5229. doi: 10.1073/pnas.75.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savage C, et al. Mutations in the Caenorhabditis elegans beta-tubulin gene mec-7: Effects on microtubule assembly and stability and on tubulin autoregulation. J Cell Sci. 1994;107:2165–2175. doi: 10.1242/jcs.107.8.2165. [DOI] [PubMed] [Google Scholar]
- 53.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang S. Stomatin Gene Family in Caenorhabditis elegans in Biological Sciences. New York: Columbia Univ Press; 2004. p. 158. [Google Scholar]
- 55.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 56.Hall DH. Electron Microscopy and Three-Dimensional Image Reconstruction in Caenorhabditis elegans: Modern Biological Analysis of an Organism. New York: Academic; 1995. pp. 394–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.