Abstract
Despite ubiquitous expression and a high level of metastasis-associated protein 1 (MTA1) coregulator, the physiological role of the MTA1 coactivator remains unknown. We found that MTA1 is a bona fide coactivator and stimulator of tyrosine hydroxylase (TH) transcription in neuronal cells and that MTA1-null mice had lower TH expression in the striatum and substantial nigra. MTA1 physically achieves these functions by interacting directly with DJ1 (Parkinson disease 7) and in turn recruits the DJ1/MTA1/RNA polymerase II complex to the bicoid binding element (BBE) in the TH promoter. Furthermore, we found that the MTA1/DJ1 complex is required for optimum stimulation of the TH expression by paired like homeodomain transcription factor (Pitx3) homeodomain transcription factor and that the MTA1/DJ1 complex is recruited to the TH gene chromatin via the direct interaction of MTA1 with Pitx3. These findings reveal a role for MTA1 as an upstream coactivator of TH and advance the notion of polygenic regulation of a disease-causing gene by coordinated interactions of three regulatory proteins.
Dynamic regulation of gene expression demands the participation of transcription factors, their coregulators, and multiprotein chromatin remodeling activity at target genes. One family of chromatin modifiers that is ubiquitously expressed is the metastasis tumor antigen (MTA) family. These family members are integral part of nucleosome remodeling and histone deacetylation complexes. MTA1, the first identified member of the MTA family, is up-regulated in a wide variety of human tumors (1, 2). MTA1 exists in corepressor or coactivator complexes containing histone deacetylase (HDAC) or RNA polymerase II (Pol II), respectively, and functions as a transcriptional coregulator to activate or repress the transcription of target genes (3, 4).
Homeobox genes encode transcription factors that have been shown to mediate key processes in development and patterning. The Pitx proteins belong to the bicoid-related subclass of paired homeodomain proteins characterized by a lysine at position 9 in the recognition helix of the homeodomain that determines the DNA-binding specificity of these proteins. A role for Pitx3 in the induction of tyrosine hydroxylase (TH), the rate-limiting enzyme for dopamine synthesis, has been suggested by the demonstration that Pitx3 can bind directly to response elements and activate the TH promoter (5). However, we are just beginning to appreciate the role of coregulators in the regulation of TH transcription by Pitx3 and to realize that Pitx3 may not act alone to stimulate TH transcription. Another candidate transcription factor that is expressed in all midbrain dopaminergic neurons is nuclear receptor-related protein 1 (Nurr1), which acts as a general TH regulator, as demonstrated by the loss of TH expression in both the substantia nigra pars compacta (SNc) and the ventral tegmental area (VTA) of Nurr1 mutant mice (6, 7). Despite these findings, we lack molecular insight into the roles played by emerging coregulators in the transcriptional control of TH deficiency, which is a prime cause of movement disorders.
Understanding the molecular basis of TH gene regulation by ubiquitous cellular factors also would be helpful in developing future strategies and modalities to treat or slow progression of the diseases associated with TH regulation. Several previous studies have attempted to identify the factors important in TH gene expression in human (8, 9), mouse (10), and rat (11) models. For example, TH expression has been shown to be regulated positively by the DJ1 coregulator (12). More recently, epigenetic profiling of the human TH promoter has suggested that chromatin remodeling could have a significant impact on conferring tissue-specific gene expression of the human TH gene (13); however, its specific role in TH transcription remains poorly understood. To elucidate these roles, we present evidence suggesting a function for the coordinated regulation of TH gene chromatin biology by the MTA1/DJ1 complex via Pitx3.
Results
DJ1 Interacts with MTA1.
While conducting a large-scale proteomic analysis of native complexes associated with coregulators (available at NURSA.org), we discovered the presence of MTA1 in complexes pulled down by DJ1 (Fig. 1A), a positive regulator of TH expression (12). Because we unexpectedly found MTA1 and DJ1 within the same complex, we proceeded to validate the interaction between MTA1 and DJ1 in the human neuroblastoma cell line SH-SY5Y by immunoprecipitating cell lysates with antibodies against MTA1 or DJ1 followed by blotting with DJ1 or MTA1 antibodies, respectively. We found a distinct interaction between MTA1 and DJ1 in vivo (Fig. 1B). We consistently found that MTA1 and DJ1 interacted in brain lysates from MTA1+/+ mice but not in those from MTA1−/− mice (Fig. 1C). Additional studies from the GST pull-down assays of 35S-labeled, in vitro-translated DJ1 and MTA1 also indicated that there is a direct interaction between these two proteins (Fig. 1D) and that DJ1 interacts with amino acids 442–542 in the C-terminal domain of MTA1 (Fig. S1).
Fig. 1.
MTA1–DJ1 interaction stimulates TH transcription. (A) Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes from nuclear extracts of HeLa cells pulled with DJ1 polyclonal antibody showing MTA1 in the same complex. (B) In vivo interaction of MTA1 and DJ1. Cell extracts from SH-SY5Y cells were subjected to Immunoprecipitation (IP) analysis with MTA1 or DJ1 antibody, followed by Western blotting with the other antibody. (C) Total brain tissue extracts from MTA1+/+ or MTA1−/− mice were tested in reciprocal immunoprecipitation/Western blotting analysis using DJ1 or MTA1 antibodies, respectively. (D) In vitro interaction of DJ1 and MTA1. The GST-DJ1 fusion protein and GST were used in a GST pull-down assay with in vitro-translated 35S-labeled full-length MTA1. (E) SH-SY5Y cells were cotransfected with MTA1 and/or DJ1 or CMV control along with full-length TH promoter-Luc, and Luc activity was measured. (Inset) Western blotting analysis for transfected myc-MTA1/Flag-DJ1 from same samples. (F) Model showing summary of results presented in this figure.
Stimulation of TH Transcription by MTA1–DJ1 Interaction.
The above findings raised the possibility that the interaction between DJ1 and MTA1 may regulate TH transcription in a cooperative manner and that the DJ1/MTA1 coregulator complex might serve as a mediator of the DJ1 regulation of TH expression. A previous study revealed that DJ1 silencing down-regulates TH protein and thus, by implication, suggests that DJ1 might be a coregulator of TH (12). For direct support of this notion, we determined that DJ1 expression stimulates TH promoter activity (Fig. 1E and Fig. S2), whereas DJ1 siRNA down-regulates TH mRNA and TH promoter activity in SH-SY5Y cells (Fig. S3). However, we unexpectedly found that, in addition to DJ1, MTA1 also stimulates TH transcription and that coexpression of MTA1 and DJ1 results in better TH transcription (Fig. 1E), presumably because of the noted direct interaction of the two coregulators (Fig. 1F).
Revelation of MTA1’s Coactivator Activity on TH Transcription.
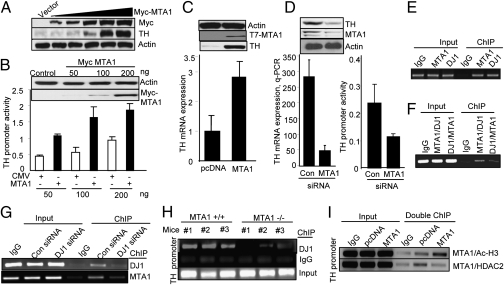
Because in the preceding set of studies the expression of MTA1 alone was accompanied by an increased TH promoter activity, we explored the possibility that MTA1 may act as a coactivator of TH. In support of this hypothesis, we found a substantial increase in TH protein levels with increasing concentrations of the myelocytomatosis viral oncogene homolog (myc)-MTA1 expression vector in SH-SY5Y cells (Fig. 2A). To determine whether the observed up-regulation of TH by MTA1 was transcriptional in nature, we next determined the effect of myc-MTA1 on TH promoter-luciferase (Luc) activity. We found a dose-dependent increase in TH transcription with increasing MTA1 concentrations in SH-SY5Y cells (Fig. 2B), suggesting that MTA1 behaves as a coactivator of the TH gene in neuroblastoma cells.
Fig. 2.
MTA1 regulation of TH expression. (A) Human neuroblastoma SH-SY5Y cells were transfected with increasing concentrations of myc-MTA1. Cell lysates were harvested and subjected to Western blotting with anti-TH, anti-myc, and anti-actin antibodies. (B) SH-SY5Y cells were transfected with increasing concentrations of myc-MTA1 along with full-length TH promoter-Luc. Cells were lysed after 48 h, and Luc activity was measured. (Insets) Expression of transfected myc-MTA1 and actin. (C) Pooled SH-SY5Y clones stably expressing pcDNA and T7-MTA1 were analyzed for the expression of T7-MTA1, TH, and actin by the Western blotting and for TH mRNA by qPCR. (D) Status of TH and MTA1 proteins, TH mRNA expression, and TH promoter activity in SH-SY5Y cells transfected with control (Con) or MTA1-siRNA. (E) Recruitment of MTA1 or DJ1 to the TH promoter region from −8,610 to −8,810 bases by ChIP assays using MTA1 or DJ1 antibodies. (F) Sequential double-ChIP analysis using indicated antibodies onto the TH promoter in SH-SY5Y cells. (G) Effect of DJ1-specific siRNA on the recruitment of MTA1 or DJ1 onto the TH promoter by ChIP. (H) ChIP analysis performed with samples from formalin-fixed total brain of MTA1+/+ and MTA1−/− mice using DJ1 antibodies or IgG. (I) Sequential double-ChIP analysis using MTA1 and followed by acetylated H3 (AcH3) antibodies or HDAC2 antibodies in SH-SY5Y clones stably expressing pcDNA and T7-MTA1. Values are the average of three independent measurements, and the SD from the mean is shown.
Results of serial deletions of a TH promoter-Luc reporter (10) revealed that MTA1 stimulation of the TH promoter activity requires a full-length promoter, and its deletion abolishes the ability of MTA1 to stimulate TH transcription (Fig. S4). To rule out the expected transient expression-linked variability in subsequent TH chromatin-remodeling studies using MTA1, we generated pooled clones of SH-SY5Y cells stably expressing either pcDNA or T7-MTA1. As expected, MTA1 expression was accompanied by increased TH protein and TH mRNA levels (Fig. 2C). Consistent with these results, we found that selective knockdown of endogenous MTA1 by RNA interference also reduced the levels of TH promoter-Luc activity, TH mRNA, and TH protein (Fig. 2D). These findings suggest that MTA1 is a coactivator of TH transcription.
MTA1 Recruitment onto TH Gene Chromatin.
To delineate the mechanism of MTA1’s regulation of TH expression, we used ChIP assays to determine whether MTA1 and DJ1 are present at the TH promoter. PCR analysis of DNA coimmunoprecipitated by MTA1 or DJ1 antibodies revealed that both MTA1 and DJ1 are recruited to TH gene chromatin (Fig. 2E). To determine the significance of the interaction between MTA1 and DJ1, we performed sequential double-ChIP assays using MTA1 and DJ1 antibodies. We found distinct corecruitment of MTA1 and DJ1, presumably as a MTA1/DJ1 complex, to the same region of the TH gene chromatin (Fig. 2F). To examine the mutual requirements of MTA1 and DJ1 for efficient interaction of these coactivators with TH gene chromatin, we examined the effect of selective siRNA-mediated depletion of MTA1 or DJ1 on DJ1 or MTA1 recruitment to TH gene chromatin in SH-SY5Y cells. We noticed a dramatic reduction in the recruitment of MTA1 or DJ1 in cells with knockdown expression of the DJ1, suggesting that both MTA1 and DJ1 are required to regulate TH gene expression (Fig. 2G). Consistent with these findings, there was significantly less recruitment of DJ1 to TH gene chromatin in brain lysates from MTA1−/− mice than in lysates from MTA1+/+ mice (Fig. 2H). These findings suggest that both MTA1 and DJ1 are required for the optimal mutual recruitment of these coactivators to the TH gene chromatin and, by implication, for the expression and function of TH.
Because MTA1 enhances TH transcription, we next performed sequential double-ChIP studies and found significantly elevated recruitment of the MTA/Pol II (an indicator of active transcription) coactivator complex onto the TH promoter in MTA1-expressing SH-SY5Y clones compared with levels recruited in control cells (Fig. S5). There also was increased recruitment of MTA1/acetylated histone 3 (H3) to the TH gene chromatin (Fig. 2I), suggesting the existence of relaxed chromatin surrounding the MTA1-targeted TH chromatin. Also consistent with these results, there was a decrease in the MTA1/HDAC2 corepressor complex from the corresponding region of the TH promoter occupied by the MTA1/Pol II complex (Fig. 2I, and Fig. S5). Overall, these results provide evidence for epigenetic changes following MTA1 recruitment onto TH gene chromatin.
MTA1 Regulation of TH Transcription Involves MTA1–Pitx3 Interaction.
Because MTA1 and DJ1 cannot bind directly to DNA, and MTA1 is recruited to the TH promoter, we next analyzed the MTA1 recruitment region of the TH promoter for possible transcriptional factors with a proven role in the transcriptional stimulation of TH. This experiment led to the observation that the MTA1-interacting region of the TH promoter contains four potential bicoid-type binding elements (BBEs) for Pitx3, a homeodomain-containing transcription factor (Fig. S6). In the light of these findings, we explored the possibility that Pitx3 may be the missing effector for the stimulation of TH transcription by MTA1 and, perhaps, by DJ1, a protein that we found to interact with MTA1.
In light of the above observations, we further explored possible interactions between MTA1, DJ1, and Pitx3 in SH-SY5Y cells. To this end, we first examined whether MTA1 or DJ1 interacts with Pitx3 in SH-SY5Y cells. Results from coimmunoprecipitation assays using MTA1 or DJ1 antibodies or IgG indicate that Pitx3 could be coimmunoprecipitated effectively with MTA1 but not with DJ1 (Fig. 3A). Furthermore, in pull-down assays, recombinant GST-MTA1 protein coprecipitated 35S-labeled Pitx3 efficiently, whereas control GST did not (Fig. S7A). To our surprise, we repeatedly found no detectable binding of recombinant GST-DJ1 with 35S-labeled Pitx3 (Fig. S7B). Pitx3 bound more efficiently to the N-terminal regions of MTA1, whereas MTA1 bound to the homeodomain of Pitx3 (Fig. S7 C and D). Overall, these findings suggest that the MTA1/DJ1 complex may be recruited to the TH gene chromatin via the direct contact of MTA1 with Pitx3.
Fig. 3.
Pitx3 mediates MTA1 and DJ1 interaction and recruitment to the TH promoter. (A) In vivo interaction of MTA1 and Pitx3 in SH-SY5Y cells by immunoprecipitation followed by Western blotting analysis. (B) Effect of Pitx3 knockdown on the ability of MTA1 or DJ1 to stimulate TH promoter Luc activity in SH-SY5Y cells. (C) Effect of DJ1 or MTA1 on the levels of TH mRNA in SH-SY5Y cells transfected with Pitx3 or control siRNAs. Values are the average of three independent measurements, and the SD from the mean is shown. (D) qPCR ChIP assay for recruitment of Pitx3 to the TH promoter region from −8,610 to −8,810. Values are the average of three independent measurements, and the SD from the mean is shown. (E and F) Effect of Pitx3 or control siRNAs on the recruitment of MTA1 or DJ1 on the TH promoter by real-time qPCR assays in SH-SY5Y cells. (G and H) Sequential double-ChIP followed by qPCR analysis performed with exponentially growing SH-SY5Y cells with Pitx3, MTA1, or DJ1 antibody. (I) Effect of MTA1 or DJ1 or both combined siRNAs on the recruitment of Pitx3 on TH promoter. (J and K) Effects of MTA1 or DJ1 and/or Pitx3 or CMV control along with full-length TH promoter-Luc activity in SH-SY5Y cells. (L) Effect of MTA1 or control siRNAs on the TH promoter-Luc activity by DJ1 or Pitx3 or together in SH-SY5Y cells. Values are the average of three independent measurements, and the SD from the mean is shown.
To understand the significance of the noted MTA1–Pitx3 interaction in the MTA1 or DJ1 regulation of TH transcription, we next determined the effect of selective siRNA-mediated knockdown of Pitx3 expression in SH-SY5Y cells on the ability of MTA1 and DJ1 to stimulate TH promoter activity and TH mRNA. Results show that, indeed, Pitx3 knockdown reduces the ability of MTA1 or DJ1 to stimulate TH promoter activity as well as TH mRNA expression (Fig. 3 B and C), suggesting that Pitx3 may be required for MTA1 stimulation of TH expression.
To understand further the significance of this finding, we examined whether Pitx3 is recruited to one or all potential BBE sites on the TH promoter using ChIP. By ChIP (Fig. S8) and by quantitative PCR (qPCR) (Fig. 3D), we found that Pitx3 is recruited selectively to BBE region II. Next we showed that Pitx3 knockdown in SH-SY5Y cells was accompanied by the reduced recruitment of MTA1 or DJ1 to the TH promoter (Fig. 3 E and F). To demonstrate that MTA1 and Pitx3 are corecruited onto the BBE II of the TH promoter, we performed a sequential double-ChIP assay in which the first ChIP used MTA1 antibody and the second ChIP used Pitx3 antibody, or vice versa. Results showed that both MTA1/Pitx3 and DJ1/Pitx3 are corecruited on the TH promoter (Fig. 3 G and H). These findings suggest that both MTA1 and DJ1 bind to the TH promoter through Pitx3, and this result probably explains the ineffectiveness of MTA1 or DJ1 in stimulating TH transcription under conditions of Pitx3 knockdown (Fig. 3 E and F).
Because Pitx3 regulates TH promoter activity through a direct interaction with the TH promoter, and chromatin modifiers play a significant role in conferring tissue-specific expression of the human TH gene (9), we believe that MTA1 and DJ1 are unrecognized coregulators supporting the stimulatory effect of Pitx3 on the TH promoter. To validate this hypothesis, we examined the effect of selective knockdown of MTA1 or DJ1 in the recruitment of Pitx3 onto TH chromatin in SH-SY5Y cells. Results showed that stable recruitment of Pitx3 onto the TH promoter decreased upon silencing of either MTA1 or DJ1, suggesting an essential coregulatory role of both MTA1 and DJ1 in TH transcription (Fig. 3I). Of interest, we found a further reduction in the recruitment of Pitx3 when MTA1 and DJ1 were knocked down simultaneously (Fig. 3I).
These findings suggest that Pitx3 is required for the efficient interaction between MTA1 or DJ1 and the TH gene chromatin and that MTA1 and DJ1 behave as coactivators of Pitx3 regulation of TH expression. To evaluate this hypothesis directly, we examined the possible cooperative effects of MTA1 or DJ1 and Pitx3 on TH promoter activity. We found that coexpression of either MTA1 or DJ1 enhances the ability of Pitx3 to stimulate the TH promoter (Fig. 3 J and K). Given that MTA1 interacts with DJ1, it is possible that both proteins are important for the optimal stimulation of TH transcription by Pitx3. We found that in the absence of MTA1 there was a moderate decline in the ability of DJ1 or Pitx3 to induce TH promoter activity, whereas MTA1 down-regulation significantly reduced the ability of the combination of DJ1 and Pitx3 to stimulate TH transcription (Fig. 3L). These findings suggest that the MTA1/DJ1 complex is required for an optimum stimulation of TH expression by Pitx3.
Pitx3 Mediates MTA1 and DJ1 Interaction with TH DNA.
Because the TH promoter region that interacts with DJ1, MTA1, and Pitx3 contains a core BBE consensus motif, we sought to determine whether the binding of MTA1/DJ1 to the TH promoter region encompassing the BBE II consensus sequence is direct or indirect by using an EMSA of nuclear extracts from SH-SY5Y cells. We found distinct protein/TH DNA complexes (Fig. 4A, lane 2) that could be supershifted by antibodies to Pitx3, MTA1, or DJ1 (Fig. 4A, lanes 3–5); coincubation of Pitx3-Ab with antibodies against MTA1 or DJ1 resulted in further supershifting of the protein/TH DNA complexes (Fig. 4A, lanes 6–8). These results suggest that all three functional proteins interact with TH DNA, presumably via MTA1, despite the lack of direct Pitx3–DJ1 binding. To demonstrate a mechanistic role of Pitx3 in recruiting MTA1 to TH DNA, we showed destabilization of the protein/DNA complex in nuclear extracts from SH-SY5Y cells with Pitx3 knockdown (Fig. 4A, lanes 10–15), again suggesting that Pitx3 mediates MTA1 and DJ1 binding to the TH promoter. Similarly, knockdown of MTA1 compromised the size of protein/DNA complexes and the ability of DJ1 or MTA1 antibodies to supershift protein/TH DNA complexes either alone or in combination with pitx3 antibody (Fig. 4B, lanes 10, 12, 14, and 15). However, Pitx3 antibody alone was able to supershift the complex, although in this case the Pitx3 antibody resulted in the formation of smaller protein/DNA complexes (Fig. 4B, compare lane 11 with 3). In contrast, knockdown of DJ1, which does not interact directly with Pitx3, had no effect on the ability of Pitx3 to form a complex with TH DNA (Fig. 4C). However, the electrophoretic mobility of the Pitx3/DNA complex was faster in the absence of DJ1 (Fig. 4C, compare lanes 4 and 12). DJ1 depletion also had no effect on the ability of MTA1 antibody to supershift the noted protein/DNA complexes (Fig. 4C, lane 11). In brief, these findings suggest that the MTA1/DJ1 complex is recruited to the TH chromatin via MTA1–Pitx3 interaction (Fig. 4D).
Fig. 4.
Mechanistic interaction of PitX3, MTA1, and DJ1 on TH promoter. (A) EMSA analysis of Pitx3, MTA1, and DJ1 binding to the BBE on the TH promoter using oligos encompassing BBE II in SH-SY5Y cells transfected with or without Pitx3 siRNA. (B) EMSA analysis of Pitx3, MTA1, and DJ1 binding to the BBE on TH promoter using oligos encompassing BBE II in SH-SY5Y cells transfected with or without MTA1 siRNA. (C) EMSA analysis of Pitx3, MTA1, and DJ1 binding to the BBE on TH promoter using oligos with or without DJ1 siRNA. (D) Summary of the findings presented here: Pitx3 is located DNA the binding site in the TH promoter and recruited a coactivator complex containing MTA1 and DJ1 through a distinct interaction with MTA1. Alteration in any one of these three gene products (MTA1, DJ1, or Pitx3) could produce variations in TH expression and a predisposition to movement disorders in genetic strains or individuals.
MTA1−/− Mice Exhibit Reduced Expression and Activity of TH.
To examine whether changes noted in the levels of TH by the status of MTA1 also are reflected in the whole-animal setting, we compared the levels of TH protein and TH mRNA in total brain from MTA1−/− and MTA1+/+ mice or of TH protein in lysates of the substantia nigra and striatum regions of brain from MTA1−/− mice with those from MTA1+/+ mice. We found a significant decrease in TH protein and mRNA levels in total brain lysates as well as in lysates (Fig. 5A) of the substantial nigra and striatum regions of brain from MTA1−/− mice compared with those from MTA1+/+ mice (Fig. 5B). We next analyzed dopamine (product of TH rate-limiting enzyme) levels in the striatum and substantia nigra in MTA1+/+ and MTA−/− mice using a conventional enzyme immunoassay (EIA). We found a significant decrease in dopamine levels in MTA1−/− mice compared with those in MTA1+/+ mice (Fig. 5C). Interestingly, there was no change in norepinephrine (NE) levels in the hippocampus and locus coeruleus of MTA1+/+ mice as compared with MTA1−/− mice (Fig. S9), indicating that dopamine biosynthesis may be selectively compromised in the substantia nigra and striatum of MTA1−/− mice. In brief, these studies provide proof-of-principle evidence supporting a causal relationship between the levels of MTA1 and reduced TH expression in cells and in animals.
Fig. 5.
MTA1−/− mice develop Parkinson-like symptoms and have reduced dopamine levels and TH. (A) Levels of TH protein and vinculin and TH mRNA in total brain from MTA1+/+ and MTA1−/− mice. (B) Levels of TH protein in lysates of the substantial nigra and striatum regions of brain from MTA+/+ mice compared with those from MTA1−/− mice. (C) Tissue dopamine levels in MTA1+/+ and MTA1−/− mice were measured using EIA kits. The bar chart shows reduced dopamine levels in striatum and substantial nigra (SN) of brain from MTA1−/− mice compared with brain from MTA1+/+ mice (n = 5 in each group). Dopamine levels are expressed as nanograms of dopamine per nanogram of protein. (D) Footprint test of an MTA1−/− mouse compared with a MTA1+/+ mouse (n = 3, 10 animals in each group). (E) Rota-rod test for MTA1+/+ and MTA1−/− mice. The bar chart shows time mice spent walking on the rotating (n = 3 groups, 10 animals in each group).
Previous expression studies showing higher levels of MTA1 mRNA in the brain (3) and our findings described above that MTA1 deficiency leads to a reduced TH expression in certain areas of the brain raise the possibility that MTA1 has a physiologic function in the brain. While maintaining the MTA1−/− mice colony, we consistently observed that MTA1−/− mice had an asymmetric hind limb movement characteristic of motion disorders. We performed a footprint test and calculated the distance between the hind paws (Fig. 5D). The footprint test reaffirmed impaired motion in MTA1−/− mice, as evidenced by greater footprint width in MTA1−/− mice than in age-and weight-matched MTA1+/+ mice. To confirm the existence of motion disorder-like symptoms, we next performed a Rota-rod test, an established method of assessing a defective motor disorder in mice. We used a range of accelerating speeds from 4–40 rpm over a period of 500 s. When MTA1−/− and MTA1+/+ mice were forced to adapt to acceleration, MTA1−/− mice were obviously impaired in their ability to remain on the rotating rod compared with MTA1+/+ mice throughout the course of repeated experiments (Fig. 5E). The observed motion impairment of the MTA1−/− mice may be caused by a defect in TH expression and dopamine production.
Discussion
Earlier studies have suggested an important role for chromatin-remodeling factors in the regulation of TH gene transcription. We now have identified MTA1 as one such factor playing an important role in TH gene regulation. This result was supported further by our finding that genetically engineered MTA1−/− mice had a reduction in TH immunoreactivity and mRNA expression. Because TH is the rate-limiting enzyme in dopamine synthesis, dopamine deficiency could arise from a reduction in the levels of tyrosine hydroxylase. Our results also suggest that MTA1 and DJ1 function as a complex and require Pitx3, a homeodomain transcription factor, to interact with TH DNA. MTA1, but not DJ1, interacts with TH DNA because of the MTA1 interaction with Pitx3. These findings, in conjunction with the observation that Pitx3 is an MTA1-interacting transcription factor, suggested that the DJ1/MTA1 complex and its interaction with Pitx3 facilitate the efficient binding and function of Pitx3 at its core DNA motif on the TH promoter and also its stimulation (Fig. 4D).
A role for Pitx3 in TH regulation had been suggested previously by the results of in vitro promoter studies, which showed that there is a response element for Pitx3 in the TH promoter to which Pitx3 binds and thus confers a transcriptional effect in vitro (5). Pitx3 may act on its own to activate the TH promoter or possibly may interact with other DNA-binding molecules such as Nurr1 to achieve this effect. One study found that Nurr1 can act cooperatively with Pitx3 to enhance the effect of Pitx3 on the TH promoter (5). The same enhancement occurs with MTA1. Mechanistic studies of the coactivator function of MTA1 revealed that MTA1 is associated with the RNA Pol II complex on the TH promoter and that HDAC2 is eliminated from this complex. Our subsequent direct promoter binding assays using nuclear extracts from SH-SY5Y cells established MTA1 as a transcriptional regulator of the human TH promoter via Pitx3. These findings reveal a previously unrecognized role for MTA1 as an upstream modifier of TH and support the notion that transcription of the disease-causing TH gene is regulated by coordinated actions of multiple coregulatory proteins.
Materials and Methods
Materials and methods are discussed in detail in SI Materials and Methods.
Generation of MTA1−/− Mice.
To generate MTA1-deficient mice, a targeting vector was designed to delete exon 2 by flanking it with LoxP splicing sites. The targeted construct was introduced into the PC3 ES cell line, and 16 positive clones were identified by Southern blotting. Two individual clones were injected into C57B6 blastocysts, male chimera mice were bred with C57B6 female mice, and germ-line transmission was confirmed by Southern blotting and PCR assay. The status of the mRNA transcripts from the Mta1 locus was verified using a pair of primers flanking exon 2.
Rota-Rod Test.
Motor coordination was studied using a five-station Rota-rod (Med-Associates). All mice were trained initially at 16 rpm for 30 s and then with accelerating speed from 4–40 rpm over the course of 5 min. Mice were given four training trials per day with an intertrial interval of 10 min. Four training trials were considered one session. The training sessions were performed over 3 consecutive days. For each training trial, mice were placed gently on the rod in the orientation opposite that of the already rotating rod so that they could acquire the skill necessary to prevent falling from the rotating rod. Mice were allowed to stay on the rod for a maximum of 500 s, which was established as the cutoff period. The length of time that each mouse was able to maintain balance on the rotating rod was recorded.
Footprint Test.
For the footprint test, mice were placed in a 15-cm wide, 1-m long corridor. The floor of this corridor was covered with white absorbent filter paper. Mice first were trained to explore the corridor. After this training, their paws were colored with eosin ink, and the width of the footprint was calculated by measuring the distance between the hind paws.
Supplementary Material
Acknowledgments
We thank Sandip Mishra for technical help during the initial stages of the project. This study was supported by National Institutes of Health Grants CA98823 (to R.K.) and HD-07857 (to B.W.O.) and by the Diana Helis Henry Medical Research Foundation (B.W.O.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101193108/-/DCSupplemental.
References
- 1.Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- 2.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 3.Mazumdar A, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 4.Gururaj AE, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci USA. 2006;103:6670–6675. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazorla P, Smidt MP, O'Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–1837. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- 6.Saucedo-Cardenas O, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetterström RH, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 8.Kessler MA, Yang M, Gollomp KL, Jin H, Iacovitti L. The human tyrosine hydroxylase gene promoter. Brain Res Mol Brain Res. 2003;112:8–23. doi: 10.1016/s0169-328x(02)00694-0. [DOI] [PubMed] [Google Scholar]
- 9.Romano G, Suon S, Jin H, Donaldson AE, Iacovitti L. Characterization of five evolutionary conserved regions of the human tyrosine hydroxylase (TH) promoter: Implications for the engineering of a human TH minimal promoter assembled in a self-inactivating lentiviral vector system. J Cell Physiol. 2005;204:666–677. doi: 10.1002/jcp.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, et al. Regulation of the tyrosine hydroxylase gene promoter by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2003;312:950–957. doi: 10.1016/j.bbrc.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Gandelman KY, Coker GT, 3rd, Moffat M, O'Malley KL. Species and regional differences in the expression of cell-type specific elements at the human and rat tyrosine hydroxylase gene loci. J Neurochem. 1990;55:2149–2152. doi: 10.1111/j.1471-4159.1990.tb05811.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhong N, et al. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem. 2006;281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 13.Romano G, Macaluso M, Lucchetti C, Iacovitti L. Transcription and epigenetic profile of the promoter, first exon and first intron of the human tyrosine hydroxylase gene. J Cell Physiol. 2007;211:431–438. doi: 10.1002/jcp.20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.