Abstract
Aptamers have emerged as promising molecular probes for in vivo cancer imaging, but the reported “always-on” aptamer probes remain problematic because of high background and limited contrast. To address this problem, we designed an activatable aptamer probe (AAP) targeting membrane proteins of living cancer cells and achieved contrast-enhanced cancer visualization inside mice. The AAP displayed a quenched fluorescence in its free state and underwent a conformational alteration upon binding to target cancer cells with an activated fluorescence. As proof of concept, in vitro analysis and in vivo imaging of CCRF-CEM cancer cells were performed by using the specific aptamer, sgc8, as a demonstration. It was confirmed that the AAP could be specifically activated by target cancer cells with a dramatic fluorescence enhancement and exhibit improved sensitivity for CCRF-CEM cell analysis with the cell number of 118 detected in 200 μl binding buffer. In vivo studies demonstrated that activated fluorescence signals were obviously achieved in the CCRF-CEM tumor sites in mice. Compared to always-on aptamer probes, the AAP could substantially minimize the background signal originating from nontarget tissues, thus resulting in significantly enhanced image contrast and shortened diagnosis time to 15 min. Furthermore, because of the specific affinity of sgc8 to target cancer cells, the AAP also showed desirable specificity in differentiating CCRF-CEM tumors from Ramos tumors and nontumor areas. The design concept can be widely adapted to other cancer cell-specific aptamer probes for in vivo molecular imaging of cancer.
Keywords: switchable aptamer probe, in vivo imaging, activatable fluorescent molecular imaging, cancer detection, cell surface protein
Development of sensitive and specific molecular probes is one of the central challenges in cancer imaging (1, 2). Aptamers are single-stranded RNA or DNA oligonucleotides with unique intramolecular conformations that hold distinct binding properties to various targets, including small molecules, proteins, and even entire organisms (3–5). As a small, polyanionic and nonimmunogenic type of probe, aptamers may exhibit faster tissue penetration and uptake, shorter residence in blood and nontarget organs, and higher ratio of target accumulation, thus affording great potential for in vivo cancer imaging (6, 7).
Design of aptamer probes for cancer imaging has primarily relied on a strategy using the so-called “always-on” probes (8), in which the reporter-bearing aptamers are bound to target cancer cells and accumulation of the reporters around cells then results in an elevated signal with reference to the surrounding environment. Various signal reporters have been adopted, including radioactive, magnetic, and fluorescent agents (9–13). Our group has demonstrated that near-infrared dye-labeled aptamers could effectively recognize cancer cells in vivo and achieve cancer imaging with high specificity (13). However, because always-on aptamer probes had constant signals, the image contrast was critically limited by a high background. And the cancer site could be clearly observed only after the physiological clearing of unbound aptamers, thus leading to a long diagnosis time, which further compromised contrast by substantial consumption of bound aptamers. Therefore, ideal aptamer probes for in vivo cancer imaging should preferably display signal alteration architectures, in which the normally quenched signal is activated only after successfully targeting cancer sites.
In the context of molecular imaging, a variety of such probes have been developed (14–21). Typical mechanisms include enzymatic cleavage of a quencher group from the fluorescence moiety using cancer-related enzymes (15–18), intracellular degradation of fluorescence-quenched protein against cancer cell receptors (19, 20), and surrounding-sensitized fluorescence labels conjugated to macromolecular ligands selectively endocytosed by cancer cells (8). However, these mechanisms cannot be utilized to develop aptamer-based activatable probes. Because the sequences of aptamer probes can be custom-designed, aptamers have the particular advantage of target recognition-triggered conformational alteration, leading, in turn, to signal alteration. This strategy has been well demonstrated with molecular beacons for nucleic acid monitoring and free protein detection (22, 23). Assuming that this strategy would work on surface membrane proteins of living cancer cells, we hypothesized that conformational alteration triggered by the specific binding of aptamers with cell membrane proteins could detect cancer cells, thus affording a substantial basis for the development of a unique kind of activatable molecular probe for in vivo cancer imaging.
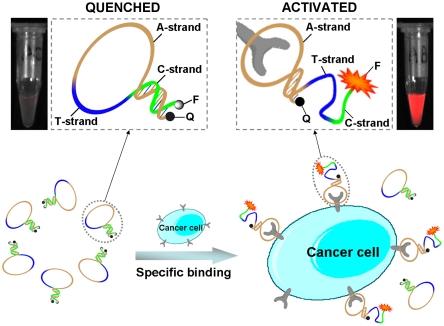
Herein we report such an activatable aptamer probe (AAP) designed based on cell membrane protein-triggered conformation alteration, as illustrated in Fig. 1. The AAP is a single-stranded oligonucleotide consisting of three fragments: a cancer-targeted aptamer sequence (A-strand), a poly-T linker (T-strand), and a short DNA sequence (C-strand) complementary to a part of the A-strand, with a fluorophore and a quencher covalently attached at either terminus. Because of hybridization of the C-strand with its complementary part of the A-strand, the AAP is hairpin structured. This conformation keeps the fluorophore in close proximity to the quencher, resulting in quenched fluorescence in the absence of a target. However, when the probe encounters the target cancer cell, it is capable of binding with protein receptors on the cell surface, causing a spontaneous conformational reorganization of the hairpin structure and, consequently, forcing the fluorophore to separate far from the quencher. Finally, a fluorescence signal is activated in response to the successful binding of the AAP to the target cancer cell. As a “signal-on” probe, the low fluorescence-quenched background may display dramatically enhanced image contrast, thus leading to a much shorter diagnosis time. Note that in our design, the AAP not only acts as a molecular recognition probe but also serves as a transducer in generating an activated fluorescence signal as a result of cell membrane protein binding events. Thus, by integrating target specificity with sensitive signal transduction, these aptamer-based probes are afforded a unique advantage.
Fig. 1.
Schematic representation of the novel strategy for in vivo cancer imaging using activatable aptamer probe (AAP) based on cell membrane protein-triggered conformation alteration. The AAP consists of three fragments: a cancer-targeted aptamer sequence (A-strand), a poly-T linker (T-strand), and a short DNA sequence (C-strand) complementary to a part of the A-strand, with a fluorophore and a quencher attached at either terminus. In the absence of a target, the AAP is hairpin structured, resulting in a quenched fluorescence. When the probe is bound to membrane receptors of the target cancer cell, its conformation is altered, thus resulting in an activated fluorescence signal.
Results and Discussion
Construction and Sequence Optimization of AAP.
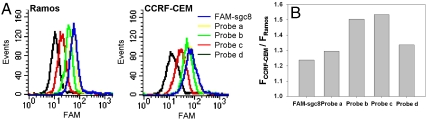
An aptamer sgc8 was used as a model system to demonstrate the feasibility of the AAP-based strategy for in vivo cancer imaging. The sgc8 was selected by cell-SELEX against human acute lymphoblastic leukemia CCRF-CEM cells (24) and identified to interact with the cell membrane protein tyrosine kinase-7 (PTK7), a protein closely associated with a number of cancers (25). The AAP for CCRF-CEM cancer imaging was constructed as a single-stranded oligonucleotide consisting of the aptamer sgc8, the T-strand, and the C-strand, with a fluorophore FAM and a quencher BHQ1 covalently attached at either terminus. To ensure that the fluorescence resonance energy transfer (FRET) between FAM and BHQ1 would be substantially eliminated after AAP activated, a total number of the nucleotides contained in the T-strand and C-strand was fixed at 25. Consequently, the signal-to-background ratio of the AAP was strongly dependent upon the C-strand sequence. More specifically, the hybridization of the C-strand with its complementary part of the A-strand resulted in low background and increased aptamer stabilization. For obtaining the AAP sequence with the highest signal-to-background ratio, four aptamer probes—probe a, b, c, and d—with different C-strand compositions were designed. As listed in Table 1, the number of nucleotides contained in C-strand was gradually increased from six in probe a to nine in probe d. Fig. 2A gives the fluorescence responses of these aptamer probes to CCRF-CEM cells with reference to the control Ramos cells. After incubation with Ramos cells, the probes displayed decreased background responses with longer C-strand complementary to the A-strand. On the other hand, with shortened C-strand, the affinity of the probes increased and the resulting signals of these probes activated by target CCRF-CEM cells were enhanced significantly. The signal-to-background ratios of these probes are given in Fig. 2B. The best signal-to-background ratio was achieved with probe c that had an eight-nucleotide sequence complementary to the A-strand. With the optimized AAP sequence, a near-infrared AAP was then constructed using a near-infrared dye, Cy5, as the reporter and BHQ2 as an efficient quencher. The utilization of the near-infrared reporter could further facilitate applications of the AAP for in vivo imaging of CCRF-CEM cancer (26).
Table 1.
All of the oligonucleotides used in this work*
| Probe | Sequence |
| Probe a | 5′-FAM-TCTAACTTTTTTTTTTTTTTTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-BHQ1-3′ |
| Probe b | 5′-FAM-TCTAACCTTTTTTTTTTTTTTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-BHQ1-3′ |
| Probe c | 5′-FAM-CTAACCGTTTTTTTTTTTTTTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-BHQ1-3′ |
| Probe d | 5′-FAM-TCTAACCGTTTTTTTTTTTTTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-BHQ1-3′ |
| Activatable aptamer probe (AAP) | 5′-Cy5-CTAACCGTTTTTTTTTTTTTTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-BHQ2-3′ |
| Control probe 1† | 5′-Cy5-ACGGTTAGTTTTTTTTTTTTTTTTTATACGGTGACTGCGCCGCCGGGAAAATACTGTCTAACCGTA-BHQ2-3′ |
| “Always-on” aptamer probe | 5′-Cy5-ATCTAACTGCTGCGCCGCCGGGAAAATACTGT ACGGTTAGA-3′ |
| Control probe 2‡ | 5′-Cy5-ATACGGTGACTGCGCCGCCGGGAAAATACTGT CTAACCGTA-3′ |
*In all sequences, the A-strand is presented in italic, the T-strand is presented underlined, and the C-strand is presented in bold.
†Control probe 1 is a negative control probe for AAP, which is constructed with the A-strand subjected to an arbitrary alteration such that it shows little affinity to target cancer cells.
‡Control probe 2 is a negative control probe for always-on aptamer probe, the sequence of which is identical with the altered aptamer sequence of control probe 1.
Fig. 2.
Sequence optimization results of the AAP using flow cytometry. (A) Flow cytometry assays of target CCRF-CEM cells or nontarget Ramos cells incubated with FAM-sgc8, probe a, probe b, probe c, and probe d, respectively. (B) The corresponding histogram of the fluorescence ratios of CCRF-CEM cells to Ramos cells for the probes.
Activation of AAP by Target Cancer Cells.
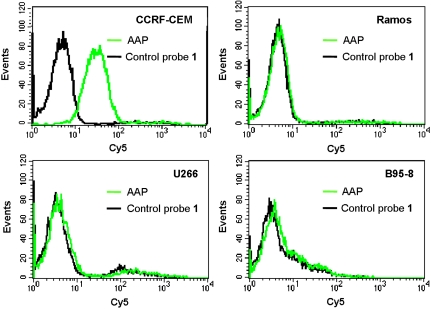
Flow cytometry assays were performed to investigate the fluorescence activation of the AAP by target cancer cells. Control probe 1, a negative control probe for the AAP, was constructed with the A-strand subjected to an arbitrary alteration such that it showed little affinity to target cancer cells. Fig. 3 depicts the fluorescence signals of the AAP and control probe 1 in response to different cell lines. It was observed that the AAP showed much higher labeling of CCRF-CEM cells than control probe 1. This revealed that the AAP was substantially activated after binding with membrane proteins of the target cancer cell with an elevated fluorescence. The activation efficiency for several nontarget cell lines, including two cancer cell lines, such as Ramos cells and U266 cells, as well as one normal cell line, such as B95-8 cells, was then investigated. We observed that fluorescence responses of the AAP to these cells did not exhibit significant difference from those obtained with control probe 1, thus showing that the AAP was not activated by these nontarget cells. This was further confirmed by flow cytometry assays of cancer cells in mouse serum with the AAP (Fig. S1). Although the surrounding environment was changed from binding buffer to mouse serum, which was more complex and unstable for the AAP, the fluorescence elevation was still detected after the AAP was activated by CCRF-CEM cells, while there was little signal enhancement observed for Ramos cells. These results implied that the AAP strategy held the desired ability to work on cell membrane proteins both in buffer and serum, and that the activation of the AAP was critically dependent on protein expression patterns of cells. Interestingly, it was found that the AAP labeled with Cy5 fluorophore and BHQ2 quencher displayed a substantially improved signal-to-background ratio (S/B ≈ 7.38) than the AAP labeled with FAM and BHQ1 (S/B ≈ 1.53), which might be attributed to the alleviation of endogenetic fluorescence of cells in the near-infrared spectral region.
Fig. 3.
Flow cytometry assays of CCRF-CEM cells, Ramos cells, U266 cells, and B95-8 cells after incubation with the AAP and control probe 1, respectively. The flow cytometry assays were performed by counting 10,000 events, and the used concentration of the probes was 25 nM.
Then, the binding assays of the AAP with CCRF-CEM cancer cells were further performed by using flow cytometry (Fig. S2). By subtracting the mean fluorescence intensity of nonspecific binding from control probe 1, the AAP was confirmed to have high affinity for CCRF-CEM cells with calculated equilibrium dissociation constants (Kd) in the nanomolar-to-picomolar range (Kd = 0.26 ± 0.02 nM). Moreover, according to the results of the binding assays, the maximum number of the AAP bound to, on average, each CCRF-CEM cell was calculated to be about 2.57 × 105.
Detection of Cancer cells with AAP.
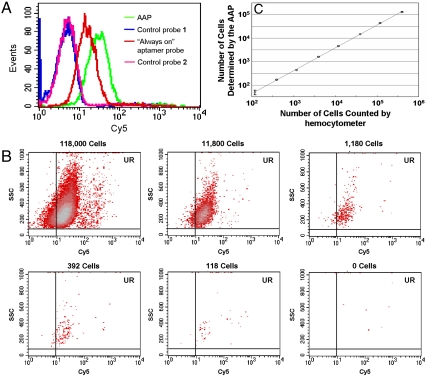
Before in vivo implementation, the AAP was tested for cancer cell detection and compared with an always-on aptamer probe that was constructed using aptamer sgc8 with a Cy5 label. As shown in Fig. 4A, after incubation with CCRF-CEM cells, the AAP achieved much higher relative fluorescence intensity, and its signal-to-background ratio was enhanced to approximately 2.5 times of that obtained by the always-on probe. It was revealed that the detection sensitivity was substantially improved by the AAP-based strategy over that of the always-on probe, resulting from the dramatically reduced background endowed by the unbound AAP in solution. This was also demonstrated through flow cytometry assays of the spiked mouse serum samples (Fig. S3).
Fig. 4.
Detection of CCRF-CEM cancer cells with the AAP. (A) Flow cytometry assay of CCRF-CEM cells detected by the AAP, in comparison with the results achieved by the “always-on” aptamer probe. The detector voltage used for control probe 1 and the AAP was 540. The detector voltage used for control probe 2 and the always-on probe was 420. (B) Flow cytometry assays of CCRF-CEM cells with decreasing cell amounts in 200 μl binding buffer using the AAP-based cancer cell detection strategy. (C) Calibration curve illustrating the relationship between the amount of CCRF-CEM cells counted by hemocytometer and the number of CCRF-CEM cells detected by flow cytometer with the AAP. Data represent mean ± standard deviation of three cell samples per group.
In order to further investigate sensitivity of the AAP for detection of CCRF-CEM cells, samples with varying CCEF-CEM cell numbers ranging from 118 to 392,000 in 200 μl binding buffer were obtained by serial dilution. To quantify target cell number, statistical analyses were performed according to the AAP-labeled events appearing in the upright (UR) region. As cell number decreased, the number of events located in the UR region decreased accordingly (Fig. 4B). Counts less than background count plus three times standard deviation were considered to be negative. Background count was determined using the same procedure without the addition of CCRF-CEM cells. For each sample, the number of CCRF-CEM cells detected by the AAP, Y, was plotted versus that measured using hemocytometer, X, as shown in Fig. 4C. The regression equation was log Y = 0.9757 × log X - 0.3321 with the smallest cell number of 118 detected in our real experiments. This low concentration was comparable to those obtained using the existing cancer detection methods that exploited aptamer-conjugated nanomaterials-based signal amplification technologies (27, 28).
In addition, the specificity of the AAP was also determined through detection of target cancer cells in mixed cell samples with different concentration ratios of CCRF-CEM to Ramos cells (Fig. S4). With the concentration ratio of CCRF-CEM to Ramos cells reduced from 9∶1 to 1∶9, the percentage of positive signals obtained using the AAP decreased accordingly from 70.36% to 14.91%. Note that, though the always-on probe could achieve an equivalent signal-to-background ratio to the AAP through the traditional sample preparation method with time-consuming washing process (Fig. S5), our assay allows a simple implementation through 15-min incubation of the AAP with the cancer cell samples before detection. This reveals that the AAP holds considerable potential as a simple, rapid, sensitive, and specific cancer cell detection strategy with no required washing step.
In Vivo Contrast-Enhanced Cancer Imaging with AAP.
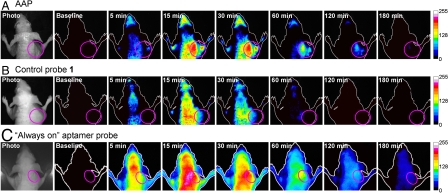
We next implemented the developed AAP for in vivo imaging of CCRF-CEM tumors implanted in nude mice. Fig. 5 displays time-dependent in vivo fluorescence images of the CCRF-CEM tumor-bearing mice after intravenous injection of 0.35 nmol labeled probes with 4.5 nmol unlabeled random oligonucleotide. The latter was injected to slow down the degradation of aptamer probes in blood. As expected, the AAP rapidly circulated within 5 min throughout the animal, and weak fluorescence signals could be seen in most parts of the body including the tumor site (Fig. 5A). At 15 min, a fluorescence image was evident in the whole body, a phenomenon that might have originated from the distribution of the AAP in subcutaneous tissues and possible degradation of the AAP in blood. More importantly, prominent fluorescence signals appeared in the tumor site at this time. This demonstrated that the AAP could be activated by target cancer cells with a faster restoration of fluorescence achieved in the tumor site than other areas. Thereafter, fluorescence signals faded continuously over most regions of the body, but a high signal contrast remained in the tumor. At 60 min, the AAP was almost cleared from blood and nontarget tissues, and the tumor was still the brightest structure visualized. Interestingly, we observed that the clearance rate of the AAP in the tumor site was much slower than that seen in other areas. This seemed to be attributed to the fact that the high affinity of the AAP to target cancer cells might protect the probes from degradation and metabolism. Even at 180 min after injection, a visible fluorescence signal was still obtained in the tumor site, again demonstrating slow clearance of the AAP in target areas.
Fig. 5.
In vivo time-dependent fluorescence imaging of CCRF-CEM tumors. CCRF-CEM tumor-bearing nude mice were intravenously injected with (A) the AAP, (B) control probe 1, and (C) the “always-on” aptamer probe, respectively. Fluorescence images of the dorsal side of live mice were then taken at several specified postinjection time points. Time of exposure for every fluorescence image was 1,000 ms in groups A and B, 100 ms in group C. The pink circle in every image locates the tumor site.
To verify that the aforementioned observations supported the feasibility of the AAP for in vivo cancer imaging, a control experiment was performed by injecting control probe 1, the fluorescence-quenched near-infrared probe, into CCRF-CEM tumor-bearing mice. As shown in Fig. 5B, control probe 1 was visualized to be distributed throughout the animal via circulation followed by a continuous clearance over most regions of the body. At 60 min, control probe 1 was almost cleared in most regions of the body, and fluorescence signals nearly vanished at 120 min. During the whole imaging process, no prominent fluorescence signal was obtained in the tumor site, indicating that control probe 1 was not accumulated or activated at the tumor site. This also strongly suggested that the fluorescence contrast achieved using the AAP did arise from specific residence or activation of the probe by the CCRF-CEM tumor. Further comparison of the AAP-based cancer imaging strategy was then performed with the always-on aptamer probe. Unlike the AAP, we observed that these always-on probes suffered high fluorescence background in the whole body from the earliest time point, even though the exposure time used for the always-on probe was as short as one tenth that of the AAP (Fig. 5C). After 15 min, fluorescence signals faded gradually over the whole body, including the tumor site. At 60 min, the tumor site was roughly distinguishable in fluorescence signals from nontarget sites with a very limited signal-to-background ratio, revealing that the high affinity of aptamer probes to target cancer cells might actually slow down their clearance in the target region. Thereafter, the probes were cleared continuously until fluorescence signals entirely disappeared in the whole body. While these findings demonstrated that the always-on aptamer probe was capable of targeting CCRF-CEM tumors in vivo, a desirable imaging contrast could not be achieved because of high fluorescence background. With distinctly different behavior, the AAP could substantially minimize the background signal originating from nontarget tissues, as often noted in this study, and it displays highly contrast-enhanced imaging from only 15 min through more than 2 h after injection.
In Vivo Specific Cancer Imaging with AAP.
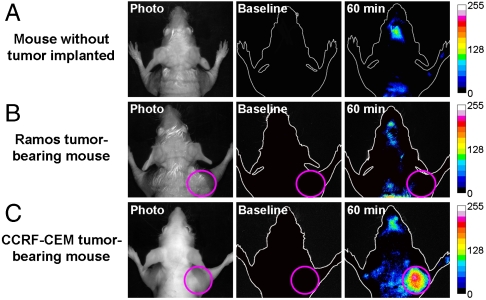
To further validate the imaging specificity, the ability of the AAP to discriminate between the CCRF-CEM tumor-bearing mice and mice with no tumor or Ramos tumor implanted was tested in vivo. According to the previous observations, a postinjection time of 60 min was selected as the optimal imaging time. As displayed in Fig. 6, a clear imaging of the CCRF-CEM tumor with substantially enhanced fluorescence signals was observed in the tumor-implanted site, the right forelimb of the tested mouse (plot C). In contrast, there was almost no fluorescence detected either in the right forelimb region of the mouse with no tumor implanted or in the Ramos tumor site (plots A and B). Although the AAP could be activated and accumulated by the target tumor, its nonspecific activation and residence in the normal tissue and nontarget tumor were minimized. These results positively support AAP as a promising specific molecular probe for in vivo cancer imaging.
Fig. 6.
In vivo specific fluorescence imaging of the CCRF-CEM tumor with the AAP. The AAP was intravenously injected into (A) the mouse with no tumor implanted, (B) the Ramos tumor-bearing mouse, and (C) the CCRF-CEM tumor-bearing mouse, respectively. Fluorescence images of the dorsal side of live mice were taken at 60-min postinjection time. Time of exposure for every fluorescence image was 1,000 ms, and the pink circles locate the tumor site.
Conclusion
We have developed a unique strategy of AAP for in vivo cancer imaging. The AAP underwent a conformational switch upon binding to proteins on target cancer cell surface with an activated fluorescence. Flow cytometry assays revealed that the AAP was specifically activated by target cancer cells and showed the improved sensitivity for detection of CCRF-CEM cells, both in buffer and serum. In vivo imaging applications demonstrated that, when compared to always-on aptamer probes, the AAP displayed substantially enhanced contrast, which will facilitate a more sensitive detection of cancer at its early stage. Furthermore, the AAP was able to give clear fluorescence imaging specific to the CCRF-CEM tumor site within 15 min after injection. This stands in contrast to always-on probes, which required a long postinjection time to obtain contrast from differentiated rates in clearing unbound and bound probes. Considering the expansion of aptamer discovery for varying cancer targets (29), the developed AAP strategy might hold great potential as a versatile molecular probe for in vivo cancer imaging with high sensitivity and specificity.
Materials and Methods
Chemicals and Materials.
All the DNA probes reported in this article were custom-designed and then synthesized by Takara Bio Inc. Sequences of the oligos are listed in Table 1. Dulbecco’s phosphate buffered saline was purchased from Sigma. Mouse serum was obtained from WACAY. All other reagents were of the highest grade available. Deionized water was obtained through the Nanopure Infinity™ ultrapure water system (Barnstead/Thermolyne Corp.). Binding buffer was prepared by adding 1 mg/mL BSA and 10% fetal bovine serum into the Dulbecco’s PBS containing 4.5 g/L glucose and 5 mM MgCl2.
Cells.
CCRF-CEM cells (T cell line, human acute lymphoblastic leukemia) were obtained from Cell Bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences. Ramos cells (B cell line, human Burkitt’s lymphoma) and U266 cells (B lymphocyte, human myeloma, plasmacytoma) were purchased from the Cancer Institute & Hospital (Chinese Academy of Medical Sciences). B95-8 cells (EBV-producing marmoset B-cell line) were provided by the Cell Center of our lab. Cells were cultured in RPMI 1640 medium supplemented with 15% fetal bovine serum (FBS) and 100 IU/mL penicillin-streptomycin and incubated at 37 °C in a humidified incubator containing 5% wt/vol CO2. The cell density was determined using a hemocytometer, and this was performed prior to any experiments.
Animals.
Male athymic BALB/c (Balb/C-nu) mice were obtained from the Shanghai SLAC Laboratory Animal Co., Ltd. (BALB/c). They were 4–6 weeks old at the start of each experiment and weighed 20–25 g. All animal operations were in accord with institutional animal use and care regulations, according to protocol No. SYXK (Xiang) 2008-0001, approved by the Laboratory Animal Center of Hunan.
Flow Cytometry Assays.
Generally, probes were incubated with 2 × 105 cells in 200 μl binding buffer at normal temperature for 15 min in the dark and then immediately determined with a FACScan cytometer (BD Biosciences) by counting 10,000 events. Especially for the detection sensitivity assay, different amounts of CCRF-CEM cells were stained by 25 nM activatable aptamer probe (AAP) in 200 μl binding buffer at normal temperature in the dark. After incubation for 15 min, the samples were immediately detected with flow cytometer at high rate by counting the AAP-labeled events appearing in the upright (UR) region for 2 min. The number of samples used to derive statistical information for each cell concentration was 3.
The binding affinity of the AAP was determined by incubating CCRF-CEM cells (approximately 1.55 × 105) on ice for 50 min in the dark with varying concentrations of the AAP in a 250-μl volume of binding buffer containing 20% FBS and 0.1 mg/ml yeast tRNA. Cells were then washed twice with 0.3 ml of the binding buffer with 0.1% sodium azide, suspended in 0.2 ml of binding buffer with 0.1% sodium azide, and subjected to flow-cytometric analysis. Control probe 1 was used as a negative control to determine nonspecific binding. All of the experiments for binding assay were repeated three times. The mean fluorescence intensity of CCRF-CEM cells labeled by the AAP was used to calculate for specific binding by subtracting the mean fluorescence intensity of nonspecific binding from control probe 1. The equilibrium dissociation constant (Kd) of the AAP–cell interaction was obtained by fitting the dependence of fluorescence intensity of specific binding on the concentration of the AAP to the equation Y = BmaxX/(Kd + X), using SigmaPlot.
In Vivo Fluorescence Imaging.
Four-week-old male BALB/c nude mice received a subcutaneous injection of 5 × 106 in vitro-propagated cancer cells into the backside. Tumors were then allowed to grow for 3–4 weeks to 1–2 cm in diameter. Before imaging, BALB/c nude mice, with or without tumors, were anesthetized with the combined use of tranquilizer and anesthetic. In detail, a 2 mg/kg dose of chlorpromazine hydrochloride was first injected intramuscularly, and several minutes later, an intraceliac injection was performed with an 80 mg/kg dose of pentobarbital sodium solution. Once the mice were anesthetized to be motionless, a 140 μl volume of physiological saline containing 0.35 nmol of labeled probes and 4.5 nmol of unlabeled random oligonucleotide was injected intravenously via the tail vein. At specified times, fluorescence images of the dorsal side of live mice were taken by a Maestro™ in vivo fluorescence imaging system (Cambridge Research & Instrumentation, Inc.). A 640 nm (± 25 nm) bandpass filter and a 680 nm longpass filter were selected to be used as the excitation filter and the emission filter, respectively. All the fluorescence images were presented after processing by the Image J software (version 1.38x).
Supplementary Material
Acknowledgments.
This work was supported by Program for Innovative Research Team of Hunan National Science Foundation (10JJ7002), International Science & Technology Cooperation Program of China (2010DFB30300), Program for Changjiang Scholar and Innovative Research Team in University Program for New Century Excellent Talents in University (NCET-06-0697, NCET-09-0338), and National Science Foundation of P. R. China (90606003, 20775021).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016197108/-/DCSupplemental.
References
- 1.Becker A, et al. Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat Biotechnol. 2001;19:327–331. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- 2.Thekkek N, Richards-Kortum R. Optical imaging for cervical cancer detection: Solutions for a continuing global problem. Nat Rev Cancer. 2008;8:725–731. doi: 10.1038/nrc2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellington A-D, Szostak J-W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Daniels D-A, Chen H, Hicke B-J, Swiderek K-M, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavitian B, et al. In vivo imaging of oligonucleotides with positron emission tomography. Nat Med. 1998;4:467–471. doi: 10.1038/nm0498-467. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt K-S, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urano Y, et al. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat Med. 2009;15:104–109. doi: 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicke B-J, et al. Tumor targeting by an aptamer. J Nucl Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 10.Wang A-Z, et al. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem. 2008;3:1311–1315. doi: 10.1002/cmdc.200800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang D-W, et al. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Sefaha K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, et al. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem-Asian J. 2010;5:2209–2213. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 14.Hama Y, Urano Y, Koyama Y, Choyke P-L, Kobayashi H. Activatable fluorescent molecular imaging of peritoneal metastases following pretargeting with a biotinylated monoclonal antibody. Cancer Res. 2007;67:3809–3817. doi: 10.1158/0008-5472.CAN-06-3794. [DOI] [PubMed] [Google Scholar]
- 15.Weissleder R, Tung C-H, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 16.Jiang T, et al. Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc Natl Acad Sci USA. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blum G, von Degenfeld G, Merchant M-J, Blau H-M, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 18.Olson E-S, et al. Activatable cell penetrating peptides linked to nanoparticles as dual probes for in vivo fluorescence and MR imaging of proteases. Proc Natl Acad Sci USA. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hama Y, et al. A target cell-specific activatable fluorescence probe for in vivo molecular imaging of cancer based on a self-quenched avidin-rhodamine conjugate. Cancer Res. 2007;67:2791–2799. doi: 10.1158/0008-5472.CAN-06-3315. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa M, et al. Fluorophore-quencher based activatable targeted optical probes for detecting in vivo cancer metastases. Mol Pharm. 2009;6:386–395. doi: 10.1021/mp800115t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa M, Kosaka N, Choyke P-L, Kobayashi H. H-type dimer formation of fluorophores: A mechanism for activatable, in vivo optical molecular imaging. ACS Chem Biol. 2009;4:535–546. doi: 10.1021/cb900089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, et al. Molecular engineering of DNA: Molecular beacons. Angew Chem Int Edit. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z, et al. Aptamer switch probe based on intramolecular displacement. J Am Chem Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 24.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shangguan D, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frangioni J-V. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Smith J, et al. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 28.Medley C-D, et al. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 29.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.