Abstract
Observing individual molecules in a complex environment by fluorescence microscopy is becoming increasingly important in biological and medical research, for which critical reduction of observation volume is required. Here, we demonstrate the use of vertically aligned silicon dioxide nanopillars to achieve below-the-diffraction-limit observation volume in vitro and inside live cells. With a diameter much smaller than the wavelength of visible light, a transparent silicon dioxide nanopillar embedded in a nontransparent substrate restricts the propagation of light and affords evanescence wave excitation along its vertical surface. This effect creates highly confined illumination volume that selectively excites fluorescence molecules in the vicinity of the nanopillar. We show that this nanopillar illumination can be used for in vitro single-molecule detection at high fluorophore concentrations. In addition, we demonstrate that vertical nanopillars interface tightly with live cells and function as highly localized light sources inside the cell. Furthermore, specific chemical modification of the nanopillar surface makes it possible to locally recruit proteins of interest and simultaneously observe their behavior within the complex, crowded environment of the cell.
Keywords: nanowire, confocal volume, sub-diffraction, nano-bio interface
The rapidly evolving field of nanotechnology creates new frontiers for biological sciences, such as quantum dots for fluorescence imaging, electrode-free nanometer light sources, nanowire-based transistor biosensors, and carbon nanotube Raman probes (1–7). In 2007, an intriguing study by Kim et al. reported that vertical silicon nanowires penetrated living mammalian cells that grew attached to the substrate without affecting the cell viability in the long term (8). Recently, Shalek et al. reported the use of vertical silicon nanowires to deliver biomolecules such as proteins and DNA plasmids into living cells that grew attached to the nanowires (9). Together with other works that show vertical nanowires can support cell culture (10–14), these observations suggest a new and exciting possibility of using vertical nanowires as a universal platform to probe intracellular molecular events.
Far-field fluorescence microscopy remains one of the most popular tools in studying interactions, dynamics, and functions in biological systems. However, for highly sensitive measurements such as single-molecule fluorescence imaging, it is essential to restrict the detection volume so that only one fluorescent molecule is observed at any given time (15–18). It is this nontrivial task to which nanotechnology can contribute, simultaneously improving access to the cell interior and limiting detection volume by virtue of the same nanoscale features. Previously, several techniques for limiting observation volumes have relied on the generation of evanescent waves, which arise when wave propagation over a boundary is forbidden so that the intensity of the electromagnetic field decays exponentially at the interface. One commonly used example of this approach is total internal reflection fluorescence (TIRF) microscopy, which generates an evanescent wave at a refractive boundary when the incident light is beyond the critical angle (19, 20). This evanescent field penetrates only approximately 100 nm into the solution and thus enables a selective visualization of fluorescence molecules near the surface. Another technique, zero-mode waveguide, utilizes subwavelength apertures in a metal film to block propagating modes of light, which produces a tightly confined evanescent field within the aperture cavity to achieve an excitation volume well below the diffraction limit (21–23).
While both TIRF and zero-mode waveguide drastically reduce the excitation volume and have proven to be valuable tools for single-molecule imaging, the volume confinement is usually restricted to a small depth along a flat surface or inside the extremely small aperture cavity. For live cell studies, this very characteristic that provides their utility also serves as a limitation: They are mainly used to probe cellular events on the plasma membrane (24–26) and do not allow deeper imaging into the cell interior. Likewise, other related methods such as the nanoprobe-based near-field scanning microscopy are subject to the same working distance limitations and additionally suffer from probe inconsistencies and incompatibility with parallel implementations (27, 28). Therefore, a detection method that affords a sub-diffraction-limited excitation volume as well as access to the cell interior will improve the sensitivity and cellular application of fluorescence measurements such as fluorescence correlation spectroscopy (29, 30), Forster resonance energy transfer (31, 32), and fluorescence fluctuation spectroscopy (33, 34).
In this work, we explore the use of vertical nanopillars to modulate light for the generation of a quickly decaying excitation field in a live cell. For this purpose, we fabricated transparent vertical silicon dioxide nanopillars in a substrate coated with a nontransparent platinum layer. With a diameter smaller than the wavelength of visible light, the transparent nanopillar restricts the propagation of light and generates an evanescence wave along its vertical surface that runs to about 1 μm deep into cell interior. Therefore, we employed the vertical nanopillars to create a highly confined observation volume that has been demonstrated to achieve single-molecule observation in high fluorescence background. In addition, we show that with specific surface modifications, silicon dioxide nanopillars can also serve as chemical probes to address cellular events with high specificity. As such, the vertical nanopillar affords a versatile experimental platform for simultaneous recruitment and sensitive optical measurements of proteins of interest in the natural cell environment.
Results
Fabrication of Nanopillar Substrates.
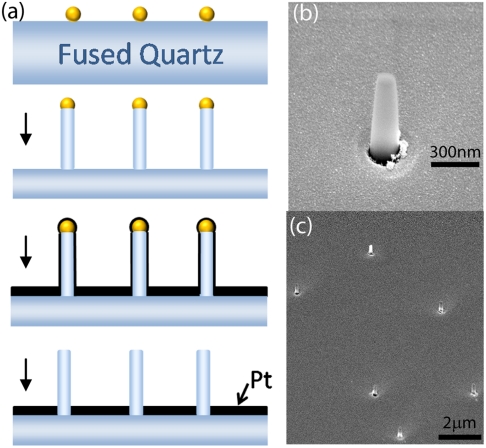
The nanopillar probes are vertical silicon dioxide nanopillars with diameters smaller than the wavelength of visible light. The SiO2 nanopillars were fabricated on fused quartz substrates with gold nanoparticles as etching masks. The fabrication diagram is schematically shown in Fig. 1A. Briefly, the gold nanoparticles were first randomly deposited on a poly-L-lysine (PLL) coated fused quartz substrate by spin coating. Exposed areas of silicon dioxide substrate not protected by gold nanoparticles were selectively and anisotropically etched by reactive ion etching (RIE) using fluorine-based gases (see Materials and Methods for detailed procedures). The substrate was then coated with a 10 nm titanium layer to facilitate attachment of the subsequent 100 nm platinum layer by electron-beam evaporation. Platinum was chosen over other metals for its ability to effectively block visible light and its biologically inert character. The gold nanoparticles, as well as the excess metal on the pillar sidewalls, were then removed by a brief etching in 2% hydrogen fluoride aqueous solution.
Fig. 1.
Fabrication of the SiO2 nanopillars. (A) Schematic illustration of the fabrication process with gold nanoparticles as etching masks. (B) Typical SEM image of a nanopillar with viewing angle at 52°. (C) SEM image of randomly dispersed nanopillars with viewing angle at 52°. The average distance between nanopillars is about 3–5 μm.
The diameter of the SiO2 nanopillars is determined by the size of the masking gold nanoparticles and the height is determined by the duration of the RIE process. Vertical silicon dioxide nanopillars with diameters ranging from 80 to 400 nm and heights from 300 nm to 1.5 μm were successfully fabricated. For subsequent experiments, we mainly used nanopillars of 150 nm in diameter and 700 nm in height. Scanning electron microscopy (SEM) images of the nanopillar substrate are shown in Fig. 1B and C. In the SEM images, the platinum substrate surface showed nanometer-scale unevenness that is typical of e-beam evaporation, while the silicon dioxide surface of the nanopillars was smooth. The typical interpillar distance was 3–5 μm, which is determined by the applied density of the gold nanoparticles.
Illumination Profile Surrounding the Nanopillar.
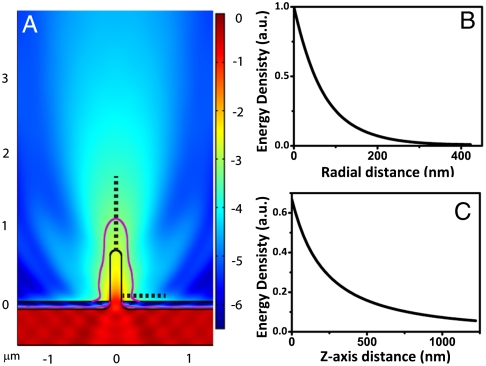
The diameter of the nanopillar (150 nm) is significantly smaller than the wavelength of probing light—this prohibits light propagation through the pillar and has a profound effect on the illumination profile. Like a waveguide, the cutoff wavelength above which no propagating modes exist in the nanopillar is dependent on the thickness of the metal coating, the size, the shape, and the material of the nanopillar. We performed a finite-element analysis simulation to numerically solve the distribution of the electromagnetic field around the pillar. The light used in the simulation was 480 nm circularly polarized light and the metal coating is 110 nm in thickness. The pillar dimensions in the simulation are the same as in our experiments—namely, 150 nm in diameter and 700 nm in height (more simulation details can be found in Materials and Methods).
A cross-section of the calculated intensity profile surrounding the nanopillar is shown in Fig. 2A. The intensity profile decays rapidly in both the axial and the radial directions as the wave propagation is truncated at the subwavelength aperture in the platinum. To avoid the interference-induced numerical instability at the platinum surface, we choose to normalize the light intensity against the value at the surface of the nanopillar, 100 nm above the platinum layer. A threshold contour of 1/e2 exhibits a shape that somewhat resembles that of candle light (red curve in Fig. 2A). In the radial direction (Fig. 2B), the light intensity decays exponentially from the surface of the nanopillar, indicating an evanescent wave at the pillar–water interface. In the axial direction (Fig. 2C), the intensity decays more slowly, penetrating to a depth of approximately 500 nm from the tip of the nanopillar. The shape of the decay, as compared with that from a comparably sized subwavelength aperture lacking the silicon dioxide pillar, indicates that the presence of the pillar aids the persistence of light along its length. This effect is most probably due to the higher refractive index of the silicon dioxide as compared to the surrounding water. We estimate the observation volume of illumination through the nanopillar to be 10-16 L. By comparison, this is an order of magnitude smaller than the excitation volume of two-photon fluorescence microscopy at 10-15 L (25).
Fig. 2.
Illumination profile surrounding a nanopillar. (A) Finite-element analysis of the intensity distribution for a nanopillar of 150 nm in diameter and 700 nm long. Purple line indicates the contour of 1/e2 intensity (the intensity at the nanopillar surface 100 nm away from the platinum substrate is designated to be 1). (B) Plot of the light intensity as a function of radial distance along the horizontal dash line in A. The light intensity decays exponentially from the surface of the nanopillar. (C) Plot of the light intensity as a function of z distance along the vertical dash line in A. The light intensity decays more slowly along the z axis, penetrating approximately 500 nm from the tip of the nanopillar.
Specific Surface Modification of SiO2 Nanopillars for Single-Molecule Imaging.
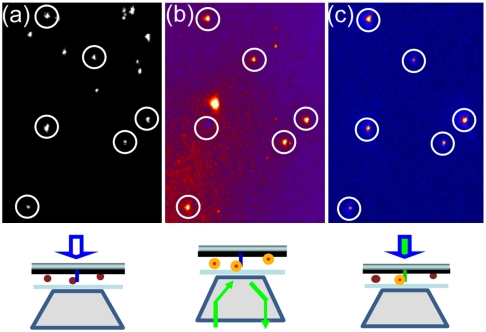
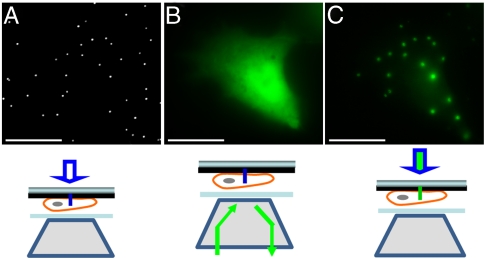
Many single-molecule studies involve immobilizing biological molecules on the surface. In order to use the evanescence wave on the nanopillar surface for single-molecule imaging, the need arose for specific surface modification of the silicon dioxide nanopillars. This was accomplished first by silanization with aminopropyltriethoxysilane (APTES), which generated a surface with terminal primary amines. These amines were then reacted with NHS-Biotin. The biotin-modified surface could now bind through a streptavidin linker to any molecule of interest. We used 605 nm streptavidin-conjugated quantum dots (QD-STA) as a model system to test the use of nanopillars for single-molecule fluorescence imaging. For imaging purposes, the nanopillar substrate was flipped over onto a coverslip so that the nanopillars were pointing downward toward the objective of an inverted microscope. Brightfield images with white-light illumination from above (white-light mode) revealed the nanopillar locations (Fig. 3A). Quantum dots were excited with a 532 nm laser either through the objective (epi-illumination mode) or through the nanopillars (nanopillar-illumination mode) (Fig. 3 B and C). Fluorescence signal was collected through the objective and detected by a cooled, back-illuminated CCD camera at 10 frames per second.
Fig. 3.
Fluorescence imaging of QD-STA using nanopillar illumination. (A) The locations of randomly distributed nanopillars are revealed by bright field imaging. (B) Epi-illumination (through the objective) excites those fluorescent QD-STAs that bind to biotinylated nanopillar surfaces and also those that are nonspecifically stuck on the platinum surface. (C) Fluorescence imaging by nanopillar illumination exclusively excites those QD-STAs that are localized on the nanopillar surfaces with extremely clean fluorescence background. The circles indicate where QD-STA colocalizes with a nanopillar. Schematic illustrations of the microscope setup for three illumination modes are sketched below each image. Note that one of the circled QD-STA is invisible in B due to QD photoblinking.
Single-molecule fluorescence imaging of QD shows that the surface modification of nanopillars is highly specific (Fig. 3). About a quarter of all nanopillars were labeled with QD (mostly singles) after incubating with 5 pM QD-STA for 10 min. Despite the nanopillar surface comprising less than 2% of the total surface area, epi-illumination revealed that the majority (> 80%) of QD-STA colocalized with the biotin-functionalized nanopillars. Nonspecifically surface-immobilized fluorescent molecules, such as those QD on the platinum surface, often create strong background for conventional fluorescence imaging. However, they were totally invisible in the nanopillar-illumination mode as shown in Fig. 3C. Compared with the epi-illumination, the nanopillar illumination gives rise to highly confined excitation volume and generates extremely low fluorescence background. The highly specific modification of the nanopillar surface also enables us to locally attach biological molecules of interest for live cell studies as shown in later sections.
Nanopillar Illumination for Single-Molecule Imaging with High Concentration of Fluorescence Molecules.
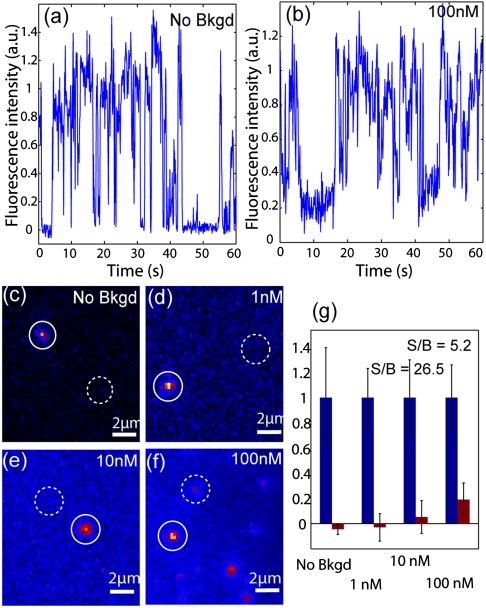
For an experimental probe of the observation volume and the concentration upper limit for single-molecule detection using nanopillar illumination, background fluorescence was created by the addition of polyethylene glycol-coated QD (QD-PEG) with the same emission wavelength. Thus, QD-STA, as the molecule of interest, was tethered to the nanopillars via biotin–streptavidin interaction as described in the last section, while QD-PEG diffused freely in solution to provide consistent background fluorescence. The intensity of the background fluorescence is proportional to the concentration of QD-PEG in solution. Of the four concentrations of QD-PEG tested (0 nM, 1 nM, 10 nM, and 100 nM), single QD-STA on the nanopillars can be clearly identified in all cases using nanopillar illumination. As shown in Fig. 4 A and B, fluorescence intensities of QD-STA show characteristic single-step photoblinking.
Fig. 4.
Nanopillar illumination enables single-molecule detection with high fluorescence background. Characteristic single-step photoblinking of QD-STA is evident both at (A) 0 nM and (B) 100 nM of QD-PEG fluorescence background. (C–F) Typical fluorescence images using nanopillar illumination show signal intensity and background intensity at each of the four QD-PEG concentrations 0 nM, 1 nM, 10 nM, and 100 nM. The signal strength is the fluorescence intensity of a QD-STA tethered to the nanopillar in the “on” state (solid circles). The background intensity is the fluorescence intensity at the nanopillar location when the attached QD-STA is in the “off” state (dotted circles). (G) Relative signal (blue) and background (red) intensities at each QD-PEG concentrations.
Fig. 4 C–F shows fluorescence images of QD-STA attached to nanopillars—one in the on state (solid circle) and the other in the photoblinking off state (dotted circle). The fluorescence intensity measured from the site of the nanopillar while the quantum dot was in the off state was taken as the background. All fluorescent intensities were baseline corrected by subtracting the signal intensity from the area of the nontransparent platinum. Therefore, the background fluorescence at the nanopillar place, when the QD-STA is in the off state, arises when the QD-PEG diffuses into the excitation volume. A summary of the average signal and background observed at each QD-PEG concentration is plotted in Fig. 4G. As expected, the background intensity at the nanopillar locations with 0 nM QD-PEG was indistinguishable from the surrounding nontransparent regions. With the addition of 1 nM QD-PEG, the background was likewise negligible. At 10 nM QD-PEG, the average background signal rose above zero but still fluctuated around the level of the surrounding area. It was only when the concentration of QD-PEG in solution reached 100 nM that the background reached a consistently positive level, at which the signal to background ratio was calculated to be approximately 5. Thus, single-molecule detection through nanopillar-illumination can be achieved at solution concentrations of up to 100 nM. This detection tolerance already renders some intracellular targets accessible.
Mammalian Cells Interfacing Tightly with the Nanopillar Substrate.
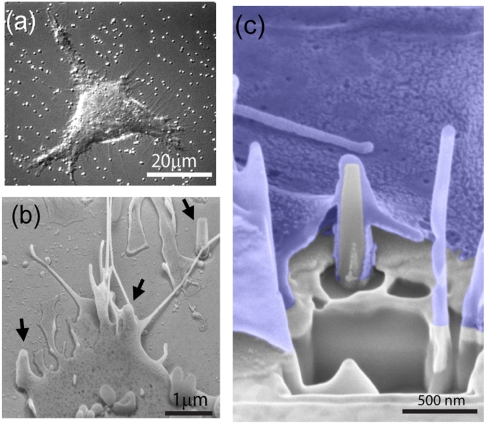
As previously stated, one of the primary motivations for developing these SiO2 nanopillar probes is to exploit their optical properties in the context of studying live cells. After coating with poly-L-lysine (PLL), nanopillar substrates have proven to be viable surfaces for culturing multiple mammalian cell types, including PC12M and Cos7 (Fig. 5A). Mammalian cells attach and spread on the nanopillar substrate within an hour of plating and are viable for weeks of culture. Cell division is not hindered by the presence of vertical nanopillars, and cell survival rate is comparable to standard cultures on flat PLL-coated glass. Furthermore, phase contrast images show that the cells indeed spread on the pillars, which are seen as localized bright spots under the white light (Fig. 5A). On average, there are 10–20 nanopillars per cell.
Fig. 5.
Mammalian cells cultured on nanopillar substrates. (A) Phase contrast image of a PC12M cell on the nanopillar substrate. Pillars can be seen as bright spots both inside and outside the cell. (B) An SEM image of a cell interacting with nanopillars. Arrows indicate locations of three nanopillars. (C) An SEM image of the interface between the cell and the nanopillar shows that the top- and bottom- cell membranes wrapping around the pillar. The cell is false-colored in blue for clarity. The interface cross-section was revealed by focused-ion-beam milling.
To examine the nanopillar–cell interface, PC12M cells cultured atop the pillars were fixed and imaged by SEM (Fig. 5B). The cells not only survived on top of the nanopillars but also engulfed them (Fig. 5 B and C). In most cases, nanopillars interfacing with cells were covered by the cell body and not easily identified under SEM. At the edge of the cell where the cell is thin and flat, tent-like structures often indicate a nanopillar below (Fig. 5B). We used focused-ion-beam milling method to cut a cross-section to expose the cell–nanopillar interface (Fig. 5C). The top cell membrane and the bottom cell membrane can be clearly seen as they wrap around the nanopillar, yielding very tight contacts between cell and pillar. Previous work by Shalek et al. demonstrated the capability of similarly structured vertical Si nanopillars to access the cell interior for the purpose of biomolecule delivery (9).
Fluorescence Imaging of Live Cells with Nanopillar Illumination.
As a demonstration of nanopillars functioning as highly localized light sources within live cells, PC12M cells were transfected with green fluorescence protein (GFP) and plated on a nanopillar substrate. The locations of all nanopillars were revealed by transillumination with white light (Fig. 6A). Excitation with a 488 nm laser by epi-illumination revealed a diffuse, bulk fluorescence over the entire cell (Fig. 6b). In the epi-illumination mode, the fluorescence intensity at nanopillar locations is comparable or slightly lower than that of its immediate surroundings, likely due to the exclusion volume of the nanopillar itself. In contrast, excitation by nanopillar illumination provided a highly localized fluorescence signal around the pillars (Fig. 6C), and the fluorescence spots colocalize perfectly and exclusively with the sites of the nanopillars inside the region of the cell. Because GFP is a cytosolic protein, the cell body that contains the bulk of the cytosol appears brighter in epi-fluorescence than the edges of the cell or cell processes. As a result, small and thin cell protrudings that contain little cytosol can be difficult to see in typical epi-illumination. This limitation does not apply to fluorescence illumination through the nanopillars, where the excitation is highly localized. As was seen in Fig. 6C, the brightness of the fluorescence excited through the pillars is less dependent on the location in the cell.
Fig. 6.
Fluorescence imaging using nanopillar illumination in live cells. (A) White-light imaging reveals the locations of nanopillars. (B) Fluorescence imaging by epi-illumination shows the cell shape of a GFP transfected cell. (C) Nanopillar illumination excites only those fluorescence molecules that are very close to nanopillars inside the cell, giving rise to fluorescence spots perfectly colocalized with the nanopillars. Scale bar : 10 μm.
Using Nanopillars to Simultaneously Recruit and Excite Molecules of Interest in the Cell.
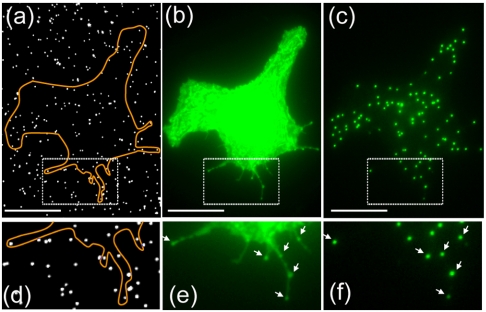
Not only can the pillars be used as localized light sources, the modification of molecules to the surface of the silicon dioxide pillars can also be used to recruit and isolate molecules of interest from within the complex, crowded environment of the cell. As a proof of principle, a biotinylated antibody against GFP was crosslinked to the biotin-modified nanopillar via the multivalent streptavidin linker. COS7 cells were transfected with a plasmid encoding a GFP-fused transmembrane protein synaptobrevin. Cells expressing GFP-synaptobrevin were then plated on the nanopillar substrates with anti-GFP antibodies linked to the pillars and allowed to attach to the surface for 3 h.
Through-the-objective fluorescent imaging of the cell revealed the shape of the cell with several cell protrudings (Fig. 7B). Locations of nanopillars that are linked with anti-GFP antibodies usually show brighter fluorescence compared with their immediate surrounding areas. This indicates that some GFP-synaptobrevin proteins were recruited to the nanopillars via GFP and anti-GFP interactions. The recruitment is more clearly seen at edges and protrudings where the overall fluorescence intensity is lower (Fig. 7E). It is interesting to note that cell protrudings seem to grow preferentially toward the nanopillars, and many protrudings end at the nanopillar locations. Nanopillar illumination shows extremely clean signal, colocalizing perfectly with the nanopillars inside the cell area (Fig. 7 C and F). The success of this scheme indicates that nanopillars not only function as localized light sources inside the cell but can also locally probe cellular events or recruit molecules of interest.
Fig. 7.
Antibody-labeled nanopillars simultaneously recruit and illuminate proteins of interest in live cells. (A) White-light imaging reveals the locations of the nanopillars modified with antibodies against GFP. (B) Fluorescence imaging by epi-illumination shows the shape of a COS7 cell expressing GFP-synaptobrevin. (C) Nanopillar illumination shows extremely clean signal, colocalizing perfectly with the nanopillars inside the cell area. (D–F) Zoom-in images show that nanopillar locations usually have brighter fluorescence compared with surrounding areas, suggesting that some GFP-synaptobrevin proteins were recruited to the nanopillars via GFP and anti-GFP interactions. Scale bar : 10 μm.
Discussion
Vertical SiO2 nanopillars provide highly localized excitation light sources for in vitro systems as well as in live cells. The sub-diffraction-limited excitation volume would be very useful for probing only a small number of fluorescence molecules in a crowded environment. For example, nanopillar probes could be used to photoactivate a small number of cell-expressed photoactivatable fluorescent proteins (35, 36), which would allow one to follow the fate or diffusion of individual molecules in live cells. We are currently working on using nanopillar illumination for local photo uncaging of glutamate (37–40) for highly localized application of neurotransmitter. Molecular specificity can be added to the optical localization by virtue of the molecular modification available to the nanopillars.
Materials and Methods
Fabrication of SiO2 Nanopillars.
Four-inch fused-silica wafers (500 μm thickness) were cleaned by standard prediffusion cleaning method (41) and subsequently coated with Poly-L-Lysine (PLL) by soaking in 1% PLL solution for 5 min. After thoroughly rinsing the wafers with DI water, gold colloidal nanoparticle solution (Ted Pella) was applied on the wafer surface and incubated at room temperature for 10 min before rinsing off. The area density of the nonspecifically stuck gold colloidal particles depends on the concentration of the colloidal solution applied. For most of our experiments, the density was controlled to be about one per 10 μm2. The wafers were then dry etched in a reactive ion etcher with a gas mixture of O2 and CHF3 at flow rates of 30 standard cubic centimeters per minute (sccm) for O2 and 50 sccm for CHF3. The pressure was kept at 40 milli-Torr and the voltage to ionize the gas mixture was maintained at 400 V. The RIE process lasted for 20–25 min at an etching rate of 35 nm/ min. Following the RIE process, a 5 min O2 plasma etching step removed the organic residue produced by the RIE etching step. Ti and Pt layers were sequentially deposited by electron-beam evaporation at 1–2 Å/s to produce a double layer of 10nmTi/100nmPt. The 10 nm Ti underlayer allowed for good adhesion of Pt overlayer to the fused quartz substrate. The 100 nm Pt layer effectively blocks > 99.9% of visible light. To remove the excess metal deposited on the top and side walls of the nanopillars, a brief hydrogen fluoride (HF) etching was applied. 2% HF solution etches SiO2 slowly and can be well controlled. About 5 nm SiO2 was removed from each nanopillar in order to lift off the excess metal on the pillar side walls.
Surface Modification of SiO2 Nanopillars.
For preparation and modification of the surface, silicon dioxide nanopillar substrates were first cleaned by oxygen plasma to remove any organic residues on the surface. The substrates were then silanized with 2% aminopropyltriethoxysilane (APTES) in acetone for 1 h, resulting in amine-terminated surface. After washing with acetone followed by isopropanol, the surfaces were dried in air. The new terminal amines were then reacted with 0.1 mg/ml NHS-Biotin (Thermo Scientific) for 1 h by diluting 1 mg/ml stock solution in DMSO into phosphate buffered saline (PBS). After thorough washing with PBS, the resulting biotin-terminated nanopillar substrates were stored in clean container for future uses. For single-molecule imaging of QD-STA, the biotin-modified substrate was first blocked with 0.1 mg/ml BSA in PBS for 1 hr before applying QD-STA at concentrations stipulated by the experiment. For high concentration background studies, QD-PEG solutions at various concentrations were supplied with 0.1 mg/ml BSA in water.
Computer Simulation of Optical Profile Surrounding the Nanopillar.
Finite-element time-domain simulations of the optical intensity near the nanopillars were carried out using Comsol (COMSOL Inc.). The light used in the simulation was 480 nm circularly polarized light. The refractive indices for fused quartz, water, and Pt were set at 1.48, 1.33, and 1.94–3.38i, respectively. The dimension of simulation area was set at 4 × 4 μm, an area large enough so that the boundary effect is not significant. The grid spacing was set between 30 and 150 nm according to the need of spatial resolution. Finer grids were used for nanopillar and surrounding areas. The adaptive mesh consists of 30,000–35,000 elements.
Imaging Setup for fluorescence Imaging with Epi- and Nanopillar Illuminations.
An inverted Nikon ECLIPSE Ti microscope was modified to incorporate both epi- and nanopillar-illumination modes. The excitation light source was a 50 mW 488 nm laser (Spectra-Physics). For epi-illumination, the excitation laser beam was introduced into a TIRF objective (1.49 NA, Nikon) that focused the light on the sample from the bottom. The beam hit the nanopillar substrate at an angle and was reflected by the Pt layer. For the nanopillar illumination, the laser beam was guided above the microscope and hit the sample from the top. The beam was focused to an approximately 100-μm diameter spot covering the field of view uniformly. In both illumination modes, the fluorescent emission was collected by the same objective, passed through a long-pass dichroic mirror and a band-pass emission filter, and collected by an EMCCD camera (Andor). For fluorescence imaging, the substrates were housed in a sealed custom-designed polydimethylsiloxane chamber to prevent evaporation of the solutions.
Scanning Electron Microscopy.
To prepare samples for SEM imaging, PC12M cells were plated on nanopillar substrates and allowed to attach for 1 d. The cells were then fixed overnight in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3), washed for 5 min in the same buffer, and postfixed for 1–2 h in 1% osmium tetroxide. After washing in DI water twice for 5 min each, the sample was dehydrated by successive exchanges to increasing concentrations of ethanol (50%, 70%, 90%, 100%, and 100% again). The sample in 100% ethanol was then dried with liquid CO2 in a critical point drier and sputter coated with a 2 nm layer of Pd/Au to enhance the contrast. The sample was imaged using a FEI Strata 235B dual-beam SEM/FIB system that combines high resolution SEM imaging and ion beam etching at the same time. FIB provides submicron dissection of the prepared samples in order to expose the cell–nanopillar cross-section.
Cell Culture on the Nanopillar Substrate.
PC12M or COS7 cells were plated on nanopillar substrates at a cell density of 106/ml and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Plasmid transfections were achieved by adding a mixture of 1 μg of DNA and 3 μl of lipofectamine to each well in a 12-well tissue culture plate. The cells were allowed to recover for 24 hrs before replated on the nanopillar substrate. To label anti-GFP antibodies to the nanopillars, the biotin-conjugated nanopillar substrates were prepared as noted previously and treated with 1 nM nonfluorescent streptavidin for 10 min. After washing out excess streptavidin, 1 nM biotin-labeled anti-GFP antibody (Invitrogen) was linked to the nanopillars via the multivalent streptavidin linker.
Acknowledgments.
This work was supported by the Bio-X Interdisciplinary Initiatives Program, National Institutes of Health Grant NS057906, a Dreyfus new faculty award, a Searle Scholar Award, and a Packard Science and Engineering Fellowship to B.C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 2.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 3.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama Y, et al. Tunable nanowire nonlinear optical probe. Nature. 2007;447:1098–1101. doi: 10.1038/nature05921. [DOI] [PubMed] [Google Scholar]
- 6.Tian B, et al. Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science. 2010;329:830–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kam NW, O’Connell M, Wisdom JA, Dai H. Carbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destruction. Proc Natl Acad Sci USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. Interfacing silicon nanowires with mammalian cells. J Am Chem Soc. 2007;129:7228–7229. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- 9.Shalek AK, et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci USA. 2010;107:1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie C, et al. Noninvasive neuron pinning with nanopillar arrays. Nano Lett. 2010;10:4020–4024. doi: 10.1021/nl101950x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallstrom W, et al. Gallium phosphide nanowires as a substrate for cultured neurons. Nano Lett. 2007;7:2960–2965. doi: 10.1021/nl070728e. [DOI] [PubMed] [Google Scholar]
- 12.Jiang K, et al. Medicinal surface modification of silicon nanowires: Impact on calcification and stromal cell proliferation. ACS Appl Mater Interfaces. 2009;1:266–269. doi: 10.1021/am800219r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi S, Yi C, Ji S, Fong CC, Yang M. Cell adhesion and spreading behavior on vertically aligned silicon nanowire arrays. ACS Appl Mater Interfaces. 2009;1:30–34. doi: 10.1021/am800027d. [DOI] [PubMed] [Google Scholar]
- 14.Turner AM, et al. Attachment of astroglial cells to microfabricated pillar arrays of different geometries. J Biomed Mater Res. 2000;51:430–441. doi: 10.1002/1097-4636(20000905)51:3<430::aid-jbm18>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 15.Moerner WE. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci USA. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sako Y, Yanagida T. Single-molecule visualization in cell biology. Nat Rev Mol Cell Biol. 2003;(Suppl):SS1–5. [PubMed] [Google Scholar]
- 17.Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 18.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 19.Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 20.Sako Y, Uyemura T. Total internal reflection fluorescence microscopy for single-molecule imaging in living cells. Cell Struct Funct. 2002;27:357–365. doi: 10.1247/csf.27.357. [DOI] [PubMed] [Google Scholar]
- 21.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 22.Moran-Mirabal JM, Craighead HG. Zero-mode waveguides: Sub-wavelength nanostructures for single molecule studies at high concentrations. Methods. 2008;46:11–17. doi: 10.1016/j.ymeth.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Samiee KT, Foquet M, Guo L, Cox EC, Craighead HG. λ-Repressor oligomerization kinetics at high concentrations using fluorescence correlation spectroscopy in zero-mode waveguides. Biophys J. 2005;88:2145–2153. doi: 10.1529/biophysj.104.052795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steyer JA, Almers W. A real-time view of life within 100 nm of the plasma membrane. Nat Rev Mol Cell Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal JK, Simon SM. Imaging single events at the cell membrane. Nat Chem Biol. 2007;3:92–98. doi: 10.1038/nchembio855. [DOI] [PubMed] [Google Scholar]
- 26.Murcia MJ, Garg S, Naumann CA. Single-molecule fluorescence microscopy to determine phospholipid lateral diffusion. Methods Mol Biol. 2007;400:277–294. doi: 10.1007/978-1-59745-519-0_19. [DOI] [PubMed] [Google Scholar]
- 27.Betzig E, Trautman JK. Near-field optics: Microscopy, spectroscopy, and surface modification beyond the diffraction limit. Science. 1992;257:189–195. doi: 10.1126/science.257.5067.189. [DOI] [PubMed] [Google Scholar]
- 28.Lewis A, et al. Near-field optics: From subwavelength illumination to nanometric shadowing. Nat Biotechnol. 2003;21:1378–1386. doi: 10.1038/nbt898. [DOI] [PubMed] [Google Scholar]
- 29.Krichevsky O, Bonnet G. Fluorescence correlation spectroscopy: The technique and its applications. Rep Prog Phys. 2002;65:251–297. [Google Scholar]
- 30.Haustein E, Schwille P. Fluorescence correlation spectroscopy: Novel variations of an established technique. Annu Rev Bioph Biom. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 31.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Z, Rao J. Biosensing and imaging based on bioluminescence resonance energy transfer. Curr Opin Biotechnol. 2009;20:37–44. doi: 10.1016/j.copbio.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blom H, Kastrup L, Eggeling C. Fluorescence fluctuation spectroscopy in reduced detection volumes. Curr Pharm Biotechno. 2006;7:51–66. doi: 10.2174/138920106775789629. [DOI] [PubMed] [Google Scholar]
- 34.Medina MA, Schwille P. Fluorescence correlation spectroscopy for the detection and study of single molecules in biology. Bioessays. 2002;24:758–764. doi: 10.1002/bies.10118. [DOI] [PubMed] [Google Scholar]
- 35.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson GH. Current Protocols in Cell Biology. New York: Wiley; 2008. Photoactivation and imaging of photoactivatable fluorescent proteins. Unit 21.6. [DOI] [PubMed] [Google Scholar]
- 37.Ellis-Davies GC. Caged compounds: Photorelease technology for control of cellular chemistry and physiology. Nat Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams SR, Tsien RY. Controlling Cell Chemistry with Caged Compounds. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 39.Callaway EM, Yuste R. Stimulating neurons with light. Curr Opin Neurobiol. 2002;12:587–592. doi: 10.1016/s0959-4388(02)00364-1. [DOI] [PubMed] [Google Scholar]
- 40.Kramer RH, Chambers JJ, Trauner D. Photochemical tools for remote control of ion channels in excitable cells. Nat Chem Biol. 2005;1:360–365. doi: 10.1038/nchembio750. [DOI] [PubMed] [Google Scholar]
- 41.Reinhardt KA, Kern W. Handbook of Silicon Wafer Cleaning Technology. 2nd Ed. Norwich, NY: William Andrew; 2008. [Google Scholar]