Abstract
The generation of the tubular network of the endoplasmic reticulum (ER) requires homotypic membrane fusion that is mediated by the dynamin-like, membrane-bound GTPase atlastin (ATL). Here, we have determined crystal structures of the cytosolic segment of human ATL1, which give insight into the mechanism of membrane fusion. The structures reveal a GTPase domain and athree-helix bundle, connected by a linker region. One structure corresponds to a prefusion state, in which ATL molecules in apposing membranes interact through their GTPase domains to form a dimer with the nucleotides bound at the interface. The other structure corresponds to a postfusion state generated after GTP hydrolysis and phosphate release. Compared with the prefusion structure, the three-helix bundles of the two ATL molecules undergo a major conformational change relative to the GTPase domains, which could pull the membranes together. The proposed fusion mechanism is supported by biochemical experiments and fusion assays with wild-type and mutant full-length Drosophila ATL. These experiments also show that membrane fusion is facilitated by the C-terminal cytosolic tails following the two transmembrane segments. Finally, our results show that mutations in ATL1 causing hereditary spastic paraplegia compromise homotypic ER fusion.
Keywords: protein structure, membrane remodeling, organelle shaping, spastic paraplegia gene 3A, endoplasmic reticulum network formation
In all eukaryotes, the endoplasmic reticulum (ER) is a continuous membrane system that comprises the nuclear envelope and a peripheral network of tubules and sheets (1, 2). The ER network is very dynamic, with tubules continuously forming and collapsing (3–5). The linkage of ER tubules into a network requires the fusion of identical membranes, but how such homotypic fusion occurs is poorly understood. By contrast, heterotypic fusion of viral and cellular membranes and of intracellular transport vesicles with target membranes has been intensively studied. In viral fusion, the membranes are pulled together by an irreversible conformational change of a single protein (6). In intracellular fusion, three t-SNARE proteins in one membrane and a v-SNARE partner in the other zipper up to form a four-helix bundle in the fused lipid bilayer, a process that is facilitated by additional proteins (7–10). The ATPase N-ethylmaleimide-sensitive fusion protein (NSF) and its cofactors then act together to disassemble the SNARE complex for the next round of fusion (11, 12).
Recent work has implicated the membrane-bound atlastin (ATL) GTPases in the homotypic fusion of ER tubules (13, 14). These proteins belong to the dynamin family of GTPases (15) and appear to exist in all metazoans, with many organisms expressing several isoforms (16). The ATLs are anchored in the membrane by two closely spaced transmembrane (TM) segments, exposing both the GTPase domain and the C-terminal tail to the cytosol (Fig. 1A). They localize to ER tubules and interact with the reticulons and DP1/Yop1p, proteins implicated in shaping ER tubules (13). A role for the ATLs in fusion is suggested by the observation that their depletion or the overexpression of dominant-negative forms reduces the interconnection of ER tubules in tissue culture cells (13). In addition, antibodies to ATL inhibit ER network formation in vitro (13). Finally, and most convincingly, proteoliposomes containing purified ATL undergo GTP-dependent fusion (14). Plant and yeast cells do not possess ATLs, but a similar GTPase, called Sey1p in Saccharomyces cerevisiae and RHD3 in Arabidopsis thaliana, may have an analogous function (13).
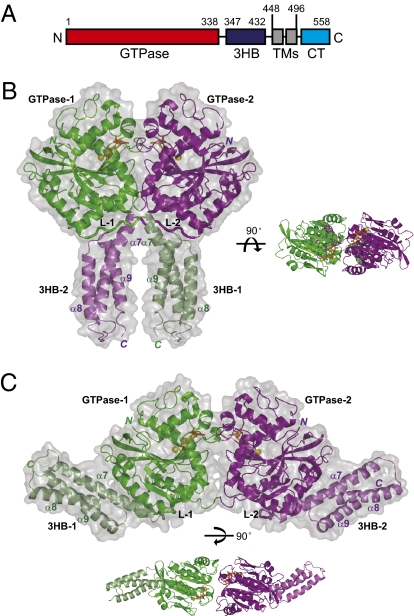
Fig. 1.
Structures of the cytosolic domain of human ATL1. (A) Scheme showing the domains of ATL1. 3HB, three-helix bundle; TMs, TM segments; CT, cytosolic tail. (B) Structure of the GDP-bound form of ATL1, corresponding to the postfusion state. The protomers in the dimer are shown in green and purple cartoon representation, superimposed on a space-filling model. The linkers (L-1 and L-2) between the GTPase domains and 3HBs (in pale colors) are highlighted. GDP is shown in orange stick representation, and magnesium ion is shown as a yellow sphere. The α-helices of the 3HBs are numbered. Right shows a different view of the dimer. (C) Structure of ATL1 crystallized with GDP and Pi, corresponding to the prefusion state. Lower shows a different view of the dimer.
Mutations in ATL1 cause the most common type of early-onset hereditary spastic paraplegia (HSP; ref. 17). The disease is characterized by progressive spasticity and weakness of the lower limbs due to a length-dependent abnormality of corticospinal axons. The expression of ATL1 with disease mutations in tissue culture cells often results in abnormally long, nonbranched ER tubules, consistent with ER fusion defects (13, 16).
Another member of the dynamin family, called mitofusin in mammals and Fzo1p in yeast, also mediates homotypic membrane fusion (18, 19); it has the same membrane topology as the ATLs and fuses outer mitochondrial membranes. Other family members, including dynamin-1, mediate the fission of membranes (20–24). However, other members, including the guanylate binding protein 1 (GBP1; refs. 25–27), the myxovirus resistance protein 1 (MxA or MX1; ref. 28), and the bacterial dynamin-like protein (BDLP; refs. 29 and 30), have poorly defined functions.
Here, we elucidate how ATL mediates homotypic membrane fusion. The proposed mechanism involves drastic conformational changes of interacting ATL molecules and explains the effect of mutations in ATL1 that cause HSP.
Results
Crystal Structures of the Cytosolic Domain of ATL1.
To investigate the mechanism of ATL1-mediated membrane fusion, we determined crystal structures of its cytosolic domain (cytATL1). Residues 18–447 (truncation before the first TM region; Fig. 1A) were expressed in Escherichia coli, purified, and crystallized in the presence of GDP. A hexagonal crystal form was used to determine a structure at 2.8-Å resolution by multi-wavelength anomalous diffraction (MAD; Table S1). Only one ATL1 molecule was present in the asymmetric unit, but a crystallographic dimer was seen along one of the twofold symmetry axes (crystal packing shown in Fig. S1A). The dimer is likely of physiological relevance, as discussed below.
CytATL1 consists of an N-terminal GTPase domain and a three-helix bundle, which are connected by a linker region (Fig. 1B). In the dimer, the GTPase domains interact with one another, such that the nucleotide binding sites face each other. The linker regions of the two ATL1 molecules cross one another, allowing the top of the three-helix bundle of one ATL1 molecule (3HB-1) to bind to the bottom of the GTPase domain of the other molecule (GTPase-2). The C termini of the two ATL1 molecules are only ∼10 Å apart, indicating that, in the full-length protein, the following TM segments would have to sit in the same membrane.
A different conformation of the cytATL1 was obtained when the protein was crystallized with GDP and high concentrations of inorganic phosphate (Pi; Fig. 1C). This structure was determined at 2.8-Å resolution from primitive orthorhombic crystals by molecular replacement, using the GTPase domain of the first structure as a search model (Table S1). A crystallographic and a noncrystallographic dimer are seen (crystal packing shown in Fig. S1B), in which the two GTPase domains interact as in the structure derived from the hexagonal crystals. In contrast to the first structure, the three-helix bundles are associated with the GTPase domain of the same molecule (3HB-1 interacts with GTPase-1; Fig. 1C) and point in opposite directions. This result implies that, with the full-length protein, the two ATL1 molecules would sit in apposing membranes. Based on the difference between the two structures, we hypothesize that the second structure corresponds to a membrane-tethered, prefusion state, and the first structure corresponds to a postfusion state in which the membranes have already fused. With this assumption, we will use the terms “prefusion” and “postfusion” structures. Although no density for Pi was observed in the prefusion structure, the conformational change between the two states is likely triggered by Pi release during the nucleotide hydrolysis cycle (see below).
Comparison with Other Dynamin-Like Proteins and GTP Hydrolysis.
The ATL1 dimer is likely of physiological significance because other dynamin-like proteins also form dimers of their GTPase domains with the nucleotides bound at the interface (Fig. S2A). ATL1 is most similar to GBP1 (Fig. S2 A and B), notably in the active sites (Fig. S3C), although dimer formation involves different interface residues. The prefusion structure of ATL1 resembles GBP1 crystallized with the nonhydrolyzable GTP analog (25), in that the helical domains point in opposite directions (Fig. S2B). This similarity suggests that the prefusion state of ATL1 is close to the GTP-bound conformation.
The GTPase domain is composed of a central β-sheet surrounded by six α-helices, and the GDP molecule is coordinated mainly by four conserved elements (Fig. S3A): the P-loop (G1 motif), the switch 1 region (G2 motif), the switch 2 region (G3 motif), and the G4 motif. The two residues of ATL1’s G4 motif (R217 and D218) are unique to the subclass of the dynamin family that includes ATLs, Sey1p, RHD3, and GBPs (13, 31). As expected, a mutation in the P loop (K80A) affects the enzymatic activity of the GTPase (Table S2). However, some uncommon residues may also be involved in GTP hydrolysis, including residue E117 in switch 1—which in human GBP1 may correspond to the catalytically important S73 residue—and Q148 in switch 2. Their mutations to Ala result in increased binding of GTPγS and reduced GTPase activity (Fig. S4A and Tables S2 and S3). R77 in ATL1 corresponds to the critical “Arg-finger” in GBP1, but it points away from GDP in our structure (Fig. S3B), suggesting that it may reorient upon GTP binding. Thus, ATL1 may undergo at least two conformational changes during the GTP hydrolysis cycle—one in the active site during hydrolysis of the γ-phosphate bond and one involving the reorientation of the three-helix bundle during Pi release.
Dimer Formation and Nucleotide Binding.
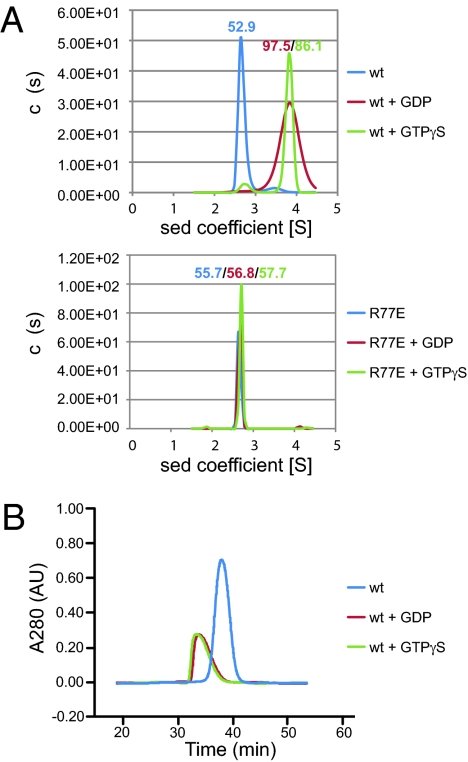
The nucleotides are fairly well buried in the structures, suggesting that nucleotide binding is linked to dimer formation. Indeed, purified cytATL1 behaved as a monomer in the absence of nucleotide, as judged by analytical ultracentrifugation (AUC; Fig. 2A, Upper) and gel filtration (Fig. 2B), whereas it formed a dimer in the presence of GDP or GTPγS. The dimer may be somewhat less stable in GDP, as indicated by the broadened peak in AUC (Fig. 2A). Mutating R77 at the interface between the protomers (Fig. S5A) to Glu abolished dimer formation, as judged by AUC (Fig. 2A, Lower), and it reduced GTPγS affinity, as measured by isothermal titration calorimetry (ITC; Fig. S6). Mutation of residue L274 at the dimer interface (Fig. S5A) to Ala reduced the association of the protomers more moderately (Fig. S7); the GTPase activity was particularly affected at low protein concentrations (Fig. S4B). A similar, concentration-dependent effect on the GTPase activity was also seen with another interface mutant (Q183; Figs. S5B and S4B). These results suggest that GTP-dependent dimer formation may link ATL molecules in apposing membranes during the first step in membrane fusion.
Fig. 2.
Nucleotide-dependent dimerization of ATL1. (A) The size of wild-type (wt) cytATL1 (theoretical molecular mass 49.8 kDa) and of the R77E mutant (both at 0.05 mM) were determined by analytical ultracentrifugation in the presence of the indicated nucleotides. The estimated molecular masses are given above the peaks (in kDa). (B) Wild-type CytATL1 was subjected to gel filtration on a Superdex-200 column in the presence of the indicated nucleotides.
Conformational Change Induced by Pi Release.
The structures suggest how Pi release may cause a conformational change of ATL1. In the prefusion structure, the GTPase domain binds to the three-helix bundle of the same ATL1 molecule. The α6-, α2-, and α3-helices of the GTPase domain surround and interact with the top of the α7-helix of the bundle (Fig. 3 A and B). Major contacts include residues L192, Y196, and L199 in α3 and M347 and L348 in α7. Residue M347 of the α7-helix also associates with M169 of the α2-helix, close to the site where α2 interacts with a kink in the α3-helix (L192). In the postfusion structure, the α3-helix is no longer kinked; it now interacts with the α7-helix in the bundle of the other protomer, using in part the same residues as in the prefusion state (L192 in α2 and M347 in α7; Fig. 3 A–C). In addition, the GTPase domain interacts with the loop between α8 and α9 of the three-helix bundle (K406 and M408). Pi release from the active sites could trigger this conformational change via movements of switch 2, which could affect the interaction of the adjacent α2-helix with the α3-helix and dislodge the α7-helix of the three-helix bundle, allowing interactions with the other protomer instead.
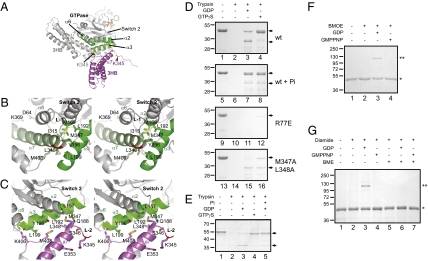
Fig. 3.
Nucleotide-dependent conformational changes of ATL1. (A) The two crystal structures were superimposed with their GTPase domains. The three-helix bundles (3HB) of the two ATL1 molecules are shown in gray and purple, respectively. Helices of the GTPase domain interacting with the 3HBs are shown in green. The bound nucleotide (in orange sticks) and the switch 2 region are indicated. Residue K345 is located in the α7-helix in the prefusion state and becomes accessible to trypsin in the postfusion state (arrowhead). (B) Stereoview of the prefusion interface between the GTPase domain and the 3HB. Interacting segments of the helices of the GTPase domain and of the α7-helix of the 3HB are shown in green, and relevant amino acids are indicated. The backbone of K345 is in red. The linker (L-1) is indicated. (C) As in B, but for the postfusion structure. (D) Confirmation of the conformational change by a trypsin-protection assay. Purified cytATL1 of wild-type (wt) or mutant human ATL1 was incubated with the indicated nucleotides (2 mM) and treated with protease. The samples in lanes 5–8 also contained 100 mM Pi. All samples were analyzed by SDS/PAGE and stained with Coomassie blue. The arrows indicate the full-length form and a protected fragment, respectively. (E) As in D, but with full-length Drosophila ATL, reconstituted into proteoliposomes. The vesicles were floated in a sucrose gradient before analysis by trypsin digestion. Pi was added at 500 mM. (F) Purified cytATL1 with a cysteine at position 357 of the 3HB was treated with the bifunctional cross-linker BMOE in the presence of the indicated nucleotides. Non-cross-linked protein (*) and cross-linked dimer (**) are indicated. (G) Purified cytATL1 with cysteines at positions 192 and 348 was incubated with the oxidant diamide in the presence of the indicated nucleotides. Where indicated, the disulfide bridge was reduced with β-mercaptoethanol (BME) before nonreducing SDS/PAGE.
A trypsin-protection assay was used to confirm the conformational change inferred from the structures. In the absence of nucleotide, wild-type cytATL1 was readily proteolyzed (Fig. 3D, lane 2 vs. 1; see Fig. S8A for controls). In the presence of GDP, a protected fragment was generated (lane 3). The fragment corresponds to the N terminus of the protein, with trypsin cleavage occurring at K345 in the linker region (confirmed by mass spectrometry analysis; data not shown); when K345 was mutated to Glu, no cleavage was observed (Fig. S8B). In the postfusion structure, the loop residue K345 is partially exposed (Fig. 3 A and C), explaining why it is accessible to trypsin.
In the presence of GTPγS, which is expected to lock ATL1 in the prefusion state, linker cleavage was largely suppressed, and instead the full cytosolic domain became resistant to trypsin digestion (Fig. 3D, lane 4). In this state, the cleavage site K345 is embedded in the α7-helix of the three-helix bundle (Fig. 3 A and B) and becomes inaccessible to trypsin. As expected, the monomeric, nucleotide-binding-deficient R77E mutant was readily degraded by trypsin, even in the presence of GDP or GTPγS (Fig. 3D, lanes 11 and 12). A similar effect was seen with a mutant, in which residues M347 and L348 were converted to Ala (Fig. 3D, lanes 15 and 16), consistent with a weakened interaction of the three-helix bundle with the GTPase domain (Fig. 3 B and C). Linker cleavage was prevented not only with GTPγS, but also when the transition state of GTP hydrolysis was mimicked by addition of GDP and AlF3 (Fig. S8C) or when the posthydrolysis state was generated by addition of GDP and Pi (Fig. 3D, lane 7). Thus, Pi release during the nucleotide hydrolysis cycle appears to trigger the transition from the prefusion to the postfusion conformation.
The same conformational change was observed with full-length Drosophila ATL in intact membranes. The full-length protein was expressed as a GST fusion in E. coli, purified in Triton X-100, and—after cleavage and removal of the GST tag—reconstituted into proteoliposomes. As with the cytosolic domain, trypsin completely degraded the protein in the absence of nucleotide (Fig. 3E, lane 2 vs. 1), but generated a fragment in the presence of GDP (lane 3). In the presence of GTPγS or GDP plus high concentrations of Pi, the protein became resistant to proteolysis (lanes 4 and 5).
To further confirm the conformational change inferred from the structures, we performed cross-linking experiments. The three-helix bundles of two cytATL1 molecules came close to one another in the presence of GDP, but not in GMPPNP or the absence of nucleotide, as demonstrated by the formation of a cross-linked dimer when the ATL mutant L357C was treated with the bifunctional cysteine cross-linker bis-maleimidoethane (BMOE; Fig. 3F, lane 3 vs. lanes 4 and 2; for position of the L357C mutation, see Fig. S9). Furthermore, as expected from the GDP structure in which residue L192 of one ATL molecule came close to L348 of the other ATL molecule (Fig. 3C), cysteines introduced at these positions can form a disulfide bridge (Fig. 3G, lane 3). Again, no cross-linking was seen with GMPPNP (lane 4) or in the absence of nucleotide (lane 2). These data strongly support the postulated conformational change.
Testing ATL Mutants in a Fusion Assay.
Next, we tested the effect of mutations on the membrane fusion activity of full-length Drosophila ATL. The proteins were reconstituted at equal concentrations into donor and acceptor proteoliposomes (Fig. S10). The donor vesicles contained lipids labeled with two fluorophores at quenching concentrations; the fusion with unlabeled acceptor vesicles resulted in dilution and dequenching. Wild-type Drosophila ATL gave efficient fusion in the presence of GTP and Mg2+ (Fig. 4A), as reported for a GST-fusion of ATL (14), although without the GST tag, the protein was significantly more active. No fusion was seen with GDP or the absence of Mg2+ (Fig. 4A). Mutant R48E—equivalent to R77E in human ATL1 (hATL1; Fig. S11)—was inactive, consistent with the defects in dimer formation and GTP binding observed with the cytosolic domain. The same effect was seen with another mutant that reduced dimer formation (L249A, corresponding to L274A in hATL1; Fig. 4A). Furthermore, the addition of the purified cytosolic domain of Drosophila ATL effectively inhibited fusion (Fig. 4B). These data support the idea that dimer formation from ATL molecules sitting in different membranes is required for membrane fusion. GTPase activity of the ATL molecules is also needed.
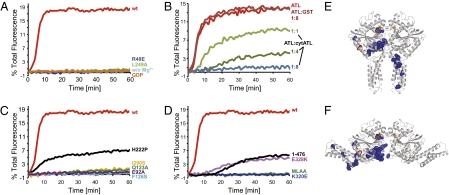
Fig. 4.
Membrane fusion with wild-type (wt) and mutant ATL. (A) Full-length wt Drosophila ATL or mutants that affect dimer formation of human cytATL1 were reconstituted at equal concentrations into donor and acceptor vesicles. Reconstitution of the proteins was efficient as shown by flotation (Fig. S10) and resulted in all ATL molecules having their cytosolic domains exposed (Fig. 3E). GTP-dependent fusion of donor and acceptor vesicles was followed by the dequenching of a fluorescent lipid present in the donor vesicles. Control experiments were performed in the absence of Mg2+ or presence of GDP instead of GTP. (B) The fusion of vesicles containing full-length wt ATL was determined in the presence of increasing concentrations of the cytosolic domain (cytATL). The molar ratios are indicated. GST was used as a control. (C) As in A, but with mutants that affect GTPase activity or cause HSP (F126S, H222P, I290S). For comparison, the data in A for wt ATL are replotted. (D) As in C, but with mutants that affect the interaction between the GTPase domain and the three-helix bundle. ATL lacking the cytosolic tail (1–476) was also tested. (E) Mutations causing HSP were mapped into the postfusion structure (blue). The effect of mutation I315S (red) can only be explained by the prefusion structure. (F) The disease mutations were mapped into the prefusion structure.
Mutations in switch 1 (E92A) or switch 2 (Q123A), corresponding to E117A and Q148A in hATL1, reduced the GTPase activity of the cytosolic domain (Fig. S4A and Table S2) and the fusion activity of the full-length protein (Fig. 4C). As observed (14), even mutants with moderately reduced GTPase activity showed drastically reduced fusion activity. Finally, mutant M322A/L323A (M347A/L348A in hATL1), in which the interaction between the three-helix bundle and the GTPase domain is disturbed, and mutant K320E (K345E in hATL1), which has high GTPase activity (Fig. S4A) but is expected to disrupt a kink between the linker and the three-helix bundle (Fig. 3C), were also defective in fusion (Fig. 4D). Mutation E328K (E353K in hATL1), which is also located in the kink, inhibited fusion less strongly (Fig. 4D). Together, these results suggest that the conclusions derived from cytATL1 are functionally significant and relevant to the fusion reaction observed with the full-length protein. We also found that the deletion of the C-terminal tail following the two TM segments (truncation at residue 476) drastically reduced, but did not abolish, fusion (Fig. 4D). Thus, the C-terminal tail also plays an important role, but whether it is involved in membrane tethering or fusion remains to be tested by more direct assays.
Disease-Causing ATL1 Mutations.
Most of the published ATL1 mutations causing HSP (see refs. 32 and 33) localize to two “hot spots” (Fig. 4 E and F): in the GTPase domain near the dimer interface and in the three-helix bundle, mostly in a region close to the GTPase domain. Several of the changes are expected to affect dimer formation and GTPase activity, or the conformational change of ATL during fusion. Disease mutations in cytATL1 had GTPase activity that varied from almost wild-type (H247P) to extremely low levels (F151S; Fig. S4C). The corresponding Drosophila ATL mutants (H222P and F126S) showed reduced fusion activity, roughly correlated with their GTPase activity (Fig. 4C). Mutation I315S (I290S in Drosophila) localizes to the periphery of the postfusion structure, but comes close to the three-helix bundle in the prefusion structure (Fig. 4 E and F, shown in red; Fig. 3B). The equivalent Drosophila mutation completely abolished fusion (Fig. 4C). Finally, several frame-shift mutations in the C-terminal tail also caused HSP (34–36), consistent with a role of the tail in fusion. Collectively, these results suggest that ATL mutations cause HSP by compromising homotypic ER fusion.
Discussion
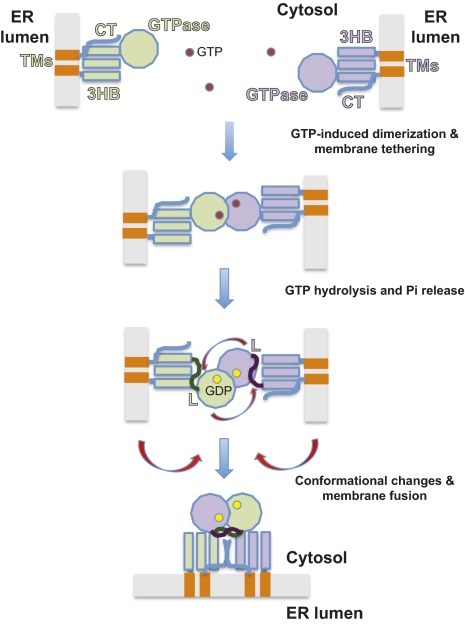
Our results suggest how ATL causes homotypic fusion of ER membranes (Fig. 5). First, two ATL molecules sitting in different membranes bind GTP and form a dimer, thereby tethering the membranes together; the three-helix bundles of the two molecules are pointing in different directions. Following GTP hydrolysis and Pi release, both ATL molecules undergo a conformational change, probably triggered by changes in the switch 2 regions that are propagated to the interaction surface between the GTPase domain and the three-helix bundle. The three-helix bundles would then be released, and the linkers may allow the dimer of GTPase domains to freely rotate. We speculate that the GTPase domain of one ATL molecule captures the three-helix bundle of the opposing ATL molecule; the resulting conformational change would pull the apposing membranes together, so that after fusion the two ATL molecules would be anchored in the same membrane (Fig. 5). This transition appears to be facilitated by the C-terminal cytosolic tails of the ATL molecules. One possibility is that the tail of one ATL molecule interacts with the three-helix bundle of the other ATL molecule. Alternatively, the tails of ATL molecules could interact with one another, either in a parallel orientation or in an anti-parallel one, as reported for the tails of mitofusins (37).
Fig. 5.
A model for ATL-mediated homotypic membrane fusion. See Discussion for details. GTP and GDP molecules are indicated as magenta and yellow spheres, respectively. G, GTPase domain, 3HB, three-helix bundle, TMs, TM domains, L, linker.
Efficient membrane fusion may require several ATL dimers to undergo the conformational change depicted in Fig. 5, perhaps by forming higher oligomers like other dynamin-like proteins, although such oligomers were not observed with cytATL. Subsequent rounds of fusion likely require the dissociation of the ATL dimers and exchange of GDP for GTP and could involve additional protein factors. ATL-mediated homotypic fusion differs from heterotypic viral fusion and SNARE-mediated fusion, in that nucleotide triphosphate hydrolysis directly drives fusion. In SNARE-mediated fusion, nucleotide hydrolysis is used to reset the fusion machinery by NSF-mediated disassembly of SNARE complexes (12), and viral fusion does not require nucleotide hydrolysis at all. A common feature of all fusion reactions is a conformational change of proteins that ultimately leads to the merging of the lipid bilayers. The mechanism proposed for ATL could also apply to Sey1p and its homologs in the fusion of yeast and plant ER membranes and to mitofusin/Fzo1p in the fusion of mitochondrial outer membranes.
While this paper was under consideration by a different journal, the same ATL structures were published (38), but the authors did not have evidence that the conformational change suggested by the structures occurred during fusion. They also did not observe ATL dimers with GDP, likely because the nucleotide was omitted from the gel filtration buffer. Our results provide strong support for a GDP-bound ATL dimer as an intermediate during the fusion reaction.
Materials and Methods
Protein Purification.
Residues 18–447 of human atlastin-1 with an N-terminal, thrombin-cleavable His6-tag were expressed in E. coli. The protein was isolated by Ni-NTA chromatography, cleaved with thrombin, and further purified by ion-exchange chromatography and gel filtration.
Crystallization.
For crystallization, 0.4 mM cytATL1 was incubated with 2 mM GDP for 2 h at room temperature. Crystallization trials were performed by the hanging-drop vapor-diffusion method at 20 °C. The hexagonal form was obtained with GDP and native or Se-Met-substituted in 0.1 M Tris·HCl (pH 8.5), 25% (wt/vol) PEG 1500, and 20% (vol/vol) glycerol. The orthorhombic form was obtained in 0.1 M Tris·HCl (pH 8.3), 25% (wt/vol) PEG 3350, and 0.1 M (NH4)2HPO4.
Data Collection and Structure Determination.
Diffraction data for both crystal forms were collected at 100 K on beamline BL17U at Shanghai Synchrotron Radiation Facility, Shanghai, China. Hexagonal crystals were used directly and orthorhombic crystals were soaked in mother liquor plus 20% (vol/vol) glycerol for a few seconds. All data sets were processed with HKL2000 and converted to CCP4 format. The structure of the Se-Met substituted protein was determined by MAD, and the structure of the protein in primitive orthorhombic crystals was determined by molecular replacement. Models were built with the Phenix program and refined and manually rebuilt against the native data set. The CNS suite and the Phenix refinement program (phenix.refine) were combined during refinement.
Analytical Ultracentrifugation and Gel Filtration.
Sedimentation velocity data were collected at a speed of 142,249 × g in an An-60 Ti rotor at 12 °C. The proteins were prepared in 50 mM Tris (pH 8.0), 5 mM MgCl2, and 1 mM DTT at 0.1–3 mg/mL Data were analyzed by the program SEDFIT (Version 11.8). Sedimentation equilibrium experiments are described in SI Materials and Methods. For gel filtration analysis, 100 μL of purified cytATL1 at 0.2 mM was incubated with or without 5 mM nucleotide and loaded onto a Superdex 200 column in buffer containing 50 mM Tris (pH 8.0), 100 mM NaCl, 5 mM MgCl2, and 1 mM nucleotide, if present.
Trypsin-Protection Assay.
Ten micrograms of cytATL1 with the indicated nucleotides was incubated with 0.6 μg of trypsin at 37 °C for 0.5 h. Trypsin digestion of full-length Drosophila ATL was done similarly with proteoliposomes (protein to lipid ratio, 1:1,000) floated in a discontinuous sucrose gradient.
Cross-Linking Assays.
For cross-linking with BMOE (Thermo Scientific), 1 μM cytATL1 was incubated with or without GDP or GMPPNP with 2 μM BMOE at room temperature for 1 h in 50 mM Hepes (pH 7.0), 100 mM NaCl, and 5 mM MgCl2. The reactions were quenched with 50 mM DTT. For cross-linking with diamide (Sigma), 5 μM cytATL1 was incubated with 50 μM diamide at room temperature for 30 min, and the reaction was quenched with 250 μM N-ethylmaleimide.
Fusion Assays.
Full-length, codon-optimized Drosophila ATL was expressed in E. coli as a GST fusion and purified on glutathione agarose, as described (14). The protein was passed through a PD-10 column, and the GST moiety was cleaved off by thrombin and removed with glutathione agarose. The cytosolic domain of Drosophila ATL (residues 1–422) was purified similarly in the absence of detergent. The generation of reconstituted liposomes at a 1:2,000 molar ratio of protein:lipid and the fusion assay were done as described (14).
Further details on methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Xue, F. Yang, and W. Shui for help with mass spectrometry; X. Yang and Z. Wang for help with ITC and AUC, respectively; A. Stein, S. Hubbard, and Q. Wang for critical reading of the manuscript; and the staff at beamline BL17U of Shanghai synchrotron radiation facility. X.L. is supported by the National Basic Research Program of China (973 Program, Grant 2010CB911800) and the National S&T Major Project on Major Infectious Diseases (Grant 2008ZX10001-010) from the Ministry of Science and Technology of the People's Republic of China. J.H. is supported by the National Basic Research Program of China (973 Program, Grant 2010CB833702) and the National Natural Science Foundation of China (Grants 30971440 and 90919009). T.A.R. and J.H. received support from the 111 Project of China (B08011). R.W.K. is supported by a European Molecular Biology Organization long-term fellowship. T.Y.L. is supported by a National Science Foundation graduate research fellowship. T.A.R. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3QNU and 3QOF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101643108/-/DCSupplemental.
References
- 1.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 2.Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 4.Du Y, Ferro-Novick S, Novick P. Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci. 2004;117:2871–2878. doi: 10.1242/jcs.01286. [DOI] [PubMed] [Google Scholar]
- 5.Prinz WA, et al. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 8.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens S, McMahon HT. Mechanisms of membrane fusion: Disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 10.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 12.Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 13.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 16.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: Clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 18.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 21.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 22.Marks B, et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 23.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashkirov PV, et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakash B, Renault L, Praefcke GJ, Herrmann C, Wittinghofer A. Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 2000;19:4555–4564. doi: 10.1093/emboj/19.17.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 28.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 29.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 30.Low HH, Sachse C, Amos LA, Löwe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 32.Smith BN, et al. Four novel SPG3A/atlastin mutations identified in autosomal dominant hereditary spastic paraplegia kindreds with intra-familial variability in age of onset and complex phenotype. Clin Genet. 2009;75:485–489. doi: 10.1111/j.1399-0004.2009.01184.x. [DOI] [PubMed] [Google Scholar]
- 33.Guelly C, et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am J Hum Genet. 2011;88:99–105. doi: 10.1016/j.ajhg.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessa A, et al. SPG3A: An additional family carrying a new atlastin mutation. Neurology. 2002;59:2002–2005. doi: 10.1212/01.wnl.0000036902.21438.98. [DOI] [PubMed] [Google Scholar]
- 35.Ivanova N, et al. Hereditary spastic paraplegia 3A associated with axonal neuropathy. Arch Neurol. 2007;64:706–713. doi: 10.1001/archneur.64.5.706. [DOI] [PubMed] [Google Scholar]
- 36.Loureiro JL, et al. Novel SPG3A and SPG4 mutations in dominant spastic paraplegia families. Acta Neurol Scand. 2009;119:113–118. doi: 10.1111/j.1600-0404.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 37.Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 38.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci U S A. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.