Abstract
InterMed is a collaboration among research groups from Stanford, Harvard, and Columbia Universities. The primary goal of InterMed has been to develop a sharable language that could serve as a standard for modeling computer-interpretable guidelines (CIGs). This language, called GuideLine Interchange Format (GLIF), has been developed in a collaborative manner and in an open process that has welcomed input from the larger community. The goals and experiences of the InterMed project and lessons that the authors have learned may contribute to the work of other researchers who are developing medical knowledge-based tools. The lessons described include (1) a work process for multi-institutional research and development that considers different viewpoints, (2) an evolutionary lifecycle process for developing medical knowledge representation formats, (3) the role of cognitive methodology to evaluate and assist in the evolutionary development process, (4) development of an architecture and (5) design principles for sharable medical knowledge representation formats, and (6) a process for standardization of a CIG modeling language.
The development and implementation of enabling tools and methods that provide ready access to knowledge and information are among the central goals of biomedical informatics. Given the immensity of the challenge, workers have increasingly recognized the need for multi-institutional collaboration in the development of such tools and methods. InterMed is an Internet-facilitated collaboration among biomedical informatics research groups at Harvard, Columbia, and Stanford Universities.1 One of the central objectives of InterMed has been to develop sets of tools and resources for disseminating clinical practice guidelines.
Clinical guidelines are tools for encouraging best practices in clinical care and are intended thereby to improve safety, quality, and cost–effectiveness. Studies have found that guideline implementations can best affect clinician behavior if guidelines can deliver patient-specific advice during the clinical encounter.2,3 For guidelines to be delivered at the point of care through decision-support systems, they must be represented in a computer-interpretable format that enables automatic inference based on patient data stored in electronic medical records (EMRs). Significant work is required to create high-quality evidence-based text guidelines, and additional work is then required to encode them as computer-interpretable guidelines (CIGs). Therefore, sharing the knowledge representation of guidelines among different medical institutions is desirable; it reduces development costs while providing consistency in guideline interpretation and potentially reducing variability in clinical practice.
Guided by that objective, the InterMed team has focused primarily on developing and evaluating a format for representing clinical guidelines that enhances their sharability and reuse across a variety of clinical settings and system architectures. This format is called GuideLine Interchange Format (GLIF). The most recent version, incorporating a number of features aimed at facilitating standardization, is known as GLIF3.
A Shared Guideline Modeling Language
Although we started our work on GLIF in 1996 with the goal that it would become a shared modeling language, our definition of “sharing” evolved over time, as monitored by an external evaluation group led by cognitive scientists from McGill University.* We initially considered sharing to be interchange, leading to the use of that word in the acronym. GLIF was intended to be a medium for translation among several guideline formalisms. At that time, the guideline application systems among which GLIF was meant to enable interchange included Medical Logic Modules (written in the Arden Syntax4), GEODE-CM,5 MBTA,6 and EON.7 Terminology used in describing guideline systems varied markedly, and different groups emphasized different functional requirements and varying aspects of computer-based guidelines in their development activities. It soon became clear that true interchange among the four formalisms could not be achieved because there were functionalities that were not supported by all four methodologies. For example, MLMs and MBTA could deliver medical advice in the form of alerts that were triggered by specific events (e.g., availability of laboratory test results), whereas the other two formalisms were not event-triggered. In addition, the different formalisms were dependent on varying expression languages when encoding decision criteria, and these languages implemented and supported differing sets of operators. Thus, instead of trying to perform bidirectional mappings between GLIF and each of the fomalisms, the emphasis was placed on developing a generic model that would capture a large common subset of functionality shared by the different models and that would facilitate sharing a guideline encoding across different institutions and software systems.
The first version of GLIF was extremely limited in capabilities and was not widely disseminated. However, based on lessons from that early work, in 1998, we published GLIF version 2 (GLIF2).8 GLIF2 enabled modeling of a guideline as a flowchart of structured steps, representing clinical actions and decisions (that were then called conditional steps). However, the attributes of structured constructs were defined only in terms of text strings that could not be parsed. As a result, such guidelines could not be used for computer-based execution that required automatic inference.
In June 1999, the next phase of InterMed began. Our goal was to expand on the early GLIF work to create a sharable guideline modeling language that would be entirely computer-interpretable and that would allow sharing of encoded guidelines by facilitating both its adaptation to local settings of different health care organizations and its integration with electronic medical records and applications such as order-entry systems.
A Work Process for Multi-institutional Development of GLIF
Throughout the InterMed effort, we sought to develop GLIF collaboratively. By involving several institutions in the InterMed collaboratory, we were able to share viewpoints and cooperate in the development of GLIF so that the end results were greater than what any one group could have accomplished on its own.9 Collaboration among geographically distributed organizations, with different goals and cultures, presents significant challenges. During the first phase of InterMed, we formulated a work process for collaboration that was based on informal introspection and more formal evaluative work. Our experience and study results suggested that occasional face-to-face meetings are crucial precursors to the effective use of distance communications technologies. They are extremely important in bridging gaps between the views of collaborators and clearing misunderstandings. They aid in establishing team spirit and mutual commitment to the project. Thus, we introduced face-to-face meetings among group members and continued to have such multi-institutional meetings about four times a year. Telephone conference calls played an important role in both task-related activities and executive (project management) activities, especially when clarifications were required. During the more intense development phases of GLIF, we had weekly conference calls involving all team members.
To ensure a collaborative process in development of the GLIF formalism, two members of the team (MP and AAB), associated with different institutions, worked full time on managing the project as well as making significant technical contributions. The project managers defined specific tasks, their priorities, and their scheduling. We established small teams of two or four participants belonging to more than one institution, so that each team had members with computer science and clinical expertise. Each team worked on specific tasks, communicating by e-mail, often several times a day, and having conference calls several times a week. Every feature of the language was added only after establishing consensus by all the InterMed members, adding to its validity and general applicability.
As the GLIF model achieved stability, we focused our effort on developing software tools. Each tool was developed by one institution, with feedback from the other InterMed members. The requirements, design, and implementation of the tools were presented to the InterMed team during monthly conference calls, using Virtual Network Computing—a remote display system that allows viewing of a computing desktop environment from anywhere on the Internet and from a wide variety of machine architectures.
To increase the validity of GLIF, we wanted to gain reactions and advice from communities beyond the InterMed collaborators. We accordingly made GLIF, its evolving documentation, and encoded guidelines available for public review on <www.glif.org>.
Lifecycle for Development of a Shared Guideline Modeling Language
Our experiences from the early phase of InterMed work taught us that many sources of information contribute requirements for a shared guideline modeling language. Moreover, as was shown through our formative evaluation of the CIG development process,10 the requirements for CIGs change over time as new information sources become available, as experience is gained, and as goals and applications of the guideline modeling language arise. We defined an evolutionary lifecycle approach by which we sought to develop the shared modeling language.11,12 In the lifecycle approach, sources of information, such as guideline modeling languages, narrative guidelines, and guideline applications, are examined to define functional requirements for sharable CIGs. The functional requirements are based on features of various existing modeling environments that are considered most essential and those most in common among models. The shared guideline modeling language then is developed to meet the functional requirements. Formative evaluation during development as well as evaluation of experience in implementation, both in GLIF and other modeling environments, shows what functionality is most successful. This, in turn, leads to revision of the functional requirements and subsequently to a modification of the feature set supported by the model. To develop GLIF3, we analyzed several sources of information, including (1) GLIF28 and examples of GLIF2-encoded guidelines; (2) other guideline modeling approaches, such as the Arden Syntax,13,14 EON,15 and PRODIGY-3,16 as well as PROforma,17 Asbru,18 GUIDE,19 and Prestige20; (3) representative guidelines included in the National Guideline Clearinghouse (www.guidelines.gov); and (4) concurrent evaluation of text-based and algorithmic guidelines encoded in GLIF by the developers as well as interpreted by the physician end users (see Cognitive Studies to Evaluate GLIF and Assist in its Evolutionary Development). Through this analysis, we identified a number of areas in which GLIF2 needed to be extended to support fully the encoding of CIGs. By March 2000, many of these requirements were already supported by the GLIF3 draft specification.21,22
In March 2000, InterMed hosted a guideline representation workshop, sponsored by the National Library of Medicine, the U.S. Army, the Agency for Healthcare Research and Quality, and the Centers for Disease Control and Prevention. An international group of 82 participants from academia, government agencies, professional organizations, health care provider organizations, and industry proposed a set of functional requirements for sharable CIGs.23 The functional requirements provided a conceptual model for the ways in which CIGs would be applied and used,11,12,24 encompassing the entire CIG lifecycle (including guideline modeling, authoring, dissemination, implementation, and use). The process we subsequently carried out, initiated by the workshop, considered additional information sources, including (1) intended users of the guideline (e.g., physicians, nurses), (2) care settings (e.g., outpatient clinic), (3) delivery platform (e.g., handheld device), (4) number of patient encounters encompassed, (5) time frame of the guideline use (e.g., emergency), (6) usage mode (e.g., within encounter), (7) application type (e.g., retrospective evaluation, chronic disease management), and (8) delivery method (e.g., alerts and reminders, interactive decision support). The conceptual guideline-use model determined the details and characteristics that would need to be captured in the CIG. We designed GLIF3 to meet these functional requirements.12,25
Cognitive Studies to Evaluate GLIF and Assist in its Evolutionary Development
We conducted a series of cognitive studies to evaluate the guideline-modeling processes. The conclusions and recommendations of these studies influenced the evolutionary design process of GLIF3.
The cognitive evaluation team (VLP and colleagues) developed and refined a theoretical and methodologic framework for analyzing the cognitive processes involved in the development and the use of clinical guidelines.10 This framework consists of formal methods from cognitive science, namely, propositional and semantic analyses that can improve the validity, usability, and comprehension of the resulting applications when used as part of the system-development process. The methods adapted and used in these studies focused on the CIG-design activities, conceptualized as a problem-solving process with partially defined initial and goal states, and with loose constraints that shape the product of design. The overall design analysis included an examination of the decisions made, the patterns of collaborative activity, the development and accomplishment of design goals, the constraints that were imposed, and the distributed cognitive collaborative effort that the researchers made to work toward refining the developed model.
With the framework that we developed, we investigated how designers and users comprehend and represent information found in clinical guidelines and subsequently utilize their representation for solving problems and making decisions.26 We found that given different domain knowledge and strategies, designers and users do not represent the information in the same way, leading to different interpretations and decisions. We investigated the influence of algorithm-based and text-based practice guidelines on clinical decision making by physicians of varying expertise levels. The results showed that both experts and nonexperts used guidelines as reminders during the problem-solving process, and that nonexperts used guidelines during the learning process as an aid to knowledge reorganization. These results were obtained regardless of whether the guideline was algorithm-based or text-based. The subjects of the study expressed their desire to use guidelines that provide faster access to pertinent information. While algorithms (flowcharts) can be read faster, they are often too rigid and not as complete as the text-based guidelines. Our results provided insight into how guidelines can be fine-tuned for different users and purposes. These empirical results, coupled with design principles from cognitive science, formed an essential part of the development process of GLIF3 and improved the validity, usability, and comprehension of the resulting knowledge representations.
In a second study, we conducted an evaluation of the cognitive processes involved in the translation of a text clinical guideline into an encoded form so that it could be shared among medical institutions. It was a comparative study at three sites (Harvard, Stanford, and Columbia) regarding the generation of individual and collaborative representations of a guideline for the management of encephalopathy using GLIF. We used process-outcome measures that we defined to compare subjects with various types of computer science and clinical expertise, and from different institutions.27 Results show that variability in strategies used by the guideline encoders was dependent on the degree of prior experience and knowledge of the domain. Differing both in content and structure, the representations developed by physicians were found to have additional information and organization not explicitly stated in the guidelines, reflecting the physicians' understanding of the underlying pathophysiology. The computer scientists developed more literal representations of the guideline; additions were limited mostly to specifications mandated by the logic of GLIF itself. Collaboration between physicians and computer scientists resulted in consistent representations that were more than the sum of the separate parts in that both domain-specific knowledge of medicine and generic knowledge of guideline structure were seamlessly integrated. Thus, due to the variability in the construction of guideline representations, we concluded that understanding the processes and limitations involved in their generation is important in developing strategies to construct shared representations that are both accurate and efficient. In addition, the encoded guidelines developed by teams that include both clinicians and experts in computer-based representations are preferable to those developed by individuals of either type working alone.
In a third investigation, InterMed investigators carried out cognitive studies to test the expressiveness and suitability of GLIF3 for encoding clinical guidelines. The aim was to assess the use of GLIF3 by individuals who were translating from text versions of clinical guidelines into an electronically encoded form.28 Specifically, we used video recording methods to investigate the encoding of two clinical guidelines into both GLIF3 and GLIF2 by biomedical informaticians. Differing in both content and structure, the representations developed in GLIF3 were found to contain a greater level of representational detail and less ambiguity than those developed in GLIF2. GLIF3 was found to be more robust than GLIF2 for representing content and logical structure of the clinical guidelines studied. This formative evaluation showed that the intended improvements in expressiveness in GLIF3 were achieved.
In a fourth study, we examined the process used by the American College of Physicians to develop clinical algorithms from narrative guidelines. We analyzed how changes progressed between subsequent versions of an algorithm. We used a classification of discrepancies between requirements, documents, and software produced29 to classify the changes between a narrative guideline and its derived clinical algorithm. Based on our analysis, we recommend procedures that could limit the number of errors produced when generating clinical algorithms. Our recommendations include (1) avoiding omission of definitions of terms, (2) validating that all information is carried from the narrative guideline to all versions of the clinical algorithm, (3) providing all the information necessary to rank treatment options, and (4) considering different clinical scenarios. Using a GLIF3 authoring tool (see Architectural Lessons) can assist guideline authors in following the first three of our recommendations.
Architectural Lessons
Based on the cognitive studies, we sought to develop a CIG modeling language that would support the specification of a CIG at different levels of abstraction. These levels would separate the conceptualization of a guideline, done by clinician modelers, from the formal computable definitions, which would later be specified by informaticians. GLIF3 supports modeling of guidelines at three levels of abstraction: a conceptual flowchart (as in GLIF2), a computable specification that can be verified for logical consistency and completeness, and an implementable specification that can be incorporated into particular institutional information systems. This last level is currently only partly developed. The separation among these three layers is important for conceptual understanding of guidelines as well as for sharing of their encoded versions, since different institutions may share encoding of the conceptual and computable levels, whereas the implementation-level specifications are likely to be different from one site to another due to local variations in terminologies, medical records, information system platforms, applications supported, and interaction conventions.
Guided by current software development methodologies, the GLIF3 model is object oriented. It consists of classes, their attributes, and the relationships among the classes. Structuring the model in this way eases conceptualization and improves data integrity. The GLIF3 model is described using class diagrams of the Unified Modeling Language (UML),30 which are the industrial standard notation for software architecture. Additional constraints on represented concepts are specified in the Object Constraint Language (OCL), a part of the UML standard.
Based on examination of conceptual entities that appear in guidelines and of constructs of guideline modeling languages, GLIF3 represents guidelines in the form of a flowchart of guideline steps. Subclasses of guideline steps are action steps and decision steps, used to represent clinical actions and decisions, respectively. Decision steps contain several decision options. Patient state steps serve as entry points into the guideline while also allowing for labeling of patient states. Branch steps and synchronization steps allow modeling of concurrent processes. In addition to the Guideline class, GLIF3 supports the use of the Macro class.31,32 Macros provide a means to specify declaratively procedural patterns that appear in guidelines in the form of single constructs but which are realized by sequences of GLIF3 steps.22 Thus, macros ease conceptualization and instantiation of procedural patterns. More details on the GLIF3 representation can be found elsewhere.33
When choosing a format in which GLIF-encoded guidelines could be stored and exchanged, we looked for a standard text-based format. We chose to use RDF (Resource Description Framework),34 which is based on the eXtensible Markup Language (XML). RDF is a foundation for processing metadata; it is an object-oriented model with well-defined semantics, and it provides interoperability among applications that exchange machine-understandable information on the Web. The GLIF3 RDF schema specifies the syntax of guidelines encoded in GLIF3. RDF files containing GLIF-encoded guidelines can be checked automatically for syntactic validity and logical consistency, when compared against this formal schema, using generic RDF validation tools.
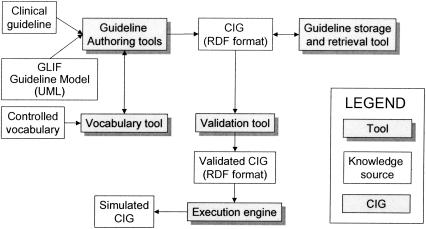
Tool support is a crucial issue for guideline development and use. We have developed tools to support authoring, viewing, retrieval, validation, and execution of GLIF-encoded guidelines (▶). When building the authoring tools for GLIF3, we had two aims. One aim was to support the specification of GLIF3 guidelines at different levels of abstraction. The other aim was to allow specialized views of guidelines as well as links to external applications. The authoring tool developed at Stanford University supports the specification of GLIF3 guidelines at different levels of abstraction. We developed this tool using Protégé-200035 and configured it in two ways: domain experts can use the first configuration for creating abstract flowcharts, while informaticians can use the other configuration to create detailed computable specifications. The first configuration allows a guideline author to specify clinical algorithms, codes for clinical terms, rules for ranking alternative treatment options written in natural language, and documentation attributes. The second configuration, which supports the computable specification, can be validated for logical consistency (see Design Principles for a Guideline Model and Exchange Format).
Figure 1.
GLIF3 tools that InterMed developed: guideline authoring tools, vocabulary tools, validation tool, execution engine, and guideline storage and retrieval tool.
A second authoring tool developed at Harvard University enables viewing the encoded guideline in both a tree view and a flowchart view. This tool has been designed so that it can be easily custom tailored for different types of guidelines and can be extended with more features, such as specialized views of guidelines or links to external applications (e.g., vocabulary databases).
The development of the two authoring tools influenced features of GLIF3. By creating the authoring tools, new types of requirements were posed for the GLIF format. Questions such as the number of guidelines per guideline file needed to be answered. The tools expressed the different perspectives of the two teams who developed the authoring tools, who had different application motivations for this work. These differences contributed to the articulation of features that would be desirable in GLIF3. We encourage teams who are creating informatics methods and tools to experiment with building different support tools to facilitate the understanding of desired features.
Another tool that influenced the design of GLIF3 was the GLIF3 GuideLine Execution Engine (GLEE).36 The development of this tool brought forth considerations relating to CIG integration into the clinical information system of a local institution. For example, during the design and implementation of GLEE we needed to address the semantics of guideline steps in GLIF. We developed rules for pairing branch and synchronization steps.†
Design Principles for a Guideline Model and Exchange Format
While developing GLIF3, we followed design principles that we derived from our past experiences as well as from formative evaluation of the process of guideline development and the role of end users. The functional requirements that were elicited at the guideline workshop also contributed to the design.
Expressiveness is the ability to encode the knowledge content of different types of guidelines. We used GLIF3 to encode 12 guidelines of different types (▶). We derived the types of guidelines from a classification scheme24 created by InterMed members, which expands the classification scheme of the National Guideline Clearinghouse (<www.guidelines.gov>). We checked that GLIF3 can express necessary components of guideline content25 by looking at: (1) structural parts of narrative guidelines: definitions, recommendations, and algorithms; and (2) decision-support tasks that guidelines involve,37 including making decisions, specifying work to be performed, data interpretation, and goal setting.
Table 1.
Classification of Guidelines Encoded in GLIF3
| Disease/Condition | Stage of Problem | Encounters | Setting | Time Frame | Computability | Reference |
|---|---|---|---|---|---|---|
| Flu | Prevention | 1 | Out | a | Algorithmic | 22 |
| Stable angina | Management | n | Out | a/c | Intermediate | 22 |
| Chronic cough | Diagnosis + management | n | Out | a | Intermediate | 22 |
| Lower back pain | Diagnosis + management | n | Out | a | Intermediate | 22 |
| Heart failure | Management | n | Out | a/c | Algorithmic | 22 |
| Depression | Management | n | Out | a | Algorithmic | 22 |
| Thyroid screening | Screening | 1 | Out | a | Algorithmic | 22 |
| Hypertension management | Management | n | Out | c | Intermediate | 42 |
| Management of acute migraines | Management | 1 | Out | a | Algorithmic | 40 |
| Prevention of migraine headache | Prevention | n | Out | c | Algorithmic | 40 |
| Management of patients following coronary-artery bypass graft surgery | Management | n | In | a | Algorithmic | 14 |
| Alzheimer's disease | Management | n | Out | c | Algorithmic | 52 |
Abbreviations: n, many; out, outpatient clinic; c, chronic; a, acute.
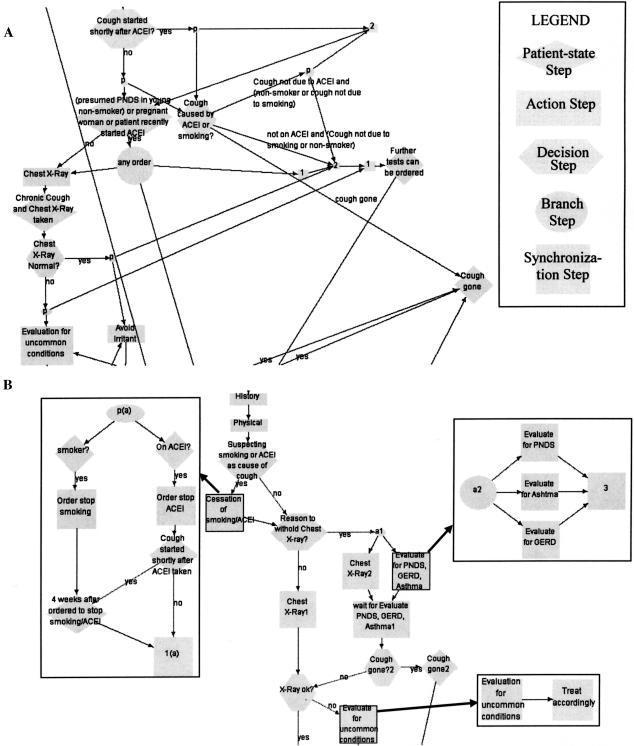
Guideline comprehension involves the ability to construct an adequate mental representation of a clinical guideline. The developer's mental model should match the guideline's process flow and recommended procedures.25 The comprehensibility of a guideline model is important for guideline authors who want to follow easily the guideline specification that they are generating, as well as for users who want to follow the guideline logic but who need to use the least number of inferences in interpreting a guideline. Comprehensibility entails visualization and readability, complexity management, and coherence facilitation. GLIF3 supports all of these types of comprehension facilitation.25 ▶ shows an example in which nesting is used to facilitate comprehension.
Figure 2.
GLIF encoding of a chronic cough management guideline. (A) A design that does not use nesting; (B) a design that uses nesting effectively to control guideline complexity by nesting three action steps into subguidelines.
Sharing of CIGs across different institutions and software systems is desirable because it can (1) provide consistency in guideline interpretation, (2) minimize misinterpretations and errors through the process of public review, and (3) reduce cost of CIG development. Sharing guidelines involves agreement on the functional requirements for guidelines that need to be supported by the model. We believe that various alternative guideline models will not converge easily, since developers' efforts are driven by different purposes or envisioned uses. It is primarily through experience with successful applications that we can expect the evolution of a common shared model so as to include features that support those applications. This involves the lifecycle process discussed in Lifecycle for Development of a Shared Guideline Modeling Language.
Sharing guidelines also involves the modeling support for adaptation of the CIGs for local settings, and its integration with specific hospital information systems environments. The local setting in which a guideline is implemented affects the way in which a guideline may be used. Local settings may differ in (1) delivery platform (e.g., handheld device), (2) mode of user interaction with the system, (3) practice environment (e.g., hospital, home), (4) lack or availability of resources, (5) local policies that may result in preference of specific treatment options, (6) differences in the physical environment (e.g., climate), or (7) differences in patient population.12 The sharable format should allow a guideline encoder to change the guideline encoding in accordance with the local setting. GLIF supports local adaptation by enumerating the details of clinical actions as subguidelines. Different subguidelines could be created for different local implementations of the same clinical action or intention. Work is currently under way to specify constraints formally on such local adaptations that satisfy intentions of guidelines.18,38
To provide patient-specific automatic decision-support services during clinical encounters, a CIG should be integrated with clinical information systems. Patient data items to which CIGs refer must be mapped to electronic medical record (EMR) entries, and guideline recommendations need to be mapped to actions of physician order-entry systems, notifications, or other procedures. In developing GLIF, we have taken the approach of defining a patient data model according to the Health Level 7 (HL7) Reference Information Model (RIM).39 The object-oriented data model of the HL7 RIM provides a declarative way of specifying medical concepts and data items that are used in a guideline. This method could facilitate mapping of concepts and data items to institutional EMRs. By relying on HL7's RIM as the basis for the patient data model of GLIF, the developers of GLIF intentionally leverage work being done by that standardization body, including standards for messaging interfaces for EMRs based on the RIM.
GLIF3 supports the use of controlled terminologies to represent the subject of each data item. This eases integration of CIGs into clinical information systems because (1) the standard terms are more precise than natural language terms, and (2) mapping of standard terms to a clinical information system can be reused by several CIGs containing the same standard terms.
Abstractions are useful to infer clinical situations (e.g., anemia) from raw data and to generalize clinical cases (e.g., diabetes mellitus) for which the guideline recommends the same action. Not all abstractions are found in standard terminologies. Moreover, they are seldom formally defined. GLIF3 uses expressions to define numeric abstractions (e.g., hypertension abstracted from blood pressure measurements) and temporal abstractions (e.g., chronic cough abstracted from episodic observations).
Other types of guideline knowledge can also be used for inference. Examples are contraindications that relate a medication to a disorder and drug interactions that relate two drugs to each other. GLIF specifies such knowledge using concept relationships (e.g., Diabetes-is-a-compelling-indication-for ACE-Inhibitor, where the concepts ACE-Inhibitor and Diabetes are linked through the relationship is-a-compelling-indication-for).
It is important to validate CIGs for logical correctness. The Stanford team used Protégé-2000 to develop a validation tool for GLIF3.40 For each attribute in a class, Protégé allows developers to define allowed data types, cardinality constraints, and lower and upper limits on numerical values. We used Protégé's constraint language to define structural integrity constraints (e.g., a branch step should not immediately be followed by a synchronization step). We used the validation tool to author guidelines and to check them for errors.40 We found errors in two of the five guidelines that the first author of this report encoded in GLIF3. These errors included (1) decision steps that were linked to fewer than two decision options, (2) synchronization steps that immediately followed branch steps, and (3) guideline steps that were not part of any algorithm. No errors were found in the other three guidelines, which were encoded after the ones in which errors were found. This may be due to experience gained by the guideline encoder.
Fostering Sharing of the GLIF3 CIG Model: Standardization Efforts
The InterMed team has sought to rely on existing standards, wherever possible, to leverage work done by others, to use mature and tested standards, and to gain acceptance of our work by vendors and future users. Before May 2001, GLIF had been representing decision logic and eligibility criteria using an expression language we developed called GEL,41 based on the Arden Syntax (an HL7 standard). Lessons learned from the development of GEL led us to understand that an object-oriented expression language would work better with object-oriented domain ontologies.41 In addition, an object-oriented language utilizes the encapsulation of data and methods that are relevant for a medical concept in a way that is not utilized by GEL. Also, an object-oriented model is extensible, so users could define new classes as well as new methods for them. We therefore decided to develop an object-oriented expression language to utilize these features. We call this language GELLO.42 The Harvard InterMed team has led its development, in collaboration with HL7, in which GELLO is being proposed as a standard.
A desired goal of InterMed has been to create a common platform for supporting the full lifecycle of guideline modeling, authoring, dissemination, implementation, and use.11,12 For such a common platform to be accepted and widely used by the CIG community, a broad spectrum of participants must have a stake in it and contribute to its further growth and development. Furthermore, the common model should be standardized and supported by tools for authoring, validation, execution, and maintenance. The InterMed group came to the conclusion that the best way to foster the long-term goal of a common CIG format that provides full lifecycle support is to have its development occur as an open process in which the broad community of modelers, developers, implementers, and other stakeholders is engaged. At the guideline workshop that was hosted by InterMed (Lifecycle for Development of a Shared Guideline Modeling Language), participants explored the issues involved in progressing toward a sharable standardized representation of clinical guidelines. Later that year, InterMed helped to establish the HL7 Clinical Guidelines Special Interest Group (CGSIG), under a reorganized Clinical Decision Support Technical Committee (CDSTC). The CDSTC also includes the Arden Syntax Special Interest Group.11 Establishment of a fully comprehensive CIG model that is accepted by the entire CGSIG is a difficult task. Therefore, the goal of the CGSIG, as well as that of the CDSTC, has become that of developing and standardizing components of CIG models on which consensus could be established among members of the CIG community. If it were possible to map large parts of the different methodologies to the common components, then sharing of significant parts of encoded guidelines across different CIG modeling methods might be feasible.
To understand which components of existing CIG modeling methods share enough similarities across the different modeling methods, InterMed initiated a collaborative study to compare six guideline-modeling methods. We initiated contact with researchers from five other groups that are developing formalisms for representing computer-interpretable guidelines.15,17,18,19,22,43 Our goal was to conduct a case study based on comparisons among GLIF3 and the other five formalisms, to find areas of commonality that would facilitate development of a shared consensus model. We found that consensus could be achieved on three components of CIG models.44 These are (1) object-oriented guideline expression language, (2) a patient data model based on a virtual medical record (VMR) that would be derived from the HL7 RIM and would specifically enable reference to the subset of EMR data needed for guideline-based decision support, and (3) guideline control flow. As part of the CDSTC, InterMed team members have been participating in standardization of these three components. We aim to continue developing and establishing those standards following HL7 methodology and in coordination with the HL7 RIM effort.
The Relationship between GLIF and Other Guideline Formalisms
Different formalisms exist for representing clinical guidelines and clinical decision rules. The Arden Syntax is suitable for representing individual decision rules in self-contained units called Medical Logic Modules (MLMs), which usually are implemented as event-driven alerts or reminders. Although the Arden Syntax has been adapted for the representation of guidelines by employing interacting MLMs,45 it does not provide adequate support for conceptualizing a multistep guideline that unfolds over time.14 The Arden Syntax, like GLIF, stresses the importance of sharing the encoded medical knowledge among different institutions and software systems. MLMs are structured such that the data declarations are separated from the MLM logic. The mappings between the institution-specific terms and the MLM's variables are specified in the data slot. In this way, the MLM logic can be shared. However, the data slot does not structure mappings of MLM data items to institutional EMRs, which impedes sharing MLMs.
GLIF tries to address the data-mapping problem by defining medical concepts in relationship to controlled terminologies and standard medical data models. In this manner, the computable specification layer of a GLIF-encoded guideline can contain concepts and data items that are not institution dependent.
GLIF is one of several formats that represent CIGs as Task-Network Models (TNMs).15,17,18,19,22,43,44 TNMs use a hierarchical decomposition of guidelines into networks of component tasks that unfold over time. Although TNM-based methodologies all use a task-network approach, the groups developing the various methodologies have adopted different approaches, reflecting their interests and expertise.44 The foci of TNM groups include (1) specification of intentions in Asbru;18 (2) integration with organizational workflow and support of decision analysis models in GUIDE;19 (3) simple scenario-based models for chronic disease management in primary care in PRODIGY;43 (4) a simple task ontology, formally grounded in the R2L logic language,46 that makes it possible to demonstrate soundness of a CIG, in PROforma17; and (5) an extensible set of models that define decision-support services that can be realized by various components such as temporal abstraction mediator and explanation generator in EON.37 InterMed team members have been focusing on developing GLIF as a representation for sharable guidelines, while learning from the experience of the other TNM groups. GLIF is also different with respect to the process in which it was developed. In this process, three aspects were most important. First, InterMed has stressed the evolutionary lifecycle approach. Second, GLIF has been developed in a collaborative way, in which each module that has been added to the language has been developed by several InterMed team members, from different InterMed sites, often after review of useful features from other modeling languages. Every feature of the language was added only after establishing consensus by all the InterMed members. Finally, we opened up the development process of GLIF, welcoming inputs from larger communities; we made GLIF, its documentation, and encoded guidelines available for public review on <www.glif.org>.
Another guideline modeling methodology, GEM,47 emphasizes the structuring of documentation attributes of guidelines. Such attributes capture information such as a guideline model's authorship, the nature of the evidence on which the guideline is based, and its intended context of use, all of which need to be made available. As with GLIF, GEM is being developed using an evolutionary lifecycle approach, although it is not aimed primarily at guideline execution.
Conclusions
CIG development takes a long time and much effort. It would be advantageous if multiple institutions were able to share CIGs that were developed at one institution or by national/international authorities such as professional specialty groups. An interchange format, which allows translation from one CIG formalism to another, could allow such sharing. Our experience with the development of GLIF has taught us that developing a bidirectional CIG interchange format is not practical due to differences in modeling goals of existing CIG formats. A different way to foster sharing is for the CIG user community to adopt a standard CIG format. This is the mode of sharing that InterMed is pursuing. GLIF was developed with the goal of it becoming a standard aggregated model that would support functionality derived from studying clinical guidelines and existing CIG modeling formalisms (the evolutionary lifecycle approach). GLIF was developed via a consensus-based multi-institutional development process that was informed by formal cognitive methodologies and was aimed at establishing credibility and validity. GLIF contains most of the important features that are needed to represent CIGs.44,48,49 These features include representation of medical actions and decisions; patient states; a sequential, parallel, and iterative control flow; a formal expression language for expressing decision and eligibility criteria; a medical ontology for defining the structure of patient data and the medical concepts to which the data refer; and support of abstractions and reasoning. Important features that are not yet supported include interfaces with medical knowledge bases and EMRs, and a formal representation of intentions that supports reasoning with intentions.
The development of a comprehensive model involves much effort, is time-consuming, and is costly. The work and investment that have gone into the development of the GLIF language will be leveraged as guidelines are implemented in GLIF and used in different medical institutions. GLIF2 has been the basis for several implementations of guideline-based applications. Researchers at the Brigham and Women's Hospital developed two applications that are based on GLIF2: (1) the BICS information system,50 which is based on simplifications and extensions to GLIF2, and (2) a Web-based application for driving clinical consultations,51 which is based on an extension to GLIF2, known as GLIF 2.5 (<http://glif.org/glif2.5_beta2.html>). GLIF 2.5 extends GLIF2 by including an expression language that is similar to Arden Syntax's logic grammar, and a simple patient data model. The Columbia team of InterMed has developed an execution engine for GLIF3.36 Nonetheless, introducing such technology in a clinical setting is a long process. To date, no computable guidelines encoded in GLIF3 have been interfaced with an existing EMR.
We have found that CIG standardization works most smoothly if focused on well-defined common components rather than on the entire modeling specification. The component-based approach involves active participation and development by all stakeholders and is based on the collective experience gained by them.
Finally, we have found that using existing standards as a starting point, while aiding in establishing credibility and consensus, does not always meet requirements. As explained in Fostering Sharing of the GLIF3 CIG Model, we tried to use the Arden Syntax as GLIF's expression language and to base GLIF's patient data model on the HL7 RIM. As we gained experience in encoding guidelines in GLIF, we found situations in which these existing standards did not match the requirements posed by real-life clinical guidelines and their implementations as decision-support systems.41 This led us to develop the GELLO expression language and to pursue the VMR model that would be derived from the HL7 RIM as a more tractable subset of EMR data needed for guideline-based decision-support. Guideline modeling has served to introduce new requirements into the HL7 standardization process, which are also being examined by the Arden Syntax group and other HL7 technical committees and interest groups. This emphasizes the evolutionary nature of standards themselves and the need for them to adapt to changing requirements.
We plan to continue the effort of developing a common CIG model under the auspices of HL7. Members of InterMed are primarily involved in formalizing (1) GELLO, (2) the VMR medical data model, and (3) representation of guideline control flow. The Columbia InterMed team is developing a decision support facility that will integrate with their existing clinical data repository and Web-based viewing system. As part of that facility, they are planning to implement GLIF-encoded guidelines in clinical settings. The Harvard group is working with the Partners HealthCare information system to explore the feasibility of using a common rules engine based on GELLO to encode the decision support logic contained in a variety of applications now encoding the knowledge in different ways.
Acknowledgments
The authors thank Elmer V. Bernstam, Ronilda Lacson, Nachman Ash, and Peter Mork for their contribution to the development of the GLIF specification and guideline encoding. Supported in part by National Library of Medicine Grant LM06594 with collaborative support from the Department of the Army and the Agency for Healthcare Research and Quality.
The group was lead by VLP, who subsequently moved to Columbia University.
More information on tools can be found at <www.smi.stanford.edu/projects/intermed-web/ToolSupport.html.>
References
- 1.Patel VL, Kaufman DR, Allen VG, Sedhortliffe EH, Cimino JJ, Greenes RA. Toward a framework for computer-mediated collaborative design in medical informatics. Methods Inf Med. 1999;38:158–76. [PubMed] [Google Scholar]
- 2.Overhage JM, Tierney WM, Zhou XH, McDonald CJ. A Randomized Trial of “Corollary orders to prevent errors of omission.”. J Am Med Inform Assoc. 1997;4:364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shea S, DuMouchel W, Bahamonde L. A meta-analysis of 16 randomized controlled trials to evaluate computer-based clinical reminder systems for preventative care in the ambulatory setting. J Am Med Inform Assoc. 1996;3:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Ludemann P, Pryor TA, Wigertz OB, Clayton PD. Rationale for the Arden syntax. Comput Biomed Res. 1994;27:291–324. [DOI] [PubMed] [Google Scholar]
- 5.Stoufflet PE, Ohno-Machado L, Deibel SRA, Lee D, Greenes RA. GEODE-CM: a state-transition framework for clinical management. Proc Annu Symp Comp Appl Med Care. 1996:924.
- 6.Barnes M, Barnett GO. An architecture for a distributed guideline server. Proc Annu Symp Comp Appl Med Care. 1995:233–7. [PMC free article] [PubMed]
- 7.Musen MA, Tu SW, Das AK, Shahar Y. EON: a component-based approach to automation of protocol-directed therapy. J Am Med Inform Assoc. 1996;3:367–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno-Machado L, Gennari JH, Murphy S, et al. The GuideLine Interchange Format: a model for representing guidelines. J Am Med Inform Assoc. 1998;5:357–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shortliffe EH, Patel VL, Cimino JJ, Barnett GO, Greenes RA. A study of collaboration among medical informatics research laboratories. Artif Intell Med. 1998;12:97–123. [DOI] [PubMed] [Google Scholar]
- 10.Patel VL, Arocha JF, Diermeier M, Shortliffe EH, Greenes RA. Methods of cognitive analysis to support the design and evaluation of biomedical systems: the case of clinical practice guidelines. J Biomed Inform. 2001;34:52–66. [DOI] [PubMed] [Google Scholar]
- 11.Greenes RA, Peleg M, Boxwala AA, Tu SW, Patel VL, Shortliffe EH. Sharable computer-based clinical practice guidelines: rationale, obstacles, approaches, and prospects. London, UK: MedInfo, 2001, pp 201–5. [PubMed]
- 12.Boxwala AA, Tu S, Zeng Q, et al. Towards a representation format for sharable clinical guidelines. J Biomed Inform. 2001;34:157–69. [DOI] [PubMed] [Google Scholar]
- 13.Clinical Decision Support & Arden Syntax Technical Committee of HL7, inventor. Arden Syntax for Medical Logic Systems, version 2.0. HL7 Press, July 7, 1999.
- 14.Peleg M, Boxwala AA, Bernstam E, Tu S, Greenes RA, Shortliffe EH. Sharable representation of clinical guidelines in GLIF: Relationship to the Arden syntax. J Biomed Inform. 2000;34:170–81. [DOI] [PubMed] [Google Scholar]
- 15.Tu SW, Musen MA. A flexible approach to guideline modeling. Proc AMIA Symp. 1999:420–4. [PMC free article] [PubMed]
- 16.Sugden B, Purves IN, Booth N, Sowerby M. The PRODIGY Project—the Interactive Development of the Release One Model. Proc AMIA Symp. 1999:359–63. [PMC free article] [PubMed]
- 17.Fox J, Rahmanzadeh A. Disseminating medical knowledge: the PROforma approach. Artif Intell Med. 1998;14:157–81. [DOI] [PubMed] [Google Scholar]
- 18.Shahar Y, Miksch S, Johnson P. The Asgaard Project: a task-specific framework for the application and critiquing of time-oriented clinical guidelines. Artif Intell Med. 1998;14:29–51. [DOI] [PubMed] [Google Scholar]
- 19.Quaglini S, Stefanelli M, Lanzola G, Caporusso V, Panzarasa S. Flexible guideline-based patient careflow systems. Artif Intell Med. 2001;22:65–80. [DOI] [PubMed] [Google Scholar]
- 20.Gordon C, Veloso M. Guidelines in healthcare: the experience of the Prestige project. Ljubljana, Slovenia: Medical Informatics Europe, 1999, pp 733–8. [PubMed]
- 21.Peleg M, Boxwala A, Ogunyemi O, Zeng Q, Tu S, Lacson R, et al. GLIF3 Technical Documentation. <http://smi-web.stanford.edu/projects/intermed-web/guidelines/GLIF1.htm>. Accessed Oct 25, 2003.
- 22.Peleg M, Boxwala A, Ogunyemi O, Zeng Q, Tu S, Lacson R, et al. GLIF3: The evolution of a guideline representation format. Proc AMIA Symp. 2000:645–9. [PMC free article] [PubMed]
- 23.InterMed. Guideline Workshop. <www.GLIF.org/workshop>. Accessed Oct 25, 2003.
- 24.Bernstam E, Ash N, Peleg M, et al. Guideline classification to assist modeling, authoring, implementation and retrieval. Proc AMIA Symp. 2000:66–70. [PMC free article] [PubMed]
- 25.Peleg M, Boxwala AA, Tu S, Greenes RA, Shortliffe EH, Patel VL. Handling expressiveness and comprehensibility requirements in GLIF3. MedInfo. 2001:241–5. [PubMed]
- 26.Patel VL, Arocha JF, Diermeier M, How J, Mottur-Pilson C. Cognitive psychological studies of representation and use of clinical practice guidelines. Int J Med Inform. 2001;63(3):147–68. [DOI] [PubMed] [Google Scholar]
- 27.Patel VL, Allen VG, Arocha JF, Shortliffe EH. Representing a clinical guideline in GLIF: individual and collaborative expertise. J Am Med Inform Assoc. 1998;5:467–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel VL, Branch T, Wang D, Peleg M, Boxwala AA. Analysis of the process of encoding guidelines: a comparison of GLIF2 and GLIF3. Methods Inf Med. 2002;41(2):102–13. [PubMed] [Google Scholar]
- 29.Knuth DA. The errors of TEX. Software-Practice and Experience. 1989;19(7):607–85. [Google Scholar]
- 30.Object Management Group. The Common Object Request Broker: Architecture and Specification; 1999. Report No.: OMG Document Number 91.12.1.
- 31.Boxwala AA, Mehta P, Peleg M, et al. Modeling guidelines using domain-level knowledge representation components. Proc AMIA Symp. 2000:974.
- 32.Choi J, Boxwala AA. Representing domain-level knowledge components using primitives. Proc AMIA Symp. 2001:879.
- 33.Boxwala AA, Peleg M, Tu S, et al. GLIF3: Representation format for sharable computer-interpretable clinical practice guidelines. Decision Systems Group, Technical Report No. TR-2003-30. [DOI] [PubMed]
- 34.W3C. Resource Description Framework (RDF). <http://www.w3.org/RDF/>. Accessed Oct 25, 2003.
- 35.Grosso WE, Eriksson H, Fergerson R, Gennari JH, Tu SW, Musen MA. Knowledge Modeling at the Millennium (The Design and Evolution of Protege-2000). 12 Banff Knowledge Acquisition for KB Systems Workshop. 1999:7-4-1 to 7-4-36.
- 36.Wang D, Shortliffe EH. GLEE—A model-driven execution system for computer-based implementation of clinical practice guidelines. Proc AMIA Symp. 2002:855–9. [PMC free article] [PubMed]
- 37.Tu SW, Musen MA. From guideline modeling to guideline execution: Defining guideline-based decision-support services. Proc AMIA Symp. 2000:863–7. [PMC free article] [PubMed]
- 38.Boxwala AA, Zeng Q, Tate D, Greenes RA, Fairchild DG. Applying axiomatic design methodology to create guidelines that are locally adaptable. Proc AMIA Symp. 2002:980.
- 39.Schadow G, Russler DC, Mead CN, McDonald CJ. Integrating medical information and knowledge in the HL7 RIM. Proc. AMIA Symp. 2000:764–8. [PMC free article] [PubMed]
- 40.Peleg M, Patel VL, Snow V, Tu S, Mottur-Pilson C, Shortliffe EH, et al. Support for guideline development through error classification and constraint checking. Proc AMIA Symp. 2002:607–11. [PMC free article] [PubMed]
- 41.Peleg M, Ogunyemi O, Tu S, et al. Using features of arden syntax with object-oriented medical data models for guideline modeling. Proc AMIA Symp. 2001:523–7. [PMC free article] [PubMed]
- 42.Ogunyemi O, Zeng Q, Boxwala A. Object-oriented guideline expression language (GELLO) specification: Brigham and Women's Hospital, Harvard Medical School, Boston; 2002. Report No. DSG-TR-2002-001.
- 43.Johnson PD, Tu SW, Booth N, Sugden B, Purves IN. Using scenarios in chronic disease management guidelines for primary care. Proc AMIA Symp. 2000:389–93. [PMC free article] [PubMed]
- 44.Peleg M, Tu SW, Bury J, et al. Comparing computer-interpretable guideline models: A case-study approach. J Am Med Inform Assoc. 2003;10(1):52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starren J, Hripcsak G, Jordan D, Allen B, Weissman C, Clayton PD. Encoding a post-operative coronary artery bypass surgery care plan in the Arden Syntax. Comput Biol Med. 1994;24(5):411–7. [DOI] [PubMed] [Google Scholar]
- 46.Fox J, Das S. Safe and sound. In: Safe and Sound. Cambridge, MA: AAAI Press, 2000.
- 47.Shiffman RN, Karras BT, Agrawal A, Chen R, Marenco L, Nath S. GEM: a proposal for a more comprehensive guideline document model using XML. J Am Med Inform Assoc. 2000;7(5):488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D, Peleg M, Tu SW, et al. Representation primitives, process models and patient data in computer-interpretable clinical practice guidelines: A Literature review of guideline representation models. Intl J Med Inform. 2002;68(1–3):59–70. [DOI] [PubMed] [Google Scholar]
- 49.Tu SW, Johnson PD, Musen MA. A typology for modeling processes in clinical guidelines and protocols. Proc AMIA Symp. 2002:1181.
- 50.Zielstorff RD, Teich JM, Paterno MD, et al. P-CAPE: a high-level tool for entering and processing clinical practice guidelines. Partners Computerized Algorithm and Editor. Proc AMIA Symp. 1998:478–82. [PMC free article] [PubMed]
- 51.Boxwala AA, Greenes RA, Deibel SR. Architecture for a multipurpose guideline execution engine. Proc AMIA Symp. 1999:701–5. [PMC free article] [PubMed]
- 52.Yin JQ, Peleg M, Boxwala AA, Greenes RA. Combining a document model and an execution model for clinical guidelines. Proc AMIA Symp. 2001:1064.