Abstract
Background
Polymorphisms of single amino acid repeats (SARPs) are a potential source of genetic variation for rapidly evolving morphological traits. Here, we characterize variation in and test for an association between SARPs and head shape, a trait under strong sexual selection, in the stalk-eyed fly, Teleopsis dalmanni. Using an annotated expressed sequence tag database developed from eye-antennal imaginal disc tissues in T. dalmanni we identified 98 genes containing nine or more consecutive copies of a single amino acid. We then quantify variation in length and allelic diversity for 32 codon and 15 noncodon repeat regions in a large outbred population. We also assessed the frequency with which amino acid repeats are either gained or lost by identifying sequence similarities between T. dalmanni SARP loci and their orthologs in Drosophila melanogaster. Finally, to identify SARP containing genes that may influence head development we conducted a two-generation association study after assortatively mating for extreme relative eyespan.
Results
We found that glutamine repeats occur more often than expected by amino acid abundance among 3,400 head development genes in T. dalmanni and D. melanogaster. Furthermore, glutamine repeats occur disproportionately in transcription factors. Loci with glutamine repeats exhibit heterozygosities and allelic diversities that do not differ from noncoding dinucleotide microsatellites, including greater variation among X-linked than autosomal regions. In the majority of cases, repeat tracts did not overlap between T. dalmanni and D. melanogaster indicating that large glutamine repeats are gained or lost frequently during Dipteran evolution. Analysis of covariance reveals a significant effect of parental genotype on mean progeny eyespan, with body length as a covariate, at six SARP loci [CG33692, ptip, band4.1 inhibitor LRP interactor, corto, 3531953:1, and ecdysone-induced protein 75B (Eip75B)]. Mixed model analysis of covariance using the eyespan of siblings segregating for repeat length variation confirms that significant genotype-phenotype associations exist for at least one sex at five of these loci and for one gene, CG33692, longer repeats were associated with longer relative eyespan in both sexes.
Conclusion
Among genes expressed during head development in stalk-eyed flies, long codon repeats typically contain glutamine, occur in transcription factors and exhibit high levels of heterozygosity. Furthermore, the presence of significant associations within families between repeat length and head shape indicates that six genes, or genes linked to them, contribute genetic variation to the development of this extremely sexually dimorphic trait.
Background
Repetitive, low complexity DNA sequences are ubiquitous in nature [1]. While these sequences are commonly utilized as markers for genetic mapping studies, few of them have been implicated as causal elements of phenotypic change. One class of repetitive sequences, known as single amino acid or codon repeats, is an exception to this pattern and has long been known to be associated with diseases of the nervous system [2,3]. Indeed, variation in the length of single amino acid tracts, often referred to as single amino acid repeat polymorphisms (SARPs), have been implicated in a variety of neuropathologies [2-5], such as Fragile-X [6], Kennedy's disease [7]. Huntington's chorea [8,9] and others [2,10,11]. In some cases, codon repeat length is positively associated with disease severity [2,12,13]. For example, a sequence of 36 or more glutamine repeats in the Huntingtin gene results in a protein product that increases neural decay [14] and causes Huntingtin's chorea [14,15]. Larger glutamine repeat tracts are associated with earlier onset and accelerated progression of the disease [16].
SARPs have also been proposed as a source of genetic variation for rapidly evolving morphological traits [17,18]. This proposition is based on the observation that trinucleotide repeats are common in eukaryotic DNA [19-21] and undergo mutation as a consequence of replication slippage more frequently than amino acid substitutions [11,22] but see [23]. Rather than being purged from the genome, long repeats are frequently conserved across vertebrates [19,24-27] and those containing glutamine or alanine tend to occur in transcription factors [28,29]. Contraction or expansion of a codon repeat in a transcription factor has the potential to modulate gene regulation in a quantitative, rather than qualitative, manner [10,30] and, therefore, result in a mutation of small effect [22]. Thus, SARPs appear to have the potential both to generate genetic variation and to enable adaptive change in morphology.
In this paper we use stalk-eyed flies as a model system [sensu [31]] to determine if SARPs could contribute to the rapid and recurrent evolution of extreme sexual dimorphism for eyestalk length in these flies [32,33]. We designed the study to address four questions: 1) Are any SARPs unequally represented among amino acids in genes expressed during eyestalk development? 2) Are any SARPs preferentially located in transcription factors? 3) Are SARPs gained or lost frequently across species? 4) Do SARPs occur in genes that influence eyestalk length? To increase the likelihood of finding genes with adaptive phenotypic effects, we focus the study on repeats with nine or more consecutive codons because long repeats typically disrupt function and should, therefore, only persist if they provide some selective advantage [17-19,24-27]. Furthermore, because replication slippage typically increases with repeat length [34], long repeats are also likely to exhibit genetic variation.
To identify genes with long repeats we use an annotated expressed sequence tag (EST) database [35] containing over 4,000 unique open reading frames derived from Teleopsis dalmanni [recently synonymized with Cyrtodiopsis, [36]] brain and eye-antennal imaginal disc tissue dissected from third-instar larvae or 1-7 day-old pupae. During this period of time these tissues develop into the adult head, eyes and brain [37]. To determine if codon repeat frequency is independent of amino acid frequency we use homologous gene regions of T. dalmanni and Drosophila melanogaster. To assess bias in gene function we compare genes with codon repeats to all genes in the T. dalmanni annotated library [35]. Using an outbred population of flies we quantify allelic diversity and heterozygosity for a sample of genes containing polyglutamine repeats and compare them to the same metrics scored on the same flies for a sample of noncoding dinucleotide microsatellites [38]. We made this comparison to determine if length variation in codon repeats is comparable to noncodon repeats, as would be expected if they mutated by a common mechanism and experienced similar constraints. We assess the frequency with which amino acid repeats may be gained or lost by finding all long repeats in homologous gene regions of both T. dalmanni and D. melanogaster and then determining if a repeat is present in the ortholog. Finally, we conduct a two-generation association study to determine if parental genotype at 32 SARP loci predicts offspring phenotype after assortatively mating by relative eyespan. To confirm significant associations at candidate loci we test for differences in eyespan among genotypes in siblings that differ in repeat length at each candidate locus.
Results
Distribution of codon repeat loci
To determine if any repeats containing nine or more codons occur out of proportion to codon abundance, we compared repeat frequencies to corresponding amino acid frequencies for genes in the T. dalmanni EST library and their homologs in D. melanogaster (Fig. 1). We found that repeat abundance varies independently of amino acid abundance in each species (T. dalmanni: χ2 = 895.9, df = 19, P < 0.0001; D. melanogaster: χ2 = 1064.6, df = 19, P < 0.0001). For both species, glutamine (Q) occurred in repeats much more often than any other amino acid (Fig. 1). In T. dalmanni, no other amino acid was over represented in repeats, but three amino acids - isoleucine (I), valine (V), and arginine (R) - formed repeats less often than expected. In D. melanogaster two other amino acids (A and G) were over represented, six amino acids (S, N, T, H, C and W) occurred in proportion to their abundance and the remaining 11 amino acids were under represented in repeats.
Figure 1.
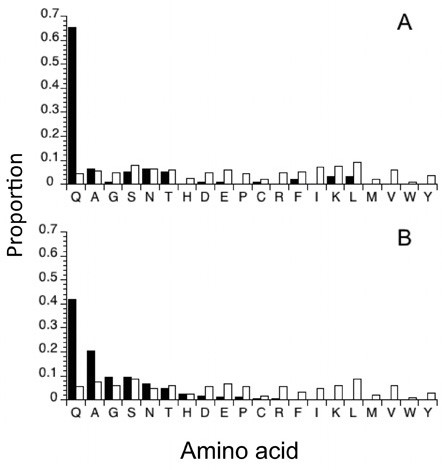
Distribution of single amino-acid repeats containing more than 8 consecutive residues (filled bars) plotted with the relative abundance of each amino acid (open bars) for two fly species. Panel A: Proportion of 98 unique open reading frames containing SARs identified in the Teleopsis dalmanni EST database. Panel B: Proportion of 343 genes containing SARs in regions of Drosophila melanogaster genes homologous to the T. dalmanni EST database.
Function of codon repeat loci
We used GeneMerge [39] to determine if the molecular functions of genes with repeats represent a nonrandom sample of the EST library. This analysis revealed that the 98 unique genes with codon repeats were more likely to exhibit DNA binding (P = 0.0021) or transcription regulator activity (P = 0.0092) after Bonferroni correction than expected. When this GeneMerge analysis was repeated using only the 64 genes that carry glutamine repeats, similar results were obtained: RNA polymerase II transcription factor activity (P = 0.0069), transcription regulator activity (P = 0.012), and DNA binding (P = 0.015) were the only molecular functions that were over-represented in the sample.
Length variation in codon repeats
To quantify standing genetic variation in codon length we genotyped a large sample of flies reared from an outbred population of T. dalmanni originally collected in 1999 in peninsular Malaysia [40,41]. Length polymorphism was detected at 25 of 32 glutamine repeat loci (Table 1). Observed heterozygosity, (average ± SE) 0.48 ± 0.03, was similar to allelic diversity, 0.53 ± 0.03, across loci. Nevertheless, after Bonferroni adjustment, goodness-of-fit tests revealed that genotype frequencies at seven loci deviated significantly from expectation (Table 1). At five loci (CG12104, CG31224, Eip75B, M-spondin, and toutatis) there was a deficiency of heterozygotes while at two loci (corto and CG10082) there were more heterozygotes than expected.
Table 1.
Heterozygosity and allelic diversity of glutamine repeat loci in T. dalmanni
| Locus (chromosome*) | Ho | He | χ2 | P | Allele No. | N |
|---|---|---|---|---|---|---|
| Band4.1 inhibitor LRP (2) | 0.59 | 0.59 | 0.00 | ns | 4 | 163 |
| Bifocal (1) | 0.73 | 0.65 | 2.75 | ns | 4 | 91 |
| Bunched (X) | 0.70 | 0.66 | 0.62 | ns | 3 | 92 |
| Cap-n-collar (2) | 0.48 | 0.50 | 0.08 | ns | 2 | 91 |
| CG10082 (2) | 0.67 | 0.53 | 7.04 | 0.0080 | 4 | 90 |
| CG10321 (2) | 0.63 | 0.60 | 0.38 | ns | 3 | 90 |
| CG10435 (2) | 0.54 | 0.41 | 0.08 | ns | 2 | 91 |
| CG12104 (1) | 0.31 | 0.45 | 7.31 | 0.0069 | 2 | 91 |
| CG17265 | - | - | - | ns | 1 | 94 |
| CG31064 (2) | 0.51 | 0.50 | 0.07 | ns | 5 | 165 |
| CG31224 (2) | 0.30 | 0.65 | 38.03 | < 0.0001 | 4 | 71 |
| CG33692 (1) | 0.61 | 0.60 | 0.16 | ns | 4 | 166 |
| CG34347 (2) | 0.60 | 0.60 | 0.00 | ns | 6 | 91 |
| CG42389 (X) | 0.56 | 0.61 | 1.87 | ns | 4 | 165 |
| CG4409 (2) | 0.35 | 0.41 | 1.13 | ns | 2 | 94 |
| CG8668 (X) | 0.68 | 0.70 | 0.28 | ns | 5 | 159 |
| Corto (2) | 0.74 | 0.67 | 4.13 | 0.042 | 5 | 155 |
| Cryptocephal (X) | 0.57 | 0.59 | 0.22 | ns | 6 | 167 |
| Cyclin-dependent kinase 8 | - | - | - | ns | 1 | 94 |
| Dachshund | - | - | - | ns | 1 | 94 |
| Dorsal switch protein 1 | - | - | - | ns | 1 | 94 |
| E5 (2) | 0.47 | 0.42 | 0.65 | ns | 2 | 86 |
| Ecdysone-induced protein 75B (1) | 0.12 | 0.47 | 43.53 | < 0.0001 | 2 | 92 |
| M-spondin (2) | 0.15 | 0.32 | 12.38 | 0.0054 | 4 | 89 |
| Mastermind (2) | 0.26 | 0.25 | 0.00 | ns | 3 | 90 |
| Mediator complex subunit 26 | - | - | - | ns | 1 | 94 |
| Ptip (1) | 0.50 | 0.53 | 0.40 | ns | 5 | 90 |
| Sine oculis-binding protein | - | - | - | ns | 1 | 94 |
| SRPK (2) | 0.60 | 0.60 | 0.00 | ns | 3 | 75 |
| Tenascin major (1) | 0.28 | 0.30 | 0.15 | ns | 2 | 92 |
| Toutatis (2) | 0.46 | 0.61 | 8.50 | 0.0063 | 7 | 167 |
| 3531953:1 (X) | 0.64 | 0.59 | 1.55 | ns | 5 | 163 |
*chromosome identity corresponds to Johns et al. (2005)
SARP loci were assigned to chromosome based on evidence of linkage to 15 noncoding microsatellite markers, which were genotyped for the same sample of flies as the SARP loci, and had previously been located on chromosomes by linkage mapping [42]. Among these flies the noncoding microsatellites had 2-6 alleles and observed heterozygosities ranging from 0.135 to 0.744. Chi-squared contingency tests on genotype counts of noncoding microsatellite markers and SARP loci revealed that eight SARP loci were associated with the first chromosome while 16 SARP loci were associated with the second chromosome and five were associated with the X chromosome (Table 1). Thus, 17% of SARP loci were found on the X chromosome, consistent with the relative size of the X estimated by other methods [43].
In a previous analysis of genetic variation among anonymous noncoding microsatellites [38], X-linked markers exhibited greater genetic variation than autosomal markers. Consequently, we compared genetic variation at glutamine repeat loci to noncoding microsatellites by type of chromosome. A two-way ANOVA on observed heterozygosity revealed that type of chromosome (F1,43 = 5.60, P = 0.023), but not type of repeat (F1,43 = 0.09, P = 0.76), was significant. Loci on the X chromosome exhibited greater heterozygosity (0.61 ± 0.05) than autosomal loci (0.46 ± 0.03). Similar results were obtained for expected heterozygosity, i.e. type of chromosome (F1,43 = 5.61, P = 0.022), but not type of repeat (F1,43 = 0.0003, P = 0.99), was significant. In contrast, the average number of alleles per locus did not depend on chromosome (F1,43 = 0.15, P = 0.70) or type of repeat (F1,51 = 0.003, P = 0.96)
We located two or more EST sequences for 12 genes that contained nine or more glutamine residues. After translating the nucleotide sequences we found variation in the length of the glutamine tract for ten of these genes. A comparison of those sequence variants with the length variants identified by PCR revealed that the length variants found among the flies that were genotyped corresponded to an ORF length as predicted by the EST sequences in all but one case (Table 2). For dorsal switch protein 1 there were two length variants among the EST sequences but only a single length variant was identified by PCR.
Table 2.
Amino acid sequence variants in the T. dalmanni EST database with length variants obtained by PCR.
| Gene | EST sequences | Repeat length | Repeat sequence | PCR product length (bp) |
|---|---|---|---|---|
| CG12104 | 1 | 14 | QQQQQQQQQQQQQQ | 192 |
| 4 | 13 | QQQQ-QQQQQQQQQ | 189 | |
| CG32133 | 2 | 14 | QQQQQQQQQQSQQQ | 214 |
| 1 | 10 | ----QQQQQQSQQQ | 202 | |
| CG4409 | 3 | 19 | QQQEQEQQQQQQQQQQQQQ | 214 |
| 6 | 16 | QQQEQEQQQQ---QQQQQQ | 205 | |
| Corto | 1 | 19 | QQQQQQQQQQQYQQQQQQQ | 496 |
| 1 | 18 | QQQQQQQ-QQQYQQQQQQQ | 493 | |
| Cryptocephal | 2 | 27 | QQQQQQQQQQQQQQQQQQQQQQQQQQQ | 227 |
| 2 | 25 | QQQQQQQQQQQQQQQQQ--QQEQQQQQ | 221 | |
| 1 | 24 | QQQQQQQQQQQQQQQQ---QQEQQQQQ | 218 | |
| 1 | 23 | QQQQQQQQQQQQQQQ----QQEQQQQQ | 215 | |
| 4 | 20 | QQQQQQQQQQQQ-------QQEQQQQQ | 206 | |
| 4 | 16 | QQQQQQQQ-----------QQQQQQQQ | 194 | |
| Dorsal switch protein 1 | 1 | 50 | QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQIQQQQQQ | 181 |
| 1 | 48 | QQQQQQQQQQ--QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQIQQQQQQ | - | |
| Mastermind | 2 | 26 | QQQQSQAQQQQQQQQQQQQQQKQQQQ | 523 |
| 1 | 25 | QQQQFQA-QQQQQQQQQQQQQKQQQQ | 520 | |
| SRPK | 1 | 30 | QQQRQQQQQQQQQFQQQQQYQQQQQYQQQQ | 172 |
| 1 | 26 | QQQRQQQQQQQ----QQQQFQQQQQYQQQQ | 160 | |
| Tenascin major | 2 | 15 | QQQQQQQQQQQQQQQ | 206 |
| 1 | 13 | QQQQQQQQQQ-QQQ | 200 |
Evolution of glutamine repeats
A total of 60 genes had a polyglutamine repeat longer than eight residues in either T. dalmanni or D. melanogaster (Table 3). The two species had similar numbers of genes with repeats (48 in T. dalmanni vs. 45 in D. melanogaster), although there were substantial differences between the species in the location of the repeats. Of the 84 total repeats found, only nine occurred in homologous regions in both species (in the genes dachshund, dorsal switch protein 1, CG17271, corto, cyclin-dependent kinase 8, mastermind, pumilio and scribbler). In contrast, 39 repeats (46%) have a homologous counterpart that contains 2 or fewer glutamines in the other species.
Table 3.
Glutamine content for aligned gene regions in D. melanogaster and T. dalmanni
| Gene name | Glutamine # | Gene name | Glutamine # | ||
|---|---|---|---|---|---|
| Dm | Td | Dm | Td | ||
| Band4.1 inhibitor LRP interactor | 7 | 9 | dikar | 16 | 2 |
| big brain | 13 | 2 | domino | 4 | 16 |
| bunched | 4 | 18 | Dorsal switch protein 1 | 22 | 36 |
| cap-n-collar | 9 | 12 | E2F transcription factor | 9 | 5 |
| CG10082 | 1 | 10* | E5 | 3 | 10 |
| CG10082 | 2 | 19** | E5 | 7 | 10 |
| CG10321 | 3 | 16 | E5 | 0 | 9 |
| CG10321 | 1 | 9 | Ecdysone-induced protein 75B | 9 | 2 |
| CG12104 | 1 | 14 | grainy head | 9 | 0 |
| CG12488 | 9 | 3 | grainy head | 9 | 1 |
| CG14023 | 16 | 1 | GUK-holder | 1 | 9 |
| CG14023 | 12 | 1 | GUK-holder | 1 | 12 |
| CG14213 | 12 | 1 | hairy | 6 | 10 |
| CG14440 | 9 | 2 | headcase | 10 | 0 |
| CG14441 | 16 | 12* | headcase | 20** | 5 |
| CG14441 | 10 | 2 | jim | 17 | 0 |
| CG14650 | 17 | 14 | La related protein | 4 | 9 |
| CG17265 | 1 | 14 | mastermind | 14 | 0 |
| CG17271 | 10 | 10 | mastermind | 17 | 7 |
| CG17446 | 21 | 9* | mastermind | 12* | 21 |
| CG17446 | 12 | 4 | mastermind | 5 | 10 |
| CG2083 | 8 | 9 | mastermind | 12* | 13 |
| CG31064 | 7 | 11 | mastermind | 14 | 14 |
| CG31738 | 0 | 15 | Mediator complex subunit 26 | 2 | 14 |
| CG32772 | 9 | 3 | milton | 0 | 9 |
| CG34114 | 9 | 1 | M-spondin | 0 | 11 |
| CG34114 | 8 | 10 | M-spondin | 6 | 9 |
| CG34347 | 0 | 11 | pipsqueak | 12 | 9* |
| CG4068 | 2 | 9 | Protein associated with topo II related - 1 | 5 | 9 |
| CG4702 | 1 | 9 | ptip | 35 | 1 |
| CG5053 | 12 | 5 | ptip | 7 | 10 |
| CG6619 | 23 | 12* | pumilio | 13 | 11 |
| CG8668 | 2 | 9 | pumilio | 12 | 15 |
| Cirl | 10 | 6 | Regena | 9 | 2 |
| corto | 17 | 0 | reversed polarity | 9 | 0 |
| corto | 8 | 10 | reversed polarity | 9 | 0 |
| corto | 11 | 10 | scribbler | 21 | 22 |
| cryptocephal | 0 | 27 | scribbler | 10 | 5 |
| C-terminal Src kinase | 9 | 4 | Sine oculis-binding protein | 0 | 12 |
| Cyclin-dependent kinase 8 | 27 | 27 | SRPK | 2 | 9 |
| dachshund | 11 | 11 | Tenascin major | 1 | 13 |
| dachshund | 15 | 4 | wallenda | 2 | 9 |
*Region does not contain a run of 9 consecutive glutamines.
**Region contains two polyglutamine repeat regions separated by a single non-glutamine amino acid.
Association of glutamine repeat length and eyespan
To identify SARP loci with potential effects on relative eyespan we tested for an association between genotype and phenotype after one generation of artificial selection by measuring relative eyespan for 587 flies, selecting extreme males and females, assortatively mating 92 pairs, and collecting their progeny. We then measured a sample of male and female progeny from each of 51 families and tested if parental genotype predicted offspring breeding value for either sex. ANCOVA on average eyespan, with body length as a covariate, for either 10 male or 10 female progeny by parental genotype at each locus revealed five autosomal loci [band4.1 inhibitor LRP interactor, ptip, CG33692, corto, and ecdysone-induced protein 75B (Eip75B)] in which P < 0.01 for at least one sex (Table 4). In addition, one of five X-linked loci [3531953:1] exhibited a nearly significant effect (P = 0.011) of male parental genotype on female phenotype (Table 5).
Table 4.
ANOVA on progeny eyespan by parent genotype for autosomal polyglutamine loci
| Female eyespan | Male eyespan | ||||
|---|---|---|---|---|---|
| Locus | F | P | F | P | N |
| Band4.1 inhibitor LRP interactor | 6.01 | 0.0002 | 4.72 | 0.0016 | 98 |
| Cap-n-collar | 0.55 | 0.58 | 0.57 | 0.57 | 89 |
| CG10082 | 1.57 | 0.18 | 1.77 | 0.13 | 88 |
| CG10321 | 0.8 | 0.49 | 1.82 | 0.13 | 88 |
| CG10435 | 0.04 | 0.85 | 0.04 | 0.84 | 89 |
| CG12104 | 3.19 | 0.046 | 2.09 | 0.13 | 89 |
| CG31064 | 0.35 | 0.93 | 0.69 | 0.68 | 98 |
| CG31224 | 1.11 | 0.37 | 1.76 | 0.10 | 73 |
| CG33692 | 2.98 | 0.011 | 3.16 | 0.0074 | 98 |
| CG34347 | 0.66 | 0.78 | 1.01 | 0.45 | 89 |
| CG4409 | 1.43 | 0.24 | 1.38 | 0.26 | 92 |
| Corto | 2.25 | 0.022 | 2.59 | 0.0087 | 96 |
| E5 | 0.85 | 0.43 | 0.90 | 0.41 | 84 |
| Ecdysone-induced protein 75B | 2.71 | 0.07 | 6.13 | 0.0032 | 91 |
| M-spondin | 0.37 | 0.83 | 0.75 | 0.56 | 87 |
| Mastermind | 1.52 | 0.21 | 2.19 | 0.08 | 88 |
| Ptip | 1.25 | 0.28 | 2.84 | 0.0079 | 88 |
| SRPK | 1.95 | 0.06 | 2.54 | 0.015 | 99 |
| Tenascin major | 0.56 | 0.57 | 1.91 | 0.15 | 90 |
| Toutatis | 1.07 | 0.40 | 1.23 | 0.28 | 89 |
Table 5.
ANOVA on progeny eyespan by parent genotype for X-linked polyglutamine loci
| Female eyespan | Male eyespan | |||||
|---|---|---|---|---|---|---|
| Locus | Parent | F | P | F | P | N |
| Bunched | Male | 0.22 | 0.80 | 0.42 | 0.66 | 45 |
| Female | 0.71 | 0.62 | 0.88 | 0.50 | 45 | |
| CG8668 | Male | 0.65 | 0.69 | 0.60 | 0.73 | 48 |
| Female | 0.64 | 0.77 | 1.18 | 0.34 | 46 | |
| CG42389 | Male | 2.04 | 0.12 | 1.87 | 0.15 | 49 |
| Female | 0.81 | 0.55 | 1.83 | 0.13 | 50 | |
| Cryptocephal | Male | 0.82 | 0.54 | 1.28 | 0.29 | 50 |
| Female | 0.33 | 0.92 | 0.71 | 0.65 | 49 | |
| 3531953:1 | Male | 4.18 | 0.011 | 3.03 | 0.039 | 48 |
| Female | 0.99 | 0.46 | 0.97 | 0.47 | 49 | |
To corroborate these putative parental genotype-offspring phenotype associations, we inspected parental genotypes to identify at least five families for each locus in which informative alleles should be segregating among siblings. At one locus, ptip, only one such family was found, so that locus was not tested further. For the other five loci, at least 30 progeny of each sex were measured, extracted and genotyped from a total of 19 informative families. A mixed model analysis of covariance, with body length as a covariate, revealed large differences in eyespan among families at all loci (Table 6) and a significant effect of progeny genotype on eyespan phentoype for at least one sex at four loci. Progeny genotype explained 7% of the phenotypic variation in both male and female eyespan for CG33692, 9% of the variance in female eyespan for corto, 6.5% of the variance in male eyespan for 3531953:1, and 2% of the variance in female eyespan for Eip75B. A plot of mean eyespan by genotype reveals that longer glutamine repeats are associated with longer relative eyespan in both sexes for CG33692 (Fig. 2).
Table 6.
Mixed model ANOVA on progeny eye span by progeny polyglutamine genotype and family
| Females | Males | |||||||
|---|---|---|---|---|---|---|---|---|
| Source of variation | df | Var Comp% | F | P | df | Var Comp% | F | P |
| Band4.1 inhibitor LRP interactor (2) | ||||||||
| Family* | 5 | 36.9 | 14.5 | < 0.0001 | 5 | 53.6 | 31.7 | < 0.0001 |
| Genotype* | 4 | 2.0 | 1.7 | 0.16 | 4 | 1.1 | 1.6 | 0.17 |
| Error | 177 | 202 | ||||||
| CG33692 (1) | ||||||||
| Family* | 5 | 37.3 | 13.2 | < 0.0001 | 5 | 38.6 | 18.2 | < 0.0001 |
| Genotype* | 7 | 5.6 | 2.7 | 0.011 | 7 | 6.8 | 3.5 | 0.0013 |
| Error | 168 | 200 | ||||||
| Corto (2) | ||||||||
| Family* | 5 | 33.7 | 10.5 | < 0.0001 | 5 | 46.1 | 18.4 | < 0.0001 |
| Genotype* | 9 | 4.9 | 2.1 | 0.035 | 9 | 1.1 | 1.3 | 0.23 |
| Error | 175 | 200 | ||||||
| Ecdysone-induced protein 75B (1) | ||||||||
| Family* | 4 | 32.9 | 14.3 | < 0.0001 | 4 | 43.3 | 19.2 | < 0.0001 |
| Genotype* | 2 | 5.4 | 4.6 | 0.012 | 2 | 1.2 | 1.8 | 0.18 |
| Error | 134 | 119 | ||||||
| 3531953:1 (X) | ||||||||
| Family* | 4 | 47.7 | 16.2 | < 0.0001 | 4 | 50.6 | 22.6 | < 0.0001 |
| Genotype* | 5 | -1.0 | 0.7 | 0.59 | 5 | 6.5 | 4.5 | 0.0049 |
| Error | 143 | 144 |
*Family and genotype are random effects and body length is a significant (not shown) covariate in all models.
Figure 2.
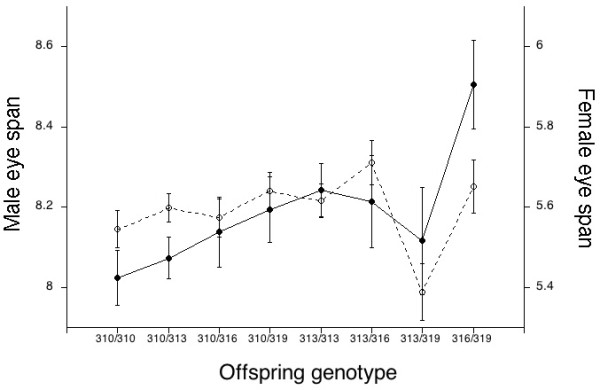
Least square adjusted mean eyespan for male (solid) and female (dashed) plotted against genotype for progeny from six families that segregated for allelic variants at CG33692.
Discussion
Distribution, variation and evolution of amino acid repeat loci
By analyzing ESTs from genes expressed during head development we find that amino acid repeats typically consist of glutamine residues, are often found in transcription factors, and exhibit high levels of polymorphism. These results are consistent with previous studies [28,44,45], which have found that glutamine repeats were the most common codon repeat in Drosophila. Finding more glutamine than any other amino acid in repeats despite using a criterion that sampled far fewer repeats, i.e. less than 25% of repeats have 9 or more codons in Drosophila [46,47], suggests that glutamine may be particularly prone to repeat formation in flies. Indeed, Faux et al. [28] found that glutamine was also the most common amino acid among repeats in Anopheles gambiae, although it was only 1/6 as common as in D. melanogaster. In contrast to flies, glutamine repeats are only the seventh most common codon repeat in mammals and sixth most common in chickens [28].
Our discovery of extensive length polymorphism for glutamine repeats is consistent with reports suggesting that CAG and CAA trinucleotide repeats are particularly prone to replication error [19,48-50]. The similarity in measures of variation between coding and noncoding repeats suggests that similar mutational processes affect both types of repeats in stalk-eyed flies. Some of this variation may not have significant phenotypic consequences since genotypic frequencies inferred from PCR products amplified from a large sample of outbred flies matched Hardy-Weinberg expectations. However, five amino-acid repeat loci contained more homozygotes than expected (CG12104, CG31224, Eip75B, M-spondin, and toutatis) while two loci (corto and CG10082) exhibited more heterozygotes than expected. An overrepresentation of heterozygotes may indicate the presence of balancing selection [51]. An excess of homozygotes could be caused by null alleles, selection, inbreeding, or population substructure [51]. Because we found no evidence of excess homozygosity among microsatellite markers typed on the same individuals and flies were sampled from a population that has been maintained in the laboratory for over 10 years, inbreeding and population substructure are unlikely explanations for genotypic differences among loci. Selection, though, could explain deviations from Hardy Weinberg if either alleles or genotypes are under selection or near loci under selection. Genotypes from at least two of these loci (corto and Eip75B) explain variation in eyespan (see below). Because we estimated genotype frequencies using flies that had been selectively chosen to differ in relative eyespan, we would expect genotype frequencies to fail to conform to Hardy Weinberg expectations whenever loci are closely associated with eyespan.
Consistent with previous reports [10,17,45,52] we find that proteins with codon repeats tend to be transcription factors or associated with gene regulation more often than expected by chance. Although transcription factors exhibit sequence conservation in their DNA binding domains [53], codon repeats tend to occur in intrinsically disordered regions, which are poorly conserved [54,55]. Thus, length variation in codon repeats is less likely to influence transcription through DNA binding activity than through other mechanisms. Nevertheless, mutational studies on at least five different proteins with conserved codon repeats have demonstrated that alteration in amino acid repeat length alters protein function [29,56].
Replication slippage provides a plausible explanation for how variation in length is generated once an amino acid repeat has formed. The origin of a repeat is, however, less obvious and likely involves other genetic mechanisms, such as unequal crossing over or gene conversion. Recently, codon repeat abundance and length has been found to be greater in genes that exhibit alternative splicing, which are also enriched for poly-Q in Drosophila [29,56]. Alternative splicing may reduce constraints by exposing multiple forms of a protein to selection. Such relaxed selection may then permit invasion and expansion of a codon repeat. The relative lack of conservation among genes containing repeats of 9 residues or more in T. dalmanni and D. melanogaster (cf. Table 3) demonstrates that glutamine repeats can be gained or lost frequently over time, which for this species pair corresponds to about 70 MY [57]. This result contrasts with Mularoni et al. [29] who found 92 repeats 8 amino acids or longer in Homo sapiens that are conserved among Eutherians and reported that conserved codon repeats tend to be longer than noncoding repeats, indicative of positive selection. These results indicate that some of the evolutionary processes influencing the origin and evolution of amino acid repeats must differ between flies and vertebrates.
Association between amino acid repeat loci and a sexually selected trait
To the extent that variation in codon repeat length alters regulation of transcription, SARPs provide a mechanism by which protein-coding regions may contribute to quantitative variation in phenotypic traits. In this study, we evaluate the possibility that length variation in loci with glutamine repeats influences the development of an unusual sexually selected and sexually dimorphic trait - relative eyespan in the stalk-eyed fly, T. dalmanni. Parental genotypes at five autosomal loci (band4.1 inhibitor LRP interactor, ptip, CG33692, corto, and Eip75B - Table 4) and one X-linked locus (3531953:1) predicted mean eyespan of progeny in at least one sex (Table 5). Several of these breeding value associations were confirmed by showing that amino acid repeat genotype predicted relative eyespan among siblings from families in which length variants were segregating at the candidate locus (Table 6). Progeny genotype explained significant variation in female eyespan for corto and Eip75B, in male eyespan for 3531953:1 and in both female and male eyespan for CG33692. Longer glutamine repeats in CG33692 are associated with longer relative eyespan in both sexes (cf. Fig. 2).
Four explanations are possible for these associations. First, associations between genotype and morphology might represent false positive results due to multiple testing or undetected population stratification. However, by first screening parent genotype against progeny breeding values and subsequently testing for associations between candidate loci genotype and progeny phenotype within nuclear families, we minimize spurious results. The apparent sex-limited genotype-phenotype associations at some loci, such as corto and Eip75B, suggest, though, that some associations are weak and deserve replication. Second, associations between genotype and morphology could result from epistatic interactions involving multiple loci. Additional breeding experiments need to be conducted to evaluate this possibility because only a limited number of genotypic combinations involving the candidate loci are represented in our breeding study. Third, associations between genotypes and morphology may be due to linkage disequilibrium. Studies on Drosophila have shown that linkage disequilibrium decays rapidly with respect to physical distance in large effective populations, but if populations are small, linkage disequilibrium may be present over longer distances [58]. It is possible, therefore, that some of the associations we found, such as that for CG33692, are caused by physical linkage to another gene that causes differences in eyestalk length. High-resolution mapping studies are needed to assess the plausibility of this suggestion. Finally, length variation in polyglutamine regions may directly influence head shape development. Ultimately, confirmation of any genotype-phenotype association will require some type of genetic manipulation, such as RNAi, that alters phenotypic expression during the appropriate developmental period.
If any of these associations do reflect causal relationships, we would expect the known biological functions of the candidate genes to be consistent with modification to head and eye development. Accordingly, three of the five candidate genes have known phenotypic effects on eye development. Unfortunately, neither the molecular function nor the biological process for CG33692 is known [59]. Two of the other candidate genes, corto and Eip75B, are known to exhibit transcription factor activity. Specifically, corto exhibits RNA polymerase II transcription factor activity, and protein binding [60,61]. Furthermore, corto acts alternatively as an enhancer of trithorax and an enhancer of polycomb. These two groups of proteins are responsible for maintaining homeotic (Hox) gene expression throughout development [62] and homeobox genes are required for Drosophila visual system development [63,64].
Ecdysone-induced protein 75B exhibits transcription factor activity as well as regulation of transcription [65] and oogenesis [66] and is part of a small group of genes whose early expression is essential for ecdysone induced changes during developmental transitions [67]. Ecdysteroids trigger major developmental transitions such as larval molting and metamorphosis in flies [68]. Furthermore, Eip75B affects eye formation in flies and moths [69,70].
The EST 3531953:1 has not yet been identified but the amino acid sequence shares structural characteristics with tousled-like kinases (Tlks). Tlks are a family of serine/threonine kinases that are involved in the cell cycle [71-75], chromatin assembly [72,75], DNA repair [76], transcription [77], and chromosome segregation [78,79]. Tlk has been directly implicated in spermatogenesis [74] and expression studies have documented a loss of tlk expression results in cell cycle arrest and apoptosis [72]. In D. melanogaster, overexpression of tlk also results in change in the texture of ommatidia and a decrease in eye size [72].
Because 3531953:1 is located on the X chromosome in T. dalmanni and exhibits structural similarity to tlks, which influence spermatogenesis and eye development, this gene is also a potential candidate for sex chromosome meiotic drive in stalk-eyed flies [40,41]. Sex chromosome meiotic drive typically occurs by differential survival of sperm [80,81] reviewed in: [82]. Male stalk-eyed flies that produce broods composed predominantly of daughters also show evidence of abnormal sperm development consistent with degenerate Y-bearing sperm [42,83]. Furthermore, the X chromosome explains over 30% of the variation in relative eyespan between lines selected for increased or decreased eyespan [43] and males that lack drive X chromosomes have longer eyestalks [42]. Thus, the possibility that 3531953:1 may provide a mechanistic link between meiotic drive and eyestalk length merits future study.
Conclusions
The effect that coding or regulatory sequence evolution has on the evolution of morphology is still contentious. Single amino acid repeat polymorphisms (SARPs) have been proposed as a genetic mechanism that can generate morphological variation [10,17,30,84-88]. This study provides several lines of support for these claims. SARPs are over-represented among genes that contain repeated glutamine residues and influence regulation of transcription. We find that glutamine repeats exhibit levels of variation comparable to anonymous dinucleotide microsatellites and can be independently gained or lost between fly species. Genotypes at five loci independently explain variation in the phenotype of a sexually selected trait, eyestalk length, and current annotation for four of those genes is consistent with a biologically important function in eyestalk development. While these putative associations are intriguing, confirmation must await fine scale mapping studies and genetic manipulations to demonstrate that allelic variation alters phenotypic expression.
Methods
Identification of repeat genes
Because EST libraries are efficient for amino acid repeat discovery [89], we searched amino acid sequences from a T. dalmanni EST database [35] for strings of nine or more consecutive amino acids. This search identified 252 ESTs. Of these, 88 carried terminal lysine or phenylalanine repeats and were excluded as cases of poly-A tails. Of the remaining 164 ESTs, 120 were identifiable on the basis of BlastX (< 1e-9) similarity to a protein in D. melangaster and represented 98 unique genes [35]. Because they are based on partial gene sequence, some annotations may change with additional sequence data.
To compare the distribution of single amino acid repeats between T. dalmanni, and Drosophila melanogaster, we created a protein database for D. melanogaster that contained only homologous regions to the T. dalmanni EST database as determined by a BlastX alignment output. A search of this database revealed 343 loci with at least one repeat containing 9 or more amino acids. Then, to determine if repeats form at random with respect to amino acid we compared the frequency of amino acids in the T. dalmanni EST database or the homologous regions for D. melanogaster to the frequency of each repeat using a chi-squared goodness of fit test. For the T. dalmanni repeat genes, we then used GeneMerge with Bonferroni adjustment [39] to determine if the molecular function represented a nonrandom sample of the putative protein-coding genes in the EST database.
Repeat length variation
To assess variation in repeat length in T. dalmanni, we attempted to genotype at least 51 male and 51 female flies reared from a large, outbred population of T. dalmanni originally collected in 1999 near Ulu Gombak in peninsular Malaysia [40,41] and subsequently maintained as a single population of over 200 individuals with approximately three overlapping generations per year. These flies were used as parents in the association study described below. We used Primer3 [90] to design polymerase chain reaction (PCR) primers that would amplify the repeat and flanking regions for 42 of the original 64 loci containing glutamine repeats. These 42 were chosen because they contained sufficient high complexity flanking regions around repeat regions to design primers. Nine of these primer sets either did not amplify a fragment of the anticipated size or produced inconsistent banding patterns. In addition, the primers for one locus (hairy) produced a fragment that was too large to genotype easily. The remaining primer sets produced reliable PCR products for 32 loci and form the basis of this study.
PCR was carried out in 10 μl reactions containing 40 ng template DNA, 1× reaction buffer, 2.5 mM MgCl2, 0.20 mM dNTPs, 2.5 pmol of each primer, and 0.5 U Taq DNA polymerase. Each amplification reaction was initiated at 95°C for 5 min; followed by 35 cycles of 95°C for 45 s, annealing temperature for 1 minute, and 72°C for 45 s; and terminated at 72°C for 10 min. Primer sequences and annealing temperatures for each locus are listed in Additional File 1, Table S1. PCR products were labeled either with a fluorescent M13 primer according to the protocol outlined in Schuelke [91] or created with primers containing fluorescently labeled nucleotides. Labeled PCR products were genotyped on an ABI 3730 DNA analyzer and products were sized using ROX500 and scored with GeneMapper 4.0 according to manufacturer (Applied Biosystems) protocols.
In addition to scoring loci for amino-acid repeat length, we also genotyped eight autosomal (chromosome 1: ms262Z, ms336, ms392, ms398; chromosome 2: ms90, ms249, ms301, ms422) and eight X-linked (ms70, ms71, ms106, ms125, ms167, ms244, ms395, ms478) noncoding dinucleotide microsatellites [38], whose genomic location had been previously determined by linkage mapping [42]. We determined chromosomal association for each codon repeat locus on the basis of significant chi-squared contingency tests between it and one or more microsatellite markers. In addition, we calculated and compared several measures of genetic variation (see below) to assess the possibility that a common mutation process, such as replication slippage, could account for length variation in both types of repeats.
Sequence analysis
To verify that variation in PCR product length was due to differences in the number of amino acids in a repeat we used Sequencher v. 4 to align and compare all loci for which we had three or more EST sequences. In addition, to confirm that amplification products contained amino acid repeats, at least one PCR product was sequenced for each locus. Sequences were obtained using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit using an ABI 3730 automatic DNA sequencer according to the manufacturer's specifications (Applied Biosystems). Sequences were cleaned, edited, and aligned using Sequencher v. 4.
Comparison of glutamine repeats between species
To determine if glutamine repeats are present in similar locations in both T. dalmanni and D. melanogaster, and therefore likely shared by a common ancestor, we used a protein database for D. melanogaster that contained only homologous regions to the T. dalmanni EST database as described above. We then searched both databases for all occurrences of more than eight consecutive glutamine residues and counted, in the other species, the total number of glutamines in the region homologous to the polyglutamine repeat. Because only a subset of the T. dalmanni EST consensus sequences have homologous sequence in D. melanogaster that is incorporated into the Blast alignment, this search resulted in fewer total polyglutamine repeats than were identified in the search of the entire EST database. However, it is important to note that analysis of these homologous gene region databases in both species eliminates much of the ascertainment bias that can potentially confound comparison between the species.
Estimating association with eyespan
To determine if variation in any of the SARP loci is associated with eyespan variation, we conducted a two-generation association experiment using an outbred laboratory population of T. dalmanni. This population was originally collected near the village of Gombak in peninsular Malaysia in 1999 and subsequently has been maintained with a population of over 200 individuals. In the first generation, we mated flies assortatively on the basis of relative eyespan in order to include alleles with extreme effects. We measured 314 females and 273 males and then selected 46 pairs with the largest and 46 pairs with the smallest eyespan to body length ratio for breeding. We used CO2 anesthesia to capture an 11× video image of each fly resting on its thoracic and orbital spines. Eyespan from the outer edges of the ommatidia, body length from the face to the wing tip, and thorax width were then measured at a resolution of 50 pixels/micrometer using Scion Image v1.59.
Breeding pairs were kept in 2.5 l clear plastic jars and 50 ml of pureed corn was provided as food and oviposition substrate twice each week for three weeks to allow progeny to develop under low competitive conditions. Fifty-one of the 92 pairs produced 20 or more progeny. After eclosion progeny were frozen at -20°C. Eyespan, body length and thorax width were measured from 10 male and 10 female progeny and used to calculate breeding values in eyespan, after adjusting for body length, for each pair. A random effects analysis of variance was used to confirm that heritable variation in eyespan was present in this sample (results not shown).
To identify loci with potential effects on relative eyespan we tested for an association between parental genotype and offspring breeding value. DNA was extracted from each parent using Chelex [92]. Parental genotypes were obtained for 47 of the 51 families at amino acid repeat loci and nocoding microsatellites [38]. For each locus we calculated observed heterozygosity, allelic diversity (i.e. expected heterozygosity) and tested for deviations from Hardy-Weinberg equilibrium expectations using a chi-squared goodness of fit test.
For autosomal loci we used analyses of covariance on eyespan, with body length as a covariate, to determine if son or daughter breeding values differed among parental genotypes at each locus. We conducted similar analyses for X-linked loci except that male and female parents were analyzed separately to account for the different patterns of inheritance of X-linked loci. Loci with significant (P ≤ 0.01) effects of parental genotype for either sex were selected for additional analysis to determine if progeny phenotypes differed among progeny genotypes within families. For each significant locus we examined the parental genotypes and attempted to identify at least five families in which the parental alleles would segregate such that the progeny would carry genotypes that would be expected to differ in eyespan. Eyespan, body length, and thorax width were then measured on a total of 956 offspring from 19 families. Progeny DNA was isolated using Chelex [92] and amplification was carried out as described above. A mixed model analysis of covariance was performed on progeny eyespan in which family and genotype were random effects and body length was a covariate for flies of each sex. We estimated variance components using restricted maximum likelihood to indicate how much of the variation in relative eyespan could be attributed to genetic variation within families.
We used JMP v5.0.1.2 (SAS Institute, 2003) for all statistical analyses.
Authors' contributions
LB conducted all genotyping and sequencing and drafted the manuscript, MP did the breeding and phenotypic measuring for the association study, RB helped to conceive of the study, identify homologous gene regions, and edit the manuscript, and GW guided the study from origin to completion, conducted many of the statistical analyses, and edited the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Primer sequences and annealing temperatures for each locus. This table contains the forward and reverse primers and annealing temperatures that were used in polymerase chain reactions. Product size range is also provided.
Contributor Information
Leanna M Birge, Email: l.birge@ucl.ac.uk.
Marie L Pitts, Email: mlpitts@wm.edu.
Baker H Richard, Email: rbaker@amnh.org.
Gerald S Wilkinson, Email: wilkinso@umd.edu.
Acknowledgements
We would like to thank Cristian Castillo-Davis for bioinformatics advice, Apurva Narechania for assistance manipulating the Blast outputs, and two anonymous reviewers for helpful comments on the manuscript. This work was supported by National Science Foundation grants DEB-0444886 and DEB-0611534 to GSW. LMB was supported by National Science Foundation grant IRFP-0913116 while writing.
References
- Richard G-F, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Ashley CT, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- Gomes-Pereira M, Foiry L, Nicole A, Huguet A, Junien C, Munnich A, G G. CAG trinucleotide repeat "big jumps": large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in Fragile × Syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- Schmidt BJ, Greenberg CR, Allingham-Hawkins DJ, Spriggs EL. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59:770–772. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Banno H, Suzuki K, Takeuchi Y, Kawashima M, Tanaka F, Adachi H, Sobue G. Molecular genetics and biomarkers of polyglutamine diseases. Curr Mol Med. 2008;8:221–234. doi: 10.2174/156652408784221298. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N. et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Nature. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Fondon JW, Hammock EAD, Hannan AJ, King DG. Simple sequence repeats: Genetic modulators of brain function and behavior. Trends Neurosci. 2008;31:328–334. doi: 10.1016/j.tins.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Harper PS, Harley HG, Reardon W, Shaw DJ. Anticipation in myotonic dystrophy: New light on an old problem. Am J Hum Genet. 1992;51:10–16. [PMC free article] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Nance MA, Myers RH. Juvenile onset Huntington's disease-clinical and research perspectives. Ment Retard Dev D R. 2001;7:153–157. doi: 10.1002/mrdd.1022. [DOI] [PubMed] [Google Scholar]
- Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. P Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yuan L, Rossiter SJ, Zuo X, Ru B, Zhong H, Han N, Jones G, Jepson PD, Zhang S. Adaptive evolution of 5'HoxD genes in the origin and diversification of the cetacean flipper. Mol Biol Evol. 2009;26:613–622. doi: 10.1093/molbev/msn282. [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Hogan CC, Nimali M. Polyglutamine expansion in Drosophila: Thermal stress and Hsp70 as selective agents. J Biosci. 2007;32:537–547. doi: 10.1007/s12038-007-0053-9. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Botas J. Mouse and fly models of neurodegeneration. Trends Genet. 2002;18:463–471. doi: 10.1016/S0168-9525(02)02729-4. [DOI] [PubMed] [Google Scholar]
- Driscoll M, Gerstbrein B. Dying for a cause: Invertebrate genetics takes on human neurodegeneration. Nat Rev Genet. 2003;4:181–194. doi: 10.1038/nrg1018. [DOI] [PubMed] [Google Scholar]
- King DG, Kashi Y. Mutation rate variation in eukaryotes: Evolutionary implications of site-specific mechanisms. Nat Rev Genet. 2007;8 doi: 10.1038/nrg2158-c1. [DOI] [Google Scholar]
- McConnell R, Middlemist S, Scala C, Strassmann JE, Queller DC. An unusually low microsatellite mutation rate in Dictyostelium discoideum. Genetics. 2007;177:1499–1507. doi: 10.1534/genetics.107.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambra R, Grimaldi B, Zamboni S, Filetici P, Macino G, Ballario P. Photomorphogenesis in the hypogeous fungus Tuber borchii: Isolation and characterization of Tbwc-1, the homologue of the blue-light photoreceptor of Neurospora crassa. Fungal Genet Biol. 2004;41:688–697. doi: 10.1016/j.fgb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Callewaert L, Christiaens V, Haelens A, Verrijdt G, Verhoeven G, Claessens F. Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem Biophys Res Commun. 2003;306:46–52. doi: 10.1016/S0006-291X(03)00902-1. [DOI] [PubMed] [Google Scholar]
- Huang H, Rambaldi I, Daniels E, Featherstone M. Expression of the Wdr9 gene and protein products during mouse development. Dev Dyn. 2003;227:608–614. doi: 10.1002/dvdy.10344. [DOI] [PubMed] [Google Scholar]
- Sheu YT, Zmuda JM, Cauley JA, Moffett SP, Rosen CJ, Ishwad C, Ferrell RE. Nuclear receptor coactivator-3 alleles are associated with serum bioavailable testosterone, insulin-like growth factor-1, and vertebral bone mass in men. J Clin Endocrin Metab. 2006;91:307–331. doi: 10.1210/jc.2005-0864. [DOI] [PubMed] [Google Scholar]
- Faux NG, Bottomley SP, Lesk AM, Irving JA, Morrison JR, de la Banda MC, Whisstock JC. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Ledda A, Toll-Riera M, Alba MM. Natural selection drives the accumulation of amino acid repeats in human proteins. Genome Research. 2010;20:745–754. doi: 10.1101/gr.101261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DG, Kashi Y. Indirect selection for mutability. Heredity. 2007;99:123–124. doi: 10.1038/sj.hdy.6800998. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS. In: Model Systems in Behavioral Ecology. Dugatkin L, editor. Princeton: Princeton University Press; 2001. Genetic consequences of sexual selection in stalk-eyed flies; pp. 72–91. [Google Scholar]
- Wilkinson GS, Dodson GN. In: The Evolution of Mating Systems in Insects and Arachnids. Choe J, Crespi B, editor. Cambridge: Cambridge University Press; 1997. Function and evolution of antlers and eye stalks in flies; pp. 310–328. full_text. [Google Scholar]
- Baker RH, Wilkinson GS. Phylogenetic analysis of eye stalk allometry and sexual dimorphism in stalk-eyed flies (Diopsidae) Evolution. 2001;55:3–32. doi: 10.1111/j.0014-3820.2001.tb00659.x. [DOI] [PubMed] [Google Scholar]
- Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RH, Morgan J, Wang X, Boore JL, Wilkinson GS. Genomic analysis of a sexually-selected character: EST sequencing and microarray analysis of eye-antennal imaginal discs in the stalk-eyed flies (Diopsidae) BMC Genomics. 2009;10:361. doi: 10.1186/1471-2164-10-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Baker R. A cladistic analysis of Diopsidae (Diptera) based on morphological and DNA sequence data. Insect Systematics & Evolution. 2002;33:325–336. [Google Scholar]
- Buschbeck EK, Roosevelt JL, Hoy RR. Eye stalks or no eye stalks: A structural comparison of pupal development in the stalk-eyed fly Cyrtodiopsis and in Drosophila. J Comp Neurol. 2001;433:486–498. doi: 10.1002/cne.1155. [DOI] [PubMed] [Google Scholar]
- Wright TF, Johns PM, Walters JR, Lerner AP, Swallow JG, Wilkinson GS. Microsatellite variation among divergent populations of stalk-eyed flies, genus Cyrtodiopsis. Genet Res. 2004;84:27–40. doi: 10.1017/S0016672304006986. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL. GeneMerge--post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Presgraves DC, Severance E, Wilkinson GS. Sex chromosome meiotic drive in stalk-eyed flies. Genetics. 1997;147:1169–1180. doi: 10.1093/genetics/147.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GS, Swallow JG, Christensen SJ, Madden K. Phylogeography of sex ratio and multiple mating in stalk-eyed flies from southeast Asia. Genetica. 2003;117:37–46. doi: 10.1023/A:1022360531703. [DOI] [PubMed] [Google Scholar]
- Johns PM, Wolfenbarger LL, Wilkinson GS. Genetic linkage between a sexually selected trait and X chromosome meiotic drive. P Roy Soc B-Biol Sci. 2005;272:2097–2103. doi: 10.1098/rspb.2005.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenbarger LL, Wilkinson GS. Sex-linked expression of a sexually selected trait in the stalk-eyed fly, Cyrtodiopsis dalmanni. Evolution. 2001;55:103–110. doi: 10.1111/j.0014-3820.2001.tb01276.x. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN. et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Salichs E, Ledda A, Mularoni L, Albà MM, de la Luna S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5:e10000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F, Roux J, Robinson-Rechav M. The expansion of amino-acid repeats is not associated to adaptive evolution in mammalian genes. BMC Genomics. 2009;10:619. doi: 10.1186/1471-2164-10-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwach P, Pophaly SD, Ganesh S. Genomic and evolutionary insights into genes encoding proteins with single amino acid repeats. Mol Biol Evol. 2006;23:1357–1369. doi: 10.1093/molbev/msk022. [DOI] [PubMed] [Google Scholar]
- Leggos D, Dalton A, Morrison PJ, DOdge A, Connart M, Kotze MJ, Rubinsztein DC. Analysis of spinocerebellar ataxia types 1, 2, 3, and 6, dentatorubral-pallidoluysian atrophy, and Friedreich's ataxia genes in spinocerebellar ataxia patients in the UK. J Med Genet. 1997;34:982–985. doi: 10.1136/jmg.34.12.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles RA, Young D, Weinrich S, Hudson J, Argyropoulos G, Ukoli F, Adams-Campell L, Dunston GM. Entent of linkage disequilbrium between the androgen receptor gene CAG and GGC repeats in human populations: Implications for prostate cancer risk. Hum Genet. 2001;109:253–261. doi: 10.1007/s004390100576. [DOI] [PubMed] [Google Scholar]
- Fujii K, Okiura T, Nishimura K, Orimoto C, Nishimukai K. Short tandem repeat typing in exon 1 of the androgen receptor gene. Leg Med (Tokyo) 2003;5:S201–S203. doi: 10.1016/s1344-6223(02)00111-6. [DOI] [PubMed] [Google Scholar]
- Short AD, Kennedy LJ, Barnes A, Fretwell N, Jones C, Thomson W, Ollier WER. Hardy-Weinberg expectations in canine breeds: Implications for genetic studies. J Hered. 2007;98:445–451. doi: 10.1093/jhered/esm020. [DOI] [PubMed] [Google Scholar]
- Gibbons JG, A R. Comparative and functional characterization of intragenic tandem repeats in 10 Aspergillus genomes. Mol Biol Evol. 2009;26:591–602. doi: 10.1093/molbev/msn277. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Hancock JM, Worthey EA, Santibanez-Koref MF. A role for selection in regulating the evolutionary emergence of disease-causing and other coding CAG repeats in humans and mice. Mol Biol Evol. 2001;18:1014–1023. doi: 10.1093/oxfordjournals.molbev.a003873. [DOI] [PubMed] [Google Scholar]
- Faux NG, Huttley GA, Mahmood K, Webb GI, de la Banda MG, Whisstock JC. RCPdb: An evolutionary classification and codon usage database for repeat-containing proteins. Genome Res. 2007;17:1118–1127. doi: 10.1101/gr.6255407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Golding GB. Genome-wide evidence for selection acting on single amino acid repeats. Genome Research. 2010;20:755–760. doi: 10.1101/gr.101246.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates DK, Wiegmann BM. In: The Evolutionary Biology of Flies. Yeates DK, Wiegmann BM, editor. New York: Columbia University Press; 2005. Phylogeny and evolution of Diptera: Recent insights and new perspectives; pp. 14–44. [Google Scholar]
- Macdonald SJ, Long AD. Identifying signatures of selection at the enhancer of split neurogenic gene complex in Drosophila. Mol Biol Evol. 2005;22:607–619. doi: 10.1093/molbev/msi046. [DOI] [PubMed] [Google Scholar]
- Mueller D, Kugler SJ, Preiss A, Maier D, Nagel AC. Genetic modifier screens on hairless gain-of function phenotypes reveal genes involved in cell differentiation, cell growth and apoptosis in Drosophila melanogaster. Genetics. 2005;171:1137–1152. doi: 10.1534/genetics.105.044453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodjabachian L, Delaage M, Maurel C, Miassod R, Jacq B, Rosset R. Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. EMBO. 1998;17:1063–1075. doi: 10.1093/emboj/17.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A, Higuet D, Rosset R, Deutsch J, Peronnet F. Corto genetically interacts with Pc-G and trx-G genes and maintains the anterior boundary of ultrabithorax expression in Drosophila larvae. Mol Genet Genomics. 2001;266:572–583. doi: 10.1007/s004380100572. [DOI] [PubMed] [Google Scholar]
- Salvaing J, Lopez A, Boivin A, Deutsch JS, Peronnet F. The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. Nucleic Acids Res. 2003;31:2873–2882. doi: 10.1093/nar/gkg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikaku MA, O'Tousa JE. Sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M. Ancient mechanisms of visual sense organ development based on comparison of the gene networks controlling larval eye, ocellus, and compound eye specification in Drosophila. Arthropod Struct Dev. 2006;35:357–378. doi: 10.1016/j.asd.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Zeitouni B, Senatore S, Severac D, Aknin C, Semeriva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS Genet. 2007;3:e174. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Freeman MR, Carlson JR, Bender M, Cooley L, Segraves WA. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 1999;126:4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- Bernardo TJ, Dubrovskaya VA, Jannat H, Maughan B, Dubrovsky EB. Hormonal regulation of the E75 gene in Drosophila: Identifying functional regulatory elements through computational and biological analysis. J Mol Biol. 2009;387:794–808. doi: 10.1016/j.jmb.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS. Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrin Met. 2000;11:276–280. doi: 10.1016/S1043-2760(00)00282-4. [DOI] [PubMed] [Google Scholar]
- Champlin DT, Truman JW. Ecdysteroids govern two phases of eye development during metamorphosis of the moth, Manduca sexta. Development. 1998;125:2009–2018. doi: 10.1242/dev.125.11.2009. [DOI] [PubMed] [Google Scholar]
- Abrell S, Carrera P, Jackle H. A modifier screen of ectopic kruppel activity identifies autosomal Drosophila chromosomal sites and genes required for normal eye development. Chromosoma. 2000;109:334–342. doi: 10.1007/s004120000092. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. Decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Carrera P, Moshkin YM, Gronke S, Sillje HH, Nigg EA, Jackle H, Karch F. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev. 2003;17:2578–2590. doi: 10.1101/gad.276703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-H, Chiang C-S, Huang H-Y, Liaw G-J. Mars and tousled-like kinase act in parallel to ensure chromosome fidelity in Drosophila. J Biomed Sci. 2009;16:51–64. doi: 10.1186/1423-0127-16-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom S, Don J. Tlk, a novel evolutionarily conserved murine serine threonine kinase, encodes multiple testis transcripts. Mol Reprod Dev. 1999;52:392–405. doi: 10.1002/(SICI)1098-2795(199904)52:4<392::AID-MRD8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW. et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–-987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair. 2009;8:87–102. doi: 10.1016/j.dnarep.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Han Z, Saam J, Adams H, Mango S, Schumacher J. The C. elegans tousled-like kinase (TLK-1) has an essential role in transcription. Curr Biol. 2003;13:1921–1929. doi: 10.1016/j.cub.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Han Z, Reifler GM, Saam SE, Mango SE, Schumacher JM. The C. elegans tousled-like kinase contributes to chromosome segregation as a substrate and regulator of the aurora B kinase. Curr Biol. 2005;15:894–904. doi: 10.1016/j.cub.2005.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie CT, Golden A. Chromosome segregation: Aurora B gets tousled. Curr Biol. 2005;15:R379–R382. doi: 10.1016/j.cub.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Lyttle TW. Cheaters sometimes prosper: Distortion of mendelian segregation by meiotic drive. Trends Genet. 1993;9:205–208. doi: 10.1016/0168-9525(93)90120-7. [DOI] [PubMed] [Google Scholar]
- Jaenike J. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 2001;32:25–49. doi: 10.1146/annurev.ecolsys.32.081501.113958. [DOI] [Google Scholar]
- Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Boston: Harvard University Press; 2006. [Google Scholar]
- Wilkinson GS, Sanchez MI. Sperm development, age and sex chromosome meiotic drive in the stalk-eyed fly, Cyrtodiopsis whitei. Heredity. 2001;87:17–24. doi: 10.1046/j.1365-2540.2001.00898.x. [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: Structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Hammock EAD, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/S0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King DG. Has simple sequence repeat mutability been selected to facilitate evolution? Isr J Ecol Evol. 2006;52:331–342. doi: 10.1560/IJEE_52_3-4_331. [DOI] [Google Scholar]
- Beldade P, Vant Hof AE, Jeronimo MA, Long AD. Microsatellite markers associated with genes expressed in developing wings of Bicyclus anynana butterflies. Mol Ecol Resour. 2009;9:1487–1497. doi: 10.1111/j.1755-0998.2009.02691.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. S K, S M, editor. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3. Woodbury: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences and annealing temperatures for each locus. This table contains the forward and reverse primers and annealing temperatures that were used in polymerase chain reactions. Product size range is also provided.


