Abstract
The study of CpG methylation of genomic DNA in neurons has emerged from the shadow of cancer biology into a fundamental investigation of neuronal physiology. This advance began with the discovery that catalytic and receptor proteins related to the insertion and recognition of this chemical mark are robustly expressed in neurons. At the smallest scale of analysis is the methylation of a single cytosine base within a regulatory cognate sequence. This singular alteration in a nucleotide can profoundly modify transcription factor binding with a consequent effect on the primary ‘transcript'. At the single promoter level, the methylation–demethylation of CpG islands and associated alterations in local chromatin assemblies creates a type of cellular ‘memory' capable of long-term regulation of transcription particularly in stages of brain development, differentiation, and maturation. Finally, at the genome-wide scale, methylation studies from post-mortem brains suggest that CpG methylation may serve to cap the genome into active and inactive territories introducing a ‘masking' function. This may facilitate rapid DNA–protein interactions by ambient transcriptional proteins onto actively networked gene promoters. Beyond this broad portrayal, there are vast gaps in our understanding of the pathway between neuronal activity and CpG methylation. These include the regulation in post-mitotic neurons of the executor proteins, such as the DNA methyltransferases, the elusive and putative demethylases, and the interactions with histone modifying enzymes.
Keywords: CpG, methylation, demethylase, epigenetics, schizophrenia, bipolar disorder
INTRODUCTION
Methylation of the 5-position of cytosines within CpG dinucleotides represents the most common covalent modification of mammalian genomic DNA. Methylated cytosine is often considered to be the fifth letter of the DNA code. This modification is thermodynamically very stable with no known enzyme capable of its direct removal. In post-mitotic neurons, methylation of the cytosine base is conceivably the longest lasting biochemical event in the body, as neuronal DNA, in most locations, will not be recycled or replicated. This can be shown in experiments in which a methyl group inserted into a hippocampal gene promoter during the first postnatal week persists unchanged into adulthood (Weaver et al, 2004). These properties are not trivial, because the brain contains higher levels of methylated cytosines compared with other organs especially in repetitive sequences that are separate from CpG islands (Ehrlich et al, 1982; Gama-Sosa et al, 1983; Wilson et al, 1987; Tawa et al, 1990; Ono et al, 1993). From an anthropological perspective, the brain manifests the greatest divergence in CpG methylation when comparing human and chimpanzee organs (fewer differences in other tissue types such as lymphocytes or liver; Enard et al (2004)).
CpG DINUCLEOTIDES AND CpG ISLANDS
In light of this modification, the CpG dinucleotide represents a singular unit of genomic architecture. Although the distribution pattern of CpG dinucleotides has not been fully decoded, they are predominantly located in a variety of repetitive and retro-viral elements in the vast intergenic regions of the genome at which locations they are invariably ‘bulk' methylated.
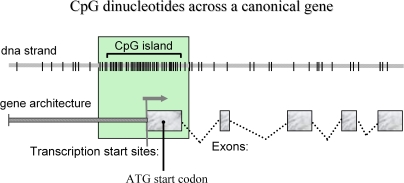
Outside repetitive elements and especially in the proximity of transcribed genes, a sizable fraction of these dinucleotides are segregated into clusters called CpG islands (Figure 1). CpG islands are noteworthy for the following reasons. First, CpG islands are predominantly hypo-methylated in all tissues at all stages of development. This differs from CpG dinucleotides located in repetitive/retroviral elements, which are 70–80% methylated. Second, 60–75% of mammalian gene promoters reside within CpG islands (Bird, 2002) in which informative methylation may occur at single CpG sites particularly within cognate sequences (single sites are recognized by the canonical methylated DNA-binding protein, MeCP2). Indeed, neuronally expressed promoters seem to more frequently reside in CpG islands (Gardiner and Frommer, 1994). Third, 50% of CpG islands are located outside of gene coding regions. Some of these islands are suspected to represent start sites for miRNA transcription (Illingworth et al, 2008).
Figure 1.
Genomic and methylomic architecture: the top helical line depicts a DNA strand of unspecified sequence that spans the promoter and upstream exons of a hypothetical gene. CpG dinucleotides are presented as short vertical bars. This assembly is superimposed on the canonical structure of the gene to illustrate spatial relationships of the genomic and methylomic maps. The cardinal element of methylomic architecture is the CpG island (ie, a region of CpG density >∼50–60%). CpG islands straddle the distal promoter region, the transcription start site, the translation start codon, and the first exon even spilling into the first intron.
In the brain, the methylation of CpG dinucleotides (mCpG) varies significantly across neuroanatomical regions (Ladd-Acosta et al, 2007). The brain also shows variability in mCpG levels across the life span, with an observed loss of overall methylation with aging (Wilson et al, 1987) but both increases and decreases at the level of individual gene promoters (Siegmund et al, 2007). This bidirectional variability across the lifespan is a simple illustration of the independence of mCpG levels relative to the minor rates of cell-division or DNA synthesis in the post-mitotic brain.
CpG METHYLATION AND GENE TRANSCRIPTION
Regulating the Message
Over the long term, CpG methylation seems uniquely adapted to maintain the permanent silencing of transcriptionally inactive promoters. Although gene activity is commonly associated with a hypo-methylated promoter, the relationship of promoter–CpG island methylation to transcription is not linear. For example, in suppressing retroviral sequences, CpG methylation occurs long after mRNA expression levels have already declined (Gautsch and Wilson, 1983). In contrast, transcriptionally active promoters seem to be resistant to CpG methylation, and while the RNA polymerase complex can bind to a fully methylated promoter, the act of transcription itself seems to initiate the demethylation process and immunizes the transcribed promoter from future re-methylation. In other words, transcription initiated demethylation creates a type of cellular memory that recalls the earlier transcription event (D'Alessio et al, 2007).
A key question regarding correlations between CpG methylation and transcription relates to the mechanism by which the methyl moiety affects gene transcription. CpG methylation seems to repel some proteins (obstruction model) and attract others (binding model). The ‘obstruction' model postulates that the presence of a methyl group at the 5′ position of cytosine in CpG dinucleotides interferes with the binding of transcription factors or the assembly of the RNA polymerase II complex. Obstruction to transcription factors by methylated CpG sites can be shown using Gel-shift assays and individual recombinant proteins in a stripped down in vitro demonstration that precludes more complex protein assemblies or interactions (Takizawa et al, 2001). Although this strongly implicates an obstructive component, experiments undertaken over 20 years ago are also consistent with the alternate model, that is, binding of restrictive proteins (Buschhausen et al, 1985; Buschhausen et al, 1987). In these papers, the investigators report on the microinjection of fully methylated Herpes Simplex thymidine kinase DNA into rat 2 TK− cells. The results have shown that for at least 48 h after microinjection the fully methylated and nonmethylated promoters were equally active in supporting thymidine kinase activity (Buschhausen et al, 1985). Moreover, re-extraction and blot analysis of the episomal DNA showed that DNA demethylation was not responsible for this activity. In a follow-up study, Buschhausen et al (1987) showed that exogenously introduced methylated DNA undergoes a transition from a naked template to one that is chromatin bound. The investigators conclude that it is the binding of various repressor proteins to the methylated CpG dinucleotides that is responsible for the time-dependent reduction in transcription. More recently, Kundakovic et al (2009) in our laboratory showed a similar result using the GAD67 promoter to drive CpG-free luciferase reporter activity in neural progenitor cells. These investigators have shown that the fully methylated promoter was expressed at the same low rate as the unmethylated promoter up until approximately 12 h after transfection. They also showed that the histone deacetylase (HDAC) inhibitor, MS-275, reversed this difference and restored robust activity to the methylated template. Presumably MS-275 facilitated the release of repressor complexes from the promoter (Kundakovic et al, 2009). Moreover, they provide ChIP evidence that at least HDAC1 may be a part of this complex. This is consistent with the known target specificity of MS-275, which includes both HDACs 1 and 2. These studies support the notion that repressor proteins are uniquely attracted to methylated CpGs (see below for MBD domain proteins) which in turn recruit HDAC enzymes to create a restrictive chromatin encasement.
DNA METHYLTRANSFERASES: INSERTING THE MARK
Regulation of Expression
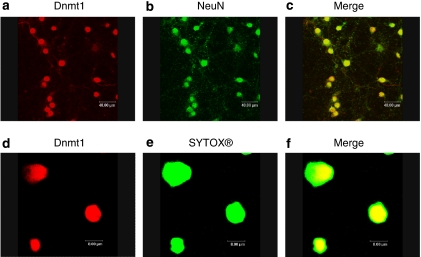
Early demonstration of an abundant expression of DNA methytransferases (Figure 2; Sharma et al, 2008) as well as vigorous demethylation activity (Brooks et al, 1996) in post-mitotic neurons, raised questions regarding the role of this methylation–demethylation enzymatic activity in cells in which DNA replication is largely irrelevant (Goto et al, 1994; Feng et al, 2005). The attachment of a methyl group to cytosine is catalyzed by two types of DNA methyltransferase activity. The ‘de novo' methyltransferases, DNMT3a and DNMT3b, catalyze the attachment of a methyl group to a previously unmethylated cytosine, essentially enhancing the informational content of the DNA strand. The ‘copying' of this modification to subsequent replicates of the DNA strand is executed by the ‘maintenance' methyltransferase DNMT1.
Figure 2.
Top panel: Dnmt1 is localized to primary neurons. Confocal laser scanning microscopy of primary cortical cultures (DIV 10) fixed with 4% paraformaldeyhde and stained with rabbit polyclonal antibody DNMT1 antibody and mouse monocloncal neuronal nuclear marker antibody (NeuN) DNMT1 appears in red (Cy5 flurophore) and NeuN appears in green (Cy2 flurophore). Percentage of colocalization was 88% ±3 for DNMT1/NeuN. ((a) DNMT1 Sigma D4567, 1: 250; (b) NeuN; Chemicon MAB677, 1: 500, (c) is merge). Bottom panel: DNMT1 colocalizes with double-stranded DNA; (d) is as above; (e) is performed by labeling double-stranded DNA with 1 μM of SYTOX. The merged images are presented in (f) (Sharma et al, 2008).
Heretofore, the identification of cis-regulatory sequences in putative promoter regions of the DNMT1 and DNMT3 genes have, without exception, been examined in dividing cells with replicating DNA such as from cancer or transformed cell lines (DNMT1; Rouleau et al, 1992; Bigey et al, 2000: DNMT3a and DNMT3b; Yanagisawa et al, 2002; Jinawath et al, 2005). Not surprisingly, there is evidence for regulation by the jun-ras-AP1 transcription factors, the development-related pathways identified by their role in cellular transformation and oncogenic programs (eg, AP1, E2F, Rb, and APC). More general transcription factors, such as Sp1 and Sp3, are also implicated in the regulation of DNMT promoters (Jinawath et al, 2005; Kimura et al, 2003; Kishikawa et al, 2002).
There are no studies deconstructing the promoter region of any of the DNMT genes in post-mitotic neurons. Nonetheless, there is clear indication of active and non-constitutive regulation of expression of these enzymes in the brain, across the lifespan and possibly across neuroanatomical regions as well. For example, there is evidence for a robust expression in neurons and perhaps oligodendrocytes, but not in astroglia (Feng et al, 2005; Veldic et al, 2004). In the adult human post-mortem brain, both DNMT1 and DNMT3a are differentially expressed across the layers of the cortex and appear specifically in GABAergic interneurons and less so in pyramidal cells (Veldic et al, 2004, 2005; Siegmund et al, 2007). In the brain, Dnmt1 is expressed from embryogenesis to adulthood. Dnmt3b is the earliest de novo enzyme to be expressed in neural progenitor tissue during early embryogenesis, but it declines as Dnmt3a expression rises (Watanabe et al, 2006; Feng et al, 2005). The expression of Dnmt3a is observed from late embryogenesis (E10) to adulthood with a peak during the early postnatal period; its expression declines to lower but detectable levels in the adult brain (Feng et al, 2005) with a reported biphasic increase in older humans (Siegmund et al, 2007). This nonconstitutive and regulated expression is supported by the observation that DNMT1 seems more highly expressed in the ‘interneurons' of the cortex (GABA interneurons in the cerebral cortex, and granular cells in the cerebellar cortex) than the ‘output' neurons, that is, pyramidal or Purkinje cells respectively (Veldic et al, 2004).
Interestingly, several studies have reported higher DNMT1 mRNA expression in schizophrenia post-mortem cortical and striatal GABAergic neurons (Veldic et al, 2004, 2005). DNMT1 expression was negatively correlated with schizophrenia candidate gene expression. DNMT3a expression is also found to be elevated in similar brain regions as DNMT1 (Zhubi et al, 2009). These findings suggest that schizophrenia may be characterized by aberrant DNA methylation, as well as perhaps a deficit in coordinating epigenetic processes in general (Gavin and Sharma, 2010). Complex alterations of the DNA methyltransferases are also reported in post-mortem brains obtained from completed suicides (Poulter et al, 2008).
Knockouts and Knockdowns of Neuronal Dnmts
Cre/loxP technology can be used to produce conditional Dnmt1 null mutants that lack exons 4 and 5 of the corresponding primary transcript (Fan et al, 2001). This deletion is engineered by cre-recombinase in either neuronal precursors (cre-recombinase under control of the Nestin or Emx1 promoter; Fan et al, 2001; Hutnick et al, 2009; Golshani et al, 2005), or in post-mitotic cortical neurons (cre-recombinase under control of the calmodulin-kinase-2a promoter (Fan et al, 2001)). Deletion of Dnmt1 in fully differentiated post-mitotic neurons, whether in vivo or in vitro, does not change DNA methylation levels, even in those neurons that have a disabled Dnmt1 gene for up to 17 months. In contrast, Dnmt1 deletion in mitotic neuronal precursors will result in hypomethylation of the succeeding neuronal progeny. These hypo-methylated neuronal progeny reveal for the first time, a profound relationship between abnormal CpG methylation and poor neuronal function and survival (Fan et al, 2001).
As an extension of this approach, a conditional Dnmt1 gene deletion limited to only the neuronal precursors of the excitatory cells of the dorsal forebrain was generated (using a Emx1-cre transgene). The resulting hypomethylation of the descendant neurons, which form the cortex of the dorsal forebrain, results in abnormal synaptic connections with incoming thalamocortical projections as measured by aberrations in several electrophysiological parameters (Golshani et al, 2005; Hutnick et al, 2009). In addition, these excitatory forebrain neurons show deficits in dendritic branching and action potential repolarization (Golshani et al, 2005). Although mutant mice are viable into adulthood, a progressive apoptotic-like degeneration of dorsal forebrain neurons develops with maturation. Additional investigations show that these phenotypes are most likely the result of hypo-methylation-mediated deregulation of genes involved in neuronal lamination and ion channel function (Hutnick et al, 2009). Curiously, in these studies, a small population of hypo-methylated neurons survived throughout the lifespan of the rodent allowing for an examination of gene expression in neurons chronically experiencing CpG hypo-methylation (Hutnick et al, 2009). Hypomethylation resulted in an upregulation of 1047 genes (6.1%) and a downregulation of 444 genes (2.6%).
Similarly, a cre-lox conditional knockout of Dnmt3a has been constructed, using a Nestin-cre transgene to drive recombination and deletion in neuronal precursors by E15 (Nguyen et al, 2007). Dnmt3a-nestin knockouts are relatively normal until 2 months of age when they begin to manifest neuromuscular abnormalities and poor survival. Neuronal loss in the hypoglossal nerve and molecular pathology of the neuromuscular endplates occurs as well (Nguyen et al, 2007, compare with Dnmt3a-Camk2a knockout in post-mitotic neurons). This conditional Dnmt3a knockout revealed no effect on promoters methylated before inactivation of Dnmt3a, since the maintenance of previous methylation patterns is the function of Dnmt1. However, in promoters experiencing active methylation–demethylation such as the Gfap promoter (glial fibrillary acidic protein specific to astrocytes), Dnmt3a participates in the remethylation phase designed to shut down the Gfap promoter in mature neurons (Nguyen et al, 2007).
A double knockout (DKO) of both Dnmt1 and Dnmt3a helps to resolve the question of redundancy between these two neuronally active Dnmts (Feng et al, 2010). Using the Camk2a-Cre transgene, Dnmt1 and Dnmt3a single (SKO) and DKOs engineered in post-mitotic neurons verify some earlier findings. SKO seem surprisingly normal in terms of brain volume, neuronal number and size, electrophysiology and behavior conditioning. In contrast, DKO manifest a series of abnormalities such as smaller hippocampal neurons (but no neuronal loss) and abnormal synaptic plasticity. Furthermore, mature neurons showed a significant loss of CpG methylation (genome-wide ∼20% as well as promoter specific) and deregulated gene expression.
It would seem then, that a mature neuron (not a neuronal precursor) that has achieved its full complement of adult methylation is resistant to losing CpG methylation, and is relatively impervious to depletions of either Dnmt1 or Dnmt3a enzyme. Indeed, a fully methylated neuron is quite viable even when it lacks Dnmt1 protein for months into adulthood (Fan et al, 2001). The depletion of both enzymes, however, has a demonstrable effect on CpG methylation and gene expression, indicating an ‘active maintenance' of this modification in the adult CNS that requires either Dnmt1 or Dnmt3a. This functional redundancy or compensation also suggests that either enzyme could perform a ‘de novo' or ‘maintenance' activity in post-mitotic cells. This was noted by Kundakovic et al (2009) who knocked-down DNMT function using siRNAs specific to each isoform. When NT2 cells were treated with an siRNA specific to DNMT1, both DNMT3a and DNMT3b were overexpressed. Similarly, DNMT3a siRNA facilitated an increase in DNMT1 and 3b protein levels. The triple DNMT1, 3a and 3b knockdown failed to decrease DNMT3a significantly from non-treated levels. Although these investigators did not examine methylation levels after knock-down, none of the siRNA treatments facilitated an increase in the RELN mRNA under study (Kundakovic et al, 2009).
The methylation of the Gfap promoter illustrates a major role of CpG methylation in the brain. Brain cells such as neurons, astrocytes, and oligodendrocytes are sequential offspring of neuronal precursor cells, and lineage specification is by a CpG methylation switch mechanism from the neurogenic pathway to the gliogenic pathway at specified times during development. Takizawa et al (2001) have shown that a single CpG site in the Gfap promoter undergoes demethylation during this switch to an astrogliogenic differentiation. Accelerated hypo-methylation of neuronal precursor cells induced by Dnmt1 knockouts results in the precocious activation of astrocyte markers (such as Gfap and S100β) and a switch from a neuronal toward an astrocyte lineage (Fan et al, 2005). Specification of lineage along the path to brain maturation can therefore be assumed to be secured in part by long-term CpG promoter methylation. An unanswered question is whether CpG methylation is also responsible for the maintenance of cellular phenotype in long-living neurons and the prevention of phenotypic ‘drift' (Laywell et al, 2005).
We have shown the effects of an in vitro Dnmt1 antisense knockdown in cortical neuron cultures on promoter methylation and mRNA expression of epigenetically regulated genes such as Reln (Noh et al, 2005). Specifically, an increase or decrease in levels of promoter methylation can be induced, using either pharmacological doses of methionine (increasing methylation) or by knocking down expression of Dnmt1 protein (decreasing methylation). The resultant increase or decrease in promoter methylation is associated with decreased or increased Reln mRNA expression, respectively.
METHYL-DNA-BINDING PROTEINS: READING THE MARK
Two fundamental protein families are known to recognize the mCpG modification. The earliest discovered group included proteins containing a ‘methyl-CpG-binding domain' or MBD of which MeCP2 was the founding protein (MeCP2, MBD 1 to 4). MeCP2 has an affinity for a single mCpG that is higher by an order of magnitude (compared with unmethylated CpG), and is expressed at a significantly higher level in the brain compared with other tissues (Nan et al, 1993). Indeed, MeCP2 seems to function as a transcriptional regulator primarily in neurons (LaSalle, 2004). In general, the increase in affinity of the MBD domain from the unmethylated to the methylated CpG varies depending on the sequence context, but can differ by over a 100-fold suggesting a great avidity to the interpretation of the methyl mark (Fraga et al, 2003). More recently, another family of proteins that uses a zinc-finger domain to bind mCpG has been reported and is represented by the protein Kaiso (Bird, 2008). Members of both families such as MeCP2 (MBD containing) and Kaiso (zinc-finger) are robustly and constitutively expressed in the adult brain (Ragione et al, 2006). It is not coincidental that both the primary ‘writer' and ‘reader' of the CpG methylation mark (eg, DNMT1 and MeCP2) are distinctly expressed in brain.
Misinterpretation of the methyl mark by these proteins, most notably by MeCP2, delays neuronal differentiation from their stem cell parentage resulting in abnormal neuronal maturation in the postnatal period. Consequently, MeCP2-null mice have profound neurological impairments (Caballero et al, 2009), and in humans, mutations in MeCP2 are responsible for the neurodevelopmental disorder Rett syndrome (Chahrour and Zoghbi, 2007 for review). MeCP2 null- neurons themselves are viable for an extended period, and the effects of mutation can be reversed using transgenic intervention (Guy et al, 2007; Jugloff et al, 2008). Surprisingly and in light of the generally repressive role of CpG methylation, attachment of MeCP2 to a gene promoter more commonly (and non-randomly) results in gene activation rather than repression (Chahrour et al, 2008). Mbd1-null mice show observable disturbances in neurogenesis, long-term potentiation and cognition (Zhao et al, 2003). In contrast, Mbd2-null and Kaiso-null mice do not seem to manifest an overtly pathological neurological phenotype.
DEMETHYLASES: REMOVING THE MARK
If CpG methylation is an attribute of a primary top-level regulator of gene expression, then it is necessary to show a facile removal of the methyl mark in real signaling time (Smith, 2000). The actual mechanism of demethylation has been the subject of much debate (Ooi and Bestor, 2008). There are two reasonable possibilities regarding the active demethylation of post-mitotic mammalian neuronal DNA. The first entails direct removal of the methyl group from 5-methylcytosine; the other suggests removal of the entire methylated cytosine base from its sugar-phosphate backbone.
The first and conceptually simplest mechanism is the direct removal of only the methyl moiety (requiring a unique ‘demethylase'). This would not disrupt the cytosine base or DNA strand, and is intuitively appealing for neuronal cellular signaling given its precision and economy. However, the elusive nature of this unidentified ‘demethylase' has prompted opinion that such an enzyme cannot be a prominent feature of mammalian cells, mitotic or post-mitotic, and demethylation must occur principally and passively in cells replicating or recycling their DNA sequences (Ooi and Bestor 2008). These reservations are also based partly on the improbable kinetics and thermodynamics of releasing a methyl group from a C–C bond. These reservations notwithstanding, demethylation of non-replicating genomes is clearly demonstrable in numerous physiological contexts such as the massive demethylation of the paternal genome (pronucleus) before the first cell division of the zygote (Mayer et al, 2000). This conundrum points to the second option, which is the removal and replacement of the methylated cytosine base in a two step deamination/base-excision mechanism engineered sequentially by a deaminase followed by a glycosylase (a methylated cytosine is deaminated to a thymine, thus allowing it to be targeted by a thymine DNA glycosylase). Recent work by Métivier et al (2008) and Kangaspeska et al (2008) reveals a methylating–demethylating cycle of periodicity in the range of 30–50 min in mitotic cells. Another study found demethylation can occur between 4 and 120 h in post-mitotic neurons (Ma et al, 2009). Demethylation in these studies is characterized by site specificity (ie, to an individual cytosine address) and is coordinated with proteins involved in the removal of methylated cytosine in the aforementioned two step enzymatic process of deamination/base-excision. In the post-mitotic neuron, mechanisms such as deamination/base-excision have long been reported, but the complexities and multistep nature of this reaction were considered inefficient to serve a role in rapid cellular signaling (Brooks et al, 1996). Additional neuronal studies manipulating DNMT enzymes with knockdowns and pharmacological inhibitors provide further support for active promoter demethylation with consequential downstream changes in gene expression (Nelson et al, 2008; Noh et al, 2005; Sharma et al, 2008). Currently, this accumulating evidence of active demethylation of CpG dinucleotides as a direct consequence of neuronal activity and cellular signaling in post-mitotic neurons (Chen et al, 2003; Martinowich et al, 2003; Miller and Sweatt, 2007; Ma et al, 2009) has yet to be reconciled with a specific enzymatic mechanism.
Recently, an alternate pathway for the reconstitution of cytosine from methylcytosine was shown in neurons, namely the conversion of the methyl group to a hydroxymethyl group. Similar to the removal of a methyl group in sterol metabolism, the hydroxymethyl moiety is liable to be expelled as formic acid (Shyadehi et al, 1996). Hydroxymethylcytosine was first detected some 35 years ago and even then was suspected to be higher in the brain than in other tissues such as the liver (Penn et al, 1972). Levels were noted to increase in the brain with development into adulthood (Penn, 1976). Conversion of the hydrophobic methyl group to the hydrophilic hydroxymethyl group on cytosine has also been shown to reduce affinity of the 5-methylcytosine-binding transcription factor MeCP2 by an order of magnitude (Valinluck et al, 2004). Indeed, oxidation of 5-methyl cytosine to 5-hydroxymethyl cytosine is the equivalent of demethylating the cytosine as far as MeCP2 affinity is concerned (Valinluck et al, 2004). Mammalian glycosylases capable of removing 5-hydroxymethyl cytosine have been identified (Tahiliani et al, 2009). A parallel report finds an elevated presence of hydroxymethyl cytosine in Purkinje neurons (Kriaucionis and Heintz, 2009).
CROSS-TALK BETWEEN DNA METHYLATION AND CHROMATIN-RELATED PROTEINS
There is substantial cross-talk between the underlying DNA methylation status and the local histone environment (closed vs open chromatin assemblies). The ability of ‘open' chromatin states to facilitate promoter demethylation also depends on transcriptional activity. The interplay between DNA methylation and histone modifications is illustrated by the complex interaction between DNA methytransferase enzymes and histone-modifiying enzymes (HDACs and HMTs; Rountree et al, 2000). For example, both DNMT1 and DNMT3a directly bind the SUV39H1 histone methyltransferase and also bind HP1 protein, the former is responsible for methylation at histone 3, lysine 9 (H3K9) and the latter serves as a high-affinity adapter to the methylated H3K9 residue (Fuks et al, 2003; Li et al, 2007). Histone acetylation with resultant upregulation of gene transcription precedes demethylation of a given promoter, suggesting that in certain types of CpG islands, transcription is the required event before the promoter sequence is demethylated (D'Alessio et al, 2007; Schmitz et al, 2009; Barreto et al, 2007). Indeed, RNAPII can bind to originally methylated DNA promoters and initiate transcription before the promoter becomes demethylated (D'Alessio et al, 2007). This relationship between histone acetylation and DNA demethylation could suggest that open chromatin makes the DNA strand accessible to a demethylating protein(s) with immediate therapeutic consequences because HDAC inhibitors are emerging in the clinical pharmacopeia. Conversely, drugs that promote the acetylation of histones (eg, HDAC inhibitors) tend to facilitate the opening of local chromatin by mediating the release of repressive methyl CpG-binding proteins (Wade et al, 1999).
The ability of the methylation machinery to target unique DNA sequences is suggested by null-mutants of other chromatin remodeling proteins, ATRX (humans) and Lsh2 proteins (mice). In animals deficient in these proteins, dramatic demethylation is limited to regions of repetitive DNA sequences (Gibbons et al, 2000; Dennis et al, 2001). Also restrictive chromatin marks such as methylated H3K9 focus CpG methylation to promoters that are silent as shown in the fungus Neurospora (Tamaru and Selker, 2001).
Recently, Miller et al (2008) showed that blocking DNMT activity in the rat hippocampus blocks behavioral memory formation. This is interesting because the DNMT-inhibitor block in memory consolidation can be rescued by inhibitors of histone deacetylation (Miller et al, 2008). This shows that there are functional consequences to the link between DNA methylation and histone acetylation. It has been suggested that HDAC inhibitors such as valproic acid and MS-275 facilitate increases in mRNA expression by increasing DNA demethylation (Kundakovic et al, 2009; Guidotti et al, 2009). In the context of schizophrenia, it was recently shown that valproic acid when co-administered with either clozapine or sulpiride facilitated chromatin remodeling through a mechanism associated with promoter demethylation (Guidotti et al, 2009).
PROMOTER REGULATION BY BULK VS SINGLE CpG METHYLATION
A distinction can be made, between the density of methylation (bulk) across a cluster of CpG dinucleotides spanning an entire repetitive element or some part of a CpG island within a promoter, vs methylation at a single CpG dinucleotide. Bulk methylation will facilitate the aggregation of repressive proteins (such as combinations of DNMTs, HDACs, and other chromatin remodeling proteins as noted above) which in turn will sequester the promoter from transcription factors. This type of repression, that is, by the overall density of methylation in the region, is largely insensitive to any uniqueness of the underlying DNA sequence and in some instances will even spill over and repress neighboring promoters. By contrast, the precise nature of the CpG methylation event is best illustrated by methylation switches on single cytosine addresses within the cognate sequence of a transcription factor. In fact, individual methylated CpG-binding proteins seem to have differences in their preference for the number, density, and distribution of methylated CpG sites (Fraga et al, 2003). For instance, MeCP2 binds strongest to a single CpG site (Nan et al, 1993) while Kaiso binds to two sequential CpG sites with a variable third site downstream (Prokhorthcouk et al, 2001), and MBD2 prefers a cluster of upto 12 methylated CpG sites (Lewis et al, 1992).
Methylation at single CpG sites within the CRE (cAMP responsive element) located in a specific Bdnf exon-promoter, or within the promoter of the rRNA gene, will shut down these genes (Iguchi-Ariga and Schaffner, 1989; Chen et al, 2003; Martinowich et al, 2003; Santoro and Grummt, 2005). Conversely, demethylation of a single CpG site in the Gfap promoter will activate gene expression and differentiation of the neuronal precursor into an astrocyte (Takizawa et al, 2001). A landmark study has shown a highly significant regulatory methylation event at a single CpG site in both the glucocorticoid and estrogen receptor gene promoters in the hippocampus as a result of behavioral stimuli, specifically maternal grooming (Weaver et al, 2004; Champagne et al, 2006). Also remarkable is the demonstration of de novo brain methylation within 1 postnatal day (Weaver et al, 2004). These results are striking given the precision and specificity of the modification. The methylation–demethylation switch at a single CpG site is induced by the behavioral paradigm and coincides with changes in mRNA expression. As a demonstration of the specificity of this mark, a second CpG dinucleotide located 5-bp upstream remains unchanged and serves as a local negative control. Further, the methylation event and its effects on transcription occur during the first few days of postnatal life, but persist into adulthood as an example of long-term molecular memory.
The distinction between bulk vs single CpG dinucleotide methylation has implications for genome-wide post-mortem brain studies using CpG immunoprecipitation approaches.
CpG METHYLATION IS MODIFIED BY NEURONAL ACTIVITY
Very little is known regarding the operational characteristics or kinetics of CpG methylation or methyltransferase catalytic activity in neurons. The expression of the DNA methyltransferases is not monotonic and can be modified by external stimuli. Dnmt1 and Dnmt3a mRNA levels are reduced by membrane depolarization, which also induces a decrease in DNA methyltransferase catalytic activity (Sharma et al, 2008). Dnmt1 is increased several fold after mild non-necrotic ischemic stress (Endres et al, 2000). Curiously, a suppression of DNA methylation in ischemic cortical tissue is associated with enhanced survival (Endres et al, 2000). Dnmt3a and Dnmt3b mRNA levels are also increased in the CA1 region of the hippocampus within 1 h of contextual fear conditioning in a rodent behavior model (Miller and Sweatt, 2007). This upregulation of Dnmt3 enzymes is associated with an increase in the promoter methylation of a memory suppressing gene suggesting regulation of neuronal gene expression by CpG methylation in real time.
Changes in CpG methylation in response to membrane depolarization, the sine qua non of neuronal functioning, are especially relevant to support a physiological role. Demethylation induced by membrane depolarization is evident from studies examining the Bdnf and Reln promoters (Chen et al, 2003; Martinowich et al, 2003; Ma et al, 2009). In the earliest such investigations conducted in primary cortical neuron cultures, KCl-induced membrane depolarization was associated with demethylation of a Bdnf promoter and induction of Bdnf mRNA expression (Chen et al, 2003; Martinowich et al, 2003). Lee et al (2008) show that increased gene expression in the context of membrane depolarization is associated with changes in DNA methylation, manifesting both increases and decreases at alternate sites along the promoter of the NR2B receptor with consequent altered binding of MeCP2 to these sites. Membrane depolarization is also capable of focusing the deamination/base-excision demethylating mechanism to promoters with mCpG sites and this is mediated by enzymes of the Gadd45 family of proteins (Ma et al, 2009). Activity-induced demethylation may be one reason why CpG islands containing active gene promoters are predominantly hypo-methylated (Schmitz et al, 2009).
Conversely, DNMT inhibitors influence membrane electrophysiology and reduce the level of mEPSCs in hippocampal neuron preparations. Nelson et al (2008) have shown that if DNA methyltransferase activity is blocked by inhibitors, then genomic DNA is demethylated as a consequence of synaptic activity (Nelson et al, 2008). Consistent with the results of Sharma et al (2008) noted above, increasing synaptic activity mimics the effects of DNMT inhibition (Nelson et al, 2008). More generally, direct manipulation of neuronal DNMT enzymes such as with DNMT inhibitors (zebularine or 5-azacytidine; Nelson et al, 2008; Levenson et al, 2006) results in effects similar to those observed with neuronal activity by causing demethylation of candidate gene promoters and increased mRNA transcription.
CpG METHYLATION AS CELLULAR MEMORY: EVIDENCE FROM BEHAVIORAL STUDIES
What is the evidence for DNA methylation in the formation of cellular memories, another hallmark of neuronal functioning as has been suggested (Crick, 1984; Holliday, 1999)? The most direct evidence for a role of CpG methylation in creating cellular memories is in primary hepatocytes wherein Thomassin et al (2001) showed that DNA demethylation of a glucocorticoid responsive sequence by glucocorticoid treatment persisted after the initial stimulus (glucocorticoids) was removed. Demethylation of the target promoter per se was not sufficient to enhance gene expression, but efficiently primed the promoter for a subsequent physiologically relevant stimulus; that is, CpG demethylation increased the speed of chromatin remodeling at the promoter (Thomassin et al, 2001). Although observed in post-mitotic hepatic cells, this is essentially a molecular equivalent to an unconditioned–conditioned stimulus paradigm.
From a broader physiological perspective, the neuron responds to stimuli–experience by modifying its mCpG profile. This is seen in studies showing changes in promoter methylation of target genes in response to environmental events. Weaver et al (2004) reported an alteration in the methylation of the glucocorticoid receptor promoter in offspring that experienced high-intensity child rearing, from a mother who may or may not be their biological parent. These changes are executed in the post-mitotic postnatal period. Miller and Sweatt (2007) observed bidirectional changes in the density of mCpG in various gene promoters thought to act in apposition during memory acquisition induced by a contextual fear conditioning paradigm. Changes in promoter methylation were specific to genes relevant to memory formation and included simultaneous increases along the protein phosphatase 1 promoter and decreases in the RELN promoter. These events are detectable within 1 h of the experimental intervention. In a further elaboration, Lubin et al (2008) show that in the same gene possessing multiple promoters (ie, Bdnf), behavioral conditioning can induce coordinate changes in both CpG demethylation and increased mRNA expression in a promoter-specific manner. Thus, context alone can demethylate Bdnf exon-promoter I and VI, whereas fear conditioning will demethylate the exon-promoter IV. These findings taken alongside the results of Weaver et al (2004, see above) suggests that CpG methylation encodes experiential memory and the effect of this modification on gene expression can be recapitulated later in life long after the original stimulus.
STUDIES IN POST-MORTEM BRAIN
Genome-Wide Patterns and the Brain Methylome: A Mask to Integrate Promoter Networks
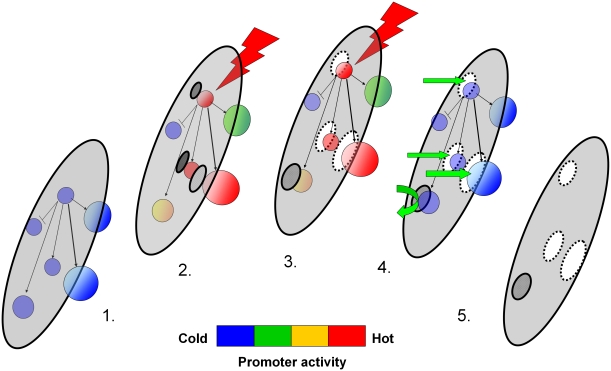
The ‘methylome' represents the total of all methylated CpG sites in the genome. The methylome in reality lies within the genome, but can be conceptualized as having separate functional properties (such as containing a two letter code, methylated or unmethylated CpGs) (Figure 3). Genome-wide studies of brain DNA that have surveyed the methylome have attempted to distinguish patterns of CpG methylation within CpG islands vs outside CpG islands that are either unique to brain tissue (vs non-brain tissue) or unique to a particular brain region. Using enzymatic digestion to isolate methylated and unmethylated DNA fractions, Rollins et al (2006) show that in the brain, the promoter and first exons are more often located in hypo-methylated CpG islands (regions that are 500-bp upstream of the first exon) while downstream exons coincide with higher methylation levels. These investigators suggest that DNA methylation ‘masks' the expanse of the genome, whereas hypo-methylated DNA coinciding with active promoters, leaves exposed only those sequences containing cis-acting regulatory elements (Figure 3). These promoter elements await interaction with ambient transcription factors, whereas the capping function of the methylome occludes degenerate sequences that may divert or delay physiologically relevant DNA–protein interactions. De Bustos et al (2009) examined DNA methylation outside CpG islands using a methylation-sensitive restriction enzyme HpaII and show that in the brain, these non-CpG island methylation sites have patterns that are different from those obtained from non-brain tissue. Ladd-Acosta et al (2007) examined CpG dinucleotide methylation at both CpG island and repetitive sequence locations and report significant differences between brain regions (cortex vs cerebellum). Alu repeats are a type of repetitive DNA element that are interspersed in both intergenic and intragenic locations (ie, common to many different genomic architectures), and contain a variable number of CpG dinucleotides. Xie et al (2009) examined CpG methylation of Alu repeats in the human cerebellum, and note a pattern of hypomethylation of CpG sites within Alu sequences especially those proximal to the TSS of neighboring genes.
Figure 3.
Masking potential of the methylome. (1) The ‘methylome' (gray disk) is embedded in the genome but is separated from the genomic promoter network (spheres). (2) Activation of the promoter net begins at a node in response to stimulation (thunderbolt) and initiates demethylation of the methylome at the location of the active promoter. (3) Demethylation is equilibrated with the intensity of promoter activity across the network. (4) Promoter network returns to baseline quiescence after end of stimulation, but is now ‘primed' for interaction with ambient DNA-binding factors (green arrows). (5) The methylome (displaced) bears the demethylated imprints or ‘mask' of the previously active promoters.
In a confluence of genetics and epigenetics, there is evidence that inherited genetic variation (such as single-nucleotide polymorphisms) may predict site-specific CpG methylation. In this framework, CpG methylation becomes a phenotypic trait that can be subjected to classical genetic analyses. A recent study applies a genome-wide association analysis to the brain methylome to associate variability in DNA sequence (SNPs) to methylation status of CpG sites, either in cis (ie, proximal along the continuous DNA strand) or trans (distant polymorphisms even on other chromosomes). Using DNA extracted from the cerebellum of 153 individuals of European ancestry and applying parallel microarray techniques, one for genotyping and the other for CpG methylation analyses, Zhang et al (2010) report on significant associations between SNP variants and methylation status at CpG sites, in either cis or trans positions. In one informative example, the investigators show the association between a distant intronic gene variant (rs2235375 associated with cleft lip and located on chromosome 1) and the methylation/expression of the IRF6 gene (transcription start site some 14 kb removed).
Studies in Psychiatric Subjects
A few studies have limited their survey to a finite number of gene promoters from human post-mortem brains with some emphasis on a candidate gene approach relevant to neuropsychiatric disorders. Siegmund et al (2007), focusing on CpG island methylation of selected CNS-related gene promoters, report that the intensity of methylation varies during the life span. Individual promoters manifest an independent methylation chronology. These changes suggest perpetual promoter methylation events across the life span, and the only diagnostic differences noted were those in clinical samples from older Alzheimer subjects. These investigators also show that higher methylation is significantly correlated to mRNA expression arguing that CpG methylation in neurons has functional consequences (Siegmund et al 2007). Mill et al (2008) likewise examined genomic DNA obtained from the frontal cortex of 125 brain samples from subjects with a diagnosis of schizophrenia, bipolar disorder, and non-psychiatric controls. DNA from these samples was enriched for the unmethylated fraction (containing CpG islands) using an enzymatic and PCR coupled method, and this enriched fraction was hybridized to a CpG island microarray. These investigators found significant evidence for a ‘modular structure' of the methylome (computed from correlated changes between promoters) from brain DNA recalling the genome-wide ‘masking' function speculated by Rollins et al (2006) (Figure 3). Mill et al (2008) suggest that the normal mCpG modularity is degraded in the major psychoses, indicating fragmented or poorly communicating promoter nets. Of clinical interest, an abundance of independent gene loci were differentially methylated between controls and psychiatric patients, factored by gender. In addition, lifetime antipsychotic use was highly correlated with hypermethylation of the MEK1 (mitogen-associated kinase-1). These large and technically sophisticated studies support a perpetually regulated methylome in the brain with implications for neuropsychiatric disorders and cognition and rehabilitation.
Single promoter studies are based on the assumption that increased methylation of a candidate gene promoter may explain the frequent finding of its reduced mRNA transcription in the diseased population. In schizophrenia, increased methylation has been reported for the RELN and COMT promoters by some but not all investigators (Grayson et al, 2005; Abdolmaleky et al, 2006, but see Tochigi et al, 2008; Dempster et al, 2006). Huang and Akbarian (2007) have directly examined the cross-talk between chromatin/histone modifications and underlying DNA methylation in the promoter of GAD1, a schizophrenia candidate gene. As predicted, DNA methylation was associated with restrictive chromatin markings in the total sample. However, when comparing diagnostic groups, the schizophrenia subjects surprisingly had lower levels of DNA methylation alongside the restrictive chromatin mark.
There is stronger consensus in the suicide literature with numerous reports of increased promoter methylation. In a recent study translating results from early developmental experiences in rodents, McGowan et al (2009) showed increased methylation at a homologous locus along the promoter of the glucocorticoid receptor in the post-mortem brains of human suicide victims who had experienced childhood abuse. Keller et al (2010) report increased promoter methylation of the BDNF IV promoter in suicide victims.
CONCLUSION
CpG methylation is a stable covalent annotation of the genome, and provides a ready mechanism for the coding of new information in the brain. This modification has long been shown to influence gene regulation. The brain is well endowed with all elements of this system, the robust expression of DNA methyltransferases, high levels of both methylated and hydroxymethylated cytosine, and the ability to modify the methylation status of address-specific cytosines. Appropriate genome-wide methylation is required for the long-term survival and functioning of neurons. In the fully matured post-mitotic state, neurons can modify methylation profiles in response to hypoxia, membrane depolarization, behavioral conditioning, and pharmacological perturbation. Methylation of address-specific cytosines is detectable within 1 h of a stimulus and its effects on regulation can be detected for an extended period, in some experiments for the natural life-span of a mammal. Misinterpretation of the mCpG annotation as with a mutated MeCP2 protein results in a uniquely abnormal brain although the mutated protein is expressed in non-brain tissue without similar pathology.
These insights notwithstanding, we know little regarding the function of the key proteins in post-mitotic neurons, such as what sustains the robust expression of DNMT enzymes in this cell type, or what are the systems for rapid and facile removal of the methyl mark in the event of demethylation. It is not clear what pathways intercede between the neuronal depolarization event and the focusing of the methylation–demethylation platform onto a given promoter address. And finally, the brain is an assembly of at least several dozen neuronal phenotypes each with a potentially different methylome. At the outset, we framed this review in the form of a question. We can now say with little reservation that given the scale of operations, from the single base to the genome, it is very likely that CpG methylation affects all three functions, message, memory, and the integration of promoter circuits by a mask-like function.
Acknowledgments
RPS is supported by PHS grant MH069839. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are thankful to Kayla Chase for helping to prepare the paper.
The authors declare that RPS, DPG, and DRG, except for income received from their primary employers, have received no financial support or compensation from any individual or corporate entity over the past 3 years for research or professional service and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene. 2000;242:407. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A. The methyl-CpG-binding protein MeCP2 and neurological disease. BiochemSoc Trans. 2008;36 (Part 4:575. doi: 10.1042/BST0360575. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G, Graessmann M, Graessmann A. Inhibition of herpes simplexthymidine kinase gene expression by DNA methylation is an indirect effect. Nucleic Acids Res. 1985;13:5503. doi: 10.1093/nar/13.15.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero MI, Hansen J, Leaford D, Pollard S, Hendrich BD. Themethyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouseembryogenesis, but play a redundant function in neural differentiation. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312 doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- D'Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol Cell Biol. 2007;27:7462. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, et al. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin. 2009;2 doi: 10.1186/1756-8935-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet. 2006;7:10. doi: 10.1186/1471-2350-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genomewide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Fassbender A, Model F, Adorján P, Pääbo S, Olek A. Differences in DNA methylation patterns between humans and chimpanzees. Curr Biol. 2004;14:R148. [PubMed] [Google Scholar]
- Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740:212. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gardiner M, Frommer M. Transcripts and CpG islands associated with the pro-opiomelanocortin gene and other neurally expressed genes. J Mol Endocrinol. 1994;12:365–382. doi: 10.1677/jme.0.0120365. [DOI] [PubMed] [Google Scholar]
- Gautsch JW, Wilson MC. Delayed de novo methylation in teratocarcinoma cells suggests additional tissue specific mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neurosci Biobehav Rev. 2010;34:882. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst. 2005;3:227. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Kundakovic M, Satta R, Grayson DR, Costa E. Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling. Trends Pharmacol Sci. 2009;30:55. doi: 10.1016/j.tips.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Is there an epigenetic component in long-term memory. J Theor Biol. 1999;200:339. doi: 10.1006/jtbi.1999.0995. [DOI] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinawath A, Miyake S, Yanagisawa Y, Akiyama Y, Yuasa Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem J. 2005;385 (Part 2:557. doi: 10.1042/BJ20040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugloff DG, Vandamme K, Logan R, Visanji NP, Brotchie JM, Eubanks JH. Targeted delivery of an Mecp2 transgene to forebrain neurons improves the behavior of female Mecp2-deficient mice. Hum Mol Genet. 2008;17:1386–1396. doi: 10.1093/hmg/ddn026. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 2002;269:2961. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Guidotti A, Grayson DR. The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes. Mol Pharmacol. 2009;75:342. doi: 10.1124/mol.108.051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM. Paradoxical role of methyl-CpG-binding protein 2 in Rett syndrome. Curr Top Dev Biol. 2004;59:61. doi: 10.1016/S0070-2153(04)59003-8. [DOI] [PubMed] [Google Scholar]
- Laywell ED, Kearns SM, Zheng T, Chen KA, Deng J, Chen HX, et al. Neuron-to-astrocyte transition: phenotypic fluidity and the formation of hybrid asterons in differentiating neurospheres. J Comp Neurol. 2005;493:321. doi: 10.1002/cne.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim W, Ham BJ, Chen W, Bear MF, Yoon BJ. Activity-dependent NR2B expression is mediated by MeCP2-dependent epigenetic regulation. Biochem Biophys Res Commun. 2008;377:930. doi: 10.1016/j.bbrc.2008.10.082. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Puri®cation, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation- related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird AP. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, et al. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proc Natl Acad Sci USA. 2005;102:1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Uehara Y, Kurishita A, Tawa R, Sakurai H. Biological significance of DNA methylation in the ageing process. Age Ageing. 1993;22:S34–S43. doi: 10.1093/ageing/22.suppl_1.s34. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Penn NW. Modification of brain deoxyribonucleic acid base content with maturation in normal and malnourished rats. Biochem J. 1976;155:709. doi: 10.1042/bj1550709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, Du L, Weaver IC, Palkovits M, Faludi G, Merali Z, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jørgensen H, Ruzov A, Wilm M, Georgiev G, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragione DR, Tiunova A, Vacca M, Strazzullo M, González E, Armstrong J, et al. The X-linked methyl binding protein gene Kaiso is highly expressed in brain but is not mutated in Rett syndrome patients. Gene. 2006;373:83. doi: 10.1016/j.gene.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau J, Tanigawa G, Szyf M. The mouse DNA methyltransferase 5'-region. A unique housekeeping gene promoter. J Biol Chem. 1992;267:7368–7377. [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:2539. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. Depolarization induces down-regulation of DNMT1 and DNMT3a mRNA in primary cortical cultures. Epigenetics. 2008;3:74. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Shyadehi AZ, Lamb DC, Kelly SL, Kelly DE, Schunck WH, Wright JN, et al. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14 alpha-demethylase of Candida albicans (other names are: lanosterol 14 alpha-demethylase, P-45014DM, and CYP51) J Biol Chem. 1996;271:12445. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SS. Gilbert's conjecture: the search for DNA (cytosine-5) demethylases and the emergence of new functions for eukaryotic DNA (cytosine-5) methyltransferases. J Mol Biol. 2000;302:1. doi: 10.1006/jmbi.2000.4046. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tawa R, Ono T, Kurishita A, Okada S, Hirose S. Changes of DNA methylationlevel during pre- and postnatal periods in mice. Differentiation. 1990;45:44–48. doi: 10.1111/j.1432-0436.1990.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinás ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20:1974. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Komori B, Sasaki T, Kato N, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63:530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience. 2006;142:727. doi: 10.1016/j.neuroscience.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- Xie H, Wang M, Bonaldo Mde F, Smith C, Rajaram V, Goldman S, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009;37:4331–4340. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa Y, Ito E, Yuasa Y, Maruyama K. The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents. Biochim Biophys Acta. 2002;1577:457. doi: 10.1016/s0167-4781(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci USA. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]