Abstract
Variants in the CHRNA5–CHRNA3–CHRNB4 gene cluster have been associated with nicotine dependence (ND) and ND-related traits. To evaluate a potential underlying mechanism for this association, we investigated the effects of 10 variants in this gene cluster and their interactive effects as a result of recent smoking on cognitive flexibility, a possible mediator of genetic effects in smokers. Cognitive flexibility of 466 European Americans (EAs; 360 current smokers) and 805 African Americans (AAs; 635 current smokers) was assessed using the Wisconsin Card Sorting Test. The main effects of variants and haplotypes and their interaction with recent smoking on cognitive flexibility were examined using multivariate analysis of variance and the haplotype analysis program HAPSTAT. In EAs, the major alleles of five variants (CHRNA5-rs3841324–22 bp-insertion-allele, CHRNA5-rs615470-C-allele, CHRNA3-rs6495307-C-allele, CHRNA3-rs2869546-T-allele, and CHRNB4-rs11637890-C-allele) were associated with significantly greater perseverative responses (P=0.003–0.017) and perseverative errors (P=0.004–0.026; recessive effect). Among EAs homozygous for the major alleles of each of these five variants, current smokers made fewer perseverative responses and perseverative errors than did past smokers. Significant interactive effects of four variants (rs3841324, rs615470, rs6495307, and rs2869546) and current smoking on cognitive flexibility were observed (perseverative responses (P=0.010–0.044); perseverative errors (P=0.017–0.050)). However, in AAs, 10 variants in this gene cluster showed no apparent effects on cognitive flexibility. These findings suggest that variation in the CHRNA5–CHRNA3–CHRNB4 gene cluster influences cognitive flexibility differentially in AAs and EAs and that current smoking moderates this effect. These findings could account in part for differences in ND risk associated with these variants in AAs and EAs.
Keywords: CHRNA5-CHRNA3-CHRNB4, cognitive flexibility, Wisconsin Card Sorting Test, gene × recent tobacco use
INTRODUCTION
Nicotinic acetylcholine receptors belong to a superfamily of ligand-gated ion channels, including GABA (A and C), serotonin, and glycine receptors. To date, nine α-nicotinic receptor subunits (α2−10) and three β-nicotinic receptor subunits (β2−4) have been identified. Nicotinic receptors are formed by pentameric combinations of α and β subunits (Gotti et al, 2006). Their structural diversity and broad expression in both the central and peripheral nervous systems suggest that these receptors may regulate neurotransmitter release from nerve terminals and participate in numerous physiological activities such as reward and cognitive functions.
Nicotinic receptors are involved in controlling dopamine release in the striatum, a region that is involved in the reward pathway, and in the development of substance dependence. Administration of nicotinic receptor antagonists, deletion of endogenous acetylcholine, or disruption of nicotinic receptor genes leads to a decreased release of dopamine (Zhou et al, 2001). Therefore, the rewarding effects of alcohol, nicotine, and other drugs of abuse, such as cocaine and opioids, may be mediated or regulated through nicotinic receptors, and variation in the genes encoding them may influence risk for substance dependence. The α5, α3, and β4 genes are in physical proximity on chromosome 15q24, forming a gene cluster (CHRNA5–CHRNA3–CHRNB4; Raimondi et al, 1992). Recently, the potential role of variants in this gene cluster in nicotine and other substance dependence and smoking-related diseases was intensively studied. Results from four genome-wide association studies provide compelling evidence that variation in a region containing the CHRNA5–CHRNA3–CHRNB4 gene cluster contributes to risk for lung cancer (Hung et al, 2008; Amos et al, 2008; Thorgeirsson et al, 2008), peripheral arterial disease (Thorgeirsson et al, 2008), or nicotine dependence (ND; Hung et al, 2008; Thorgeirsson et al, 2008; Berrettini et al, 2008). Candidate gene studies have shown that variants in this gene cluster (eg, the nonsynonymous variant rs16969968 in CHRNA5, and the 3′UTR variant rs578776 and the exon 5 variant rs1051730 in CHRNA3) contribute to the risk of nicotine, alcohol, and/or cocaine dependence (Saccone et al, 2007; Bierut et al, 2008; Grucza et al, 2008; Berrettini et al, 2008; Weiss et al, 2008; Wang et al, 2009).
Nicotinic receptors also participate in neuronal differentiation and synaptic plasticity, which are important for the neurochemical foundation of learning and memory. Studies have shown that stimulation of nicotinic receptors has a role, either directly or by interaction with other neurotransmitters, in several executive functions such as response inhibition, attention, and working memory (Rezvani and Levin, 2001). The prefrontal cortex (PFC) is involved in working memory, and plasticity of excitatory synaptic transmission within the PFC is an important cellular mechanism of memory. Nicotinic receptors are expressed in two classes of GABA-mediated interneurons in the PFC (McGehee, 2007). These receptors may participate in some fast excitatory neurotransmission and regulate the release of neurotransmitters such as glutamate and GABA. Cholinergic innervation of the PFC from basal forebrain nuclei may affect PFC microcircuitry by activation of these nicotinic receptors, which can enhance working memory and attention.
Some nicotinic receptor gene variants have been shown to influence cognitive function. For example, the C-allele of the α4-nicotinic receptor gene (CHRNA4) C1545T polymorphism (rs1044396) affected the strength of attentional scaling through an additive model, with no association found for the dopamine β-hydroxylase (DBH) gene G444A polymorphism (rs1108580), suggesting that CHRNA4 C1545T selectively contributes to individual differences in visuospatial attention (Greenwood et al, 2005). Two other studies investigated the association between a polymorphism in the exon 2 and intron 2 junction of CHRNA4 (rs6090384) with attention, one showing a strong association of this polymorphism with severe inattention defined by latent class analysis (Todd et al, 2003), although the other study did not support the finding (Lee et al, 2008). The importance of other nicotinic receptor genes, including the CHRNA5–CHRNA3–CHRNB4 gene cluster, in modulating cognitive function remains to be determined.
This study investigated whether variation in the CHRNA5–CHRNA3–CHRNB4 gene cluster could affect cognitive flexibility (measured by the Wisconsin Card Sorting Test or WCST). We chose to focus on cognitive flexibility, as it can be affected by tobacco use. There is evidence that nicotine administration can produce short-term enhancement of attention and memory (Ernst et al, 2001), and smoking cessation in adolescent smokers can lead to acute impairment of verbal and working memory (Jacobsen et al, 2005). We also analyzed the effect of tobacco use and the interaction of tobacco use and genetic variation on cognitive flexibility. As nongenetic factors such as age, sex, ancestry proportion, and education may influence cognitive function, their confounding effects were also considered.
MATERIALS AND METHODS
Subjects
We studied 466 European Americans (EAs) and 805 African Americans (AAs). Of them, 419 EAs and 674 AAs were included in our recent study that examined the influence of variation in the WW and C2 domain containing one gene (WWC1or KIBRA) on cognitive flexibility (Zhang et al, 2009). All subjects were originally recruited for genetic association studies of drug or alcohol dependence. They were interviewed using an electronic version of the semistructured assessment for drug dependence and alcoholism (SSADDA) instrument (Pierucci-Lagha et al, 2005). Information on sex, age, ethnicity, years of education, and the recency of tobacco use was collected at the baseline interview. There were 271 male EAs (58.8%) and 436 male AAs (54.2%). The average age (±SD) was 40 (±12) years for EAs and 41 (±10) years for AAs. EAs received 13±3 years (mean±SD) of education and AAs received 12±2 years (mean±SD) of education. All subjects reported a lifetime history of tobacco use, which was quantified as a tobacco recency score (1: last smoked within 2 weeks (360 EAs and 635 AAs); 2: last smoked in the past 2–4 weeks (five EAs and four AAs); 3: last smoked in the past 1–6 months (10 EAs and 7 AAs); 4: last smoked in the past 6–12 months (1 EA and 10 AAs); 5: last smoked over 1 year ago (90 EAs and 149 AAs)). In all, 409 EAs (87.7%) and 712 AAs (88.4%) had a lifetime DSM-IV (American Psychiatric Association, 1994) diagnosis of substance (alcohol, cocaine, opioid, or ND) use. Subjects affected with major psychotic disorders (eg, schizophrenia, schizoaffective disorder, or bipolar disorder I) were excluded. They were recruited from the University of Connecticut Health Center (Farmington, CT, USA), the Yale University School of Medicine APT Foundation (New Haven, CT, USA), or from the University of Pennsylvania Medical Center (Philadelphia, PA, USA). The institutional review board at each institution approved the study protocol. All subjects provided written informed consent after receiving a complete description of the study. Characteristics of the participants in this study are presented in Table 1.
Table 1. Characteristics of Study Subjects and Recency of Tobacco Use.
| European Americans (EAs) | African Americans (AAs) | |
|---|---|---|
| Number of subjects | 466 | 805 |
| Males (%) | 271 (58.8%) | 436 (54.2%) |
| Age (years±SD) | 40 (±12) | 41 (±10) |
| Education (years±SD) | 13 (±3) | 12 (±2) |
| Recency of tobacco use | ||
| ⩽2 Weeks (current user) | 360 (77.3%) | 635 (78.9%) |
| 2–4 Weeks | 5 (1.1%) | 4 (0.5%) |
| 1 Month–6 months | 10 (2.1%) | 7 (0.9%) |
| 6 Moths–1 year | 1 (0.2%) | 10 (1.2%) |
| >1 Year | 90 (19.3%) | 149 (18.5%) |
| Multiple substance dependence | 409 (87.7%) | 712 (88.4%) |
| Alcohol dependence | 286 (61.4%) | 522 (64.8%) |
| Cocaine dependence | 292 (62.7%) | 552 (68.6%) |
| Opioid dependence | 223 (47.8%) | 160 (19.9%) |
| Nicotine dependence | 297 (63.7%) | 480 (59.6%) |
WCST Assessment of Cognitive Flexibility
Cognitive flexibility is the human ability to adapt one's cognitive processing strategies to face new and unexpected conditions in the environment (Canas et al, 2003). To evaluate whether cognitive flexibility is influenced by specific genetic factors and/or tobacco use, we used the 128-card computerized version of the WCST (Heaton and PAR Staff, 1999). The WCST is a complex test that involves multiple cognitive processes (eg, problem solving, set shifting, working memory, and attention). During the test, subjects are required to match response cards to four stimulus cards on three dimensions (color, form, or number) by pressing one of four number keys on the computer keyboard. The participant was required to determine which sorting principle was correct and when the principle would shift during the test. The computerized version of the WCST continues until all 128 cards are sorted, which differs from the traditional WCST in which the test ends after six correct categories are completed (Robinson et al, 1980).
In this study, three indices of the WCST were used to assess each individual's cognitive flexibility: percentage of perseverative responses (%PR), percentage of perseverative errors (%PE), and percentage of non-perseverative errors (%N-PE). Factor analysis of the WCST has shown that perseverative errors could be the most useful outcome measure in assessing executive function (Greve et al, 2005). Higher values of %PR, %PE, and/or %N-PE are indicative of poorer WCST performance and less cognitive flexibility.
DNA, Markers, and Genotyping
DNA was obtained from immortalized cell lines or directly from blood or saliva. Nine single-nucleotide polymorphisms (SNPs) and one 22-bp insertion/deletion (indel) polymorphism covering the CHRNA5–CHRNA3–CHRNB4 gene cluster region with an average intermarker distance of 8620 bp were selected from the ABI SNPbrowser (De La Vega et al, 2006) or the NCBI SNP database (http://www.ncbi.nim.nih.gov/projects/SNP). Detailed information on these SNPs is summarized in Table 2. SNPs were genotyped with a fluorogenic 5′ nuclease assay (TaqMan) method (Shi et al, 1999), using the ABI PRISM 7900 Sequence Detection System (ABI, Foster City, CA, USA). The 22-bp indel marker rs3841324 was genotyped by directly resolving the PCR products on agarose gel as described previously (Sherva et al, 2010).
Table 2. Information of 10 Genetic Variants in the CHRNA5-CHRNA3-CHRNB4 Gene Cluster.
| SNP | rs# | Gene | Location | Chr. 15 Positiona | Variation | MAFb (EAs) | MAFb (AAs) |
|---|---|---|---|---|---|---|---|
| SNP1 | rs3841324 | CHRNA5 | 5′ Near gene | 76644868… | Del/ | Del (0.384) | Del (0.208) |
| 76644889 | CTATTTCCCTCTGGCCCCGCCC | ||||||
| SNP2 | rs684513 | CHRNA5 | Intron 1 | 76645455 | C/G | G (0.221) | G (0.199) |
| SNP3 | rs16969968 | CHRNA5 | Exon 5 | 76669980 | Asn(A)/Asp(G) | A (0.348) | A (0.045) |
| SNP4 | rs615470 | CHRNA5 | Exon 6 (3′UTR) | 76673043 | C/T | T (0.311) | T (0.393) |
| SNP5 | rs578776 | CHRNA3 | Exon 6 (3′UTR) | 76675455 | C/T | T (0.311) | C (0.442) |
| SNP6 | rs6495307 | CHRNA3 | Intron 5 | 76677376 | C/T | T (0.387) | T (0.413) |
| SNP7 | rs1051730 | CHRNA3 | Exon 5 | 76681394 | Gly(C)/Gly(T) | T (0.355) | T (0.097) |
| SNP8 | rs3743078 | CHRNA3 | Intron 4 | 76681814 | C/G | C (0.265) | G (0.391) |
| SNP9 | rs2869546 | CHRNA3 | Intron 4 | 76694400 | C/T | C (0.338) | C (0.376) |
| SNP10 | rs11637890 | CHRNB4 | 5′ Near gene | 76722474 | C/G | G (0.354) | G (0.150) |
Chromosome positions are based on Homo sapiens chromosome 15 genomic contig NT_010194.16.
Minor allele frequency in African Americans (AAs) and European Americans (EAs) included in this study.
Statistical Analysis
Linkage disequilibrium (LD) between markers was computed using the software program Haploview (Barrett et al, 2005), and haplotype blocks were defined according to the criteria of Gabriel et al (2002). Because 7 of the 10 markers (rs3841324, rs16969968, rs615470, rs578776, rs1051730, rs3743078, and rs11637890) showed a significant difference (P<0.001) in their allele frequencies between EAs and AAs and the self-reported population was validated using 38 ancestry informative markers (Yang et al, 2005a, 2005b; Luo et al, 2005) by the program STRUCTURE 2.3.3 (Falush et al, 2003), the genetic effects of all variants and their interaction with recent tobacco use on cognitive flexibility were analyzed separately in the two population groups. A multivariate analysis of variance was performed using the general linear model procedure in the SPSS16.0 software package (SPSS, Chicago, Illinois). WCST indices (%PR, %PE, and %N-PE) were treated as dependent variables, marker genotypes were treated as independent variables, and nongenetic factors (sex, age, ancestry proportion, recency of tobacco use, and years of education) were incorporated in the model as covariates. Interactive effects of genotypes and tobacco recency on cognitive flexibility were examined as well. The influence of continuous variables (age and years of education) on cognitive flexibility was analyzed by correlational analyses in SPSS 16.0. Mean ancestry proportions of AAs or EAs in the lowest and highest quartiles of WCST scores were compared using the unpaired t-test to determine whether cognitive flexibility was greatly influenced by variation in the genetic admixture of the study subjects.
In addition, the joint effect of markers in haplotype blocks on cognitive flexibility was analyzed using the program HAPSTAT (Lin et al, 2005), a general likelihood-based approach that infers the effect of haplotypes and haplotype × environment on a disease phenotype. The effect of haplotypes and haplotype × tobacco recency on cognitive flexibility was denoted as ‘estimates.' Positive values of ‘estimates' reflect more perseverative responses, perseverative errors, or non-perseverative errors (ie, poorer WCST performance), whereas negative values of ‘estimates' reflect fewer perseverative responses, perseverative errors, or non-perseverative errors (ie, better WCST performance). In the haplotype analysis, the three WCST domains (%PR, %PE, and %N-PE) were considered as dependent variables, haplotypes were viewed as independent variables, and sex, age, ancestry proportion, education, and recency of tobacco use were treated as covariates.
RESULTS
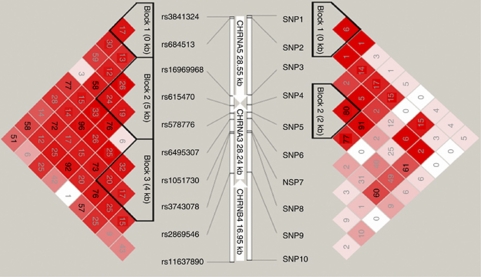
LD patterns across the gene cluster are shown in Figure 1. In EAs, the 10 markers showed a high correlation and the first 8 markers (four in CHRNA5 and four in the 3′ end of CHRNA3) were distributed in three haplotype blocks on the basis of complete LD within the blocks. In AAs, two haplotype blocks were observed (two CHRNA5 markers (rs3841324 and rs684513) were in one block and CHRNA5-rs615470 and CHRNA3-rs578776 were in another block). CHRNA5-rs16969968 was highly correlated with markers in these two haplotype blocks. The remaining four CHRNA3 SNPs (rs6495307, rs1051730, rs3743078, and rs2869546) and CHRNB4-rs11637890 were statistically independent.
Figure 1.
Haplotype blocks and linkage disequilibrium (LD) patterns for 10 CHRNA5–CHRNA3–CHRNB4 variants in European Americans (EAs) and African Americans (AAs). Numbers in the squares are r-squared measures of pairwise marker–marker correlations (r2 × 100). The haplotype block was defined by the criteria of Gabriel et al. (2002) and indicated in black triangles.
Consistent with our recent study (Zhang et al, 2009), the present study showed age to be strongly inversely correlated with cognitive flexibility in both AAs and EAs, such that older individuals made more perseverative responses (EA: r=0.20, P<0.001; AA: r=0.22, P<0.001), perseverative errors (EA: r=0.20, P<0.001; AA: r=0.23, P<0.001), and non-perseverative errors (EA: r=0.21, P<0.001; AA: r=0.14, P<0.001). However, the length of education was directly correlated with cognitive flexibility, but the effect was stronger in AAs than in EAs. Thus, more educated subjects made fewer perseverative responses (EA: r=−0.08, P=0.105; AA: r=−0.128, P<0.001), perseverative errors (EA: r=−0.03, P=0.132; AA: r=−0.137, P<0.001), and non-perseverative errors (EA: r=−0.06, P=0.177; AA: r=−0.180, P<0.001). Recent tobacco use was associated with poorer performance on two WCST domains (ie, significantly greater perseverative responses and perseverative errors) in AAs (%PR: F(1, 804)=9.36, P=0.002; %PE: F(1, 804)=8.65, P=0.003). No influence of recent tobacco use on cognitive flexibility was observed in EAs. No sex effect on cognitive flexibility was observed in either EAs or AAs. No significant difference in ancestry proportions was observed between EAs and AAs who had the lowest and highest quarters of WCST scores. This implies that variation of ancestry proportions in the study subjects did not significantly confound the genetic effect of nicotinic receptor genes on cognitive flexibility (Table 3).
Table 3. Comparison of Ancestry Proportions of Subjects with the Lowest and Highest Quarters of WCST scores.
| WCST scores (%) | European Americans (EAs) European ancestry proportion mean±(SEM) | African Americans (AAs) African ancestry proportion mean±(SEM) |
|---|---|---|
| %PR lowest quarter | 0.984 (±0.004) (n=116) | 0.961 (±0.005) (n=201) |
| %PR highest quarter | 0.988 (±0.004) (n=116) | 0.964 (±0.006) (n=201) |
| t=0.58 | t=0.33 | |
| df=230 | df=400 | |
| P=0.559 | P=0.738 | |
| %PE lowest quarter | 0.984 (±0.004) (n=116) | 0.962 (±0.005) (n=201) |
| %PE highest quarter | 0.987 (±0.004) (n=116) | 0.961 (±0.006) (n=201) |
| t=0.50 | t=0.04 | |
| df=230 | df=400 | |
| P=0.617 | P=0.964 | |
| %P-NE lowest quarter | 0.984 (±0.004) (n=116) | 0.959 (±0.005) (n=201) |
| %P-NE highest quarter | 0.970 (±0.008) (n=116) | 0.958 (±0.007) (n=201) |
| t=1.47 | t=0.10 | |
| df=230 | df=400 | |
| P=0.143 | P=0.916 |
%PR lowest quarter, %PE lowest quarter and %P-NE lowest quarter: Subjects with the lowest quarter of WCST scores.
%PR highest quarter, %PE highest quarter and %P-NE highest quarter: Subjects with the highest quarter of WCST scores.
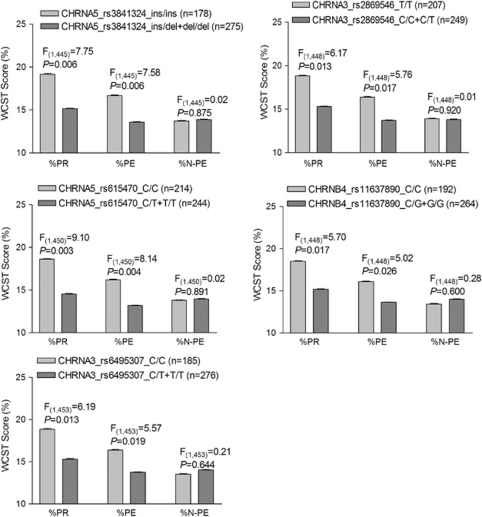
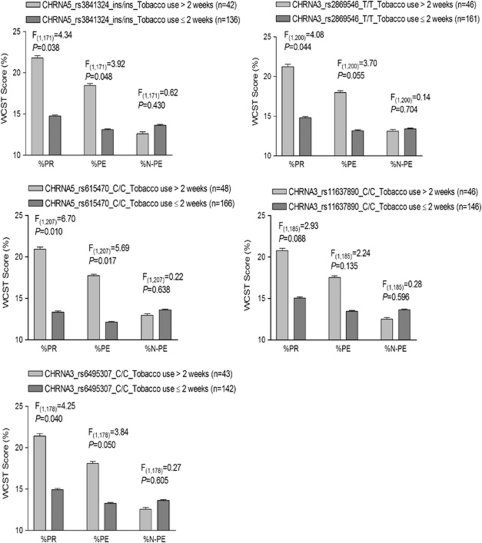
The influence of variants and of variant × recent tobacco use on cognitive flexibility is described in Tables 4 and 5. In EAs (see Table 4), 5 of the 10 markers (CHRNA5-rs3841324, CHRNA5-rs615470, CHRNA3-rs6495307, CHRNA3-rs2869546, and CHRNB4-rs11637890) showed statistically significant effects on two WCST domains (%PR: P=0.023, 0.010, 0.046, 0.046, and 0.050, respectively; %PE: P=0.025, 0.017, 0.059, 0.056, and 0.083, respectively). Moreover, EAs with minor alleles of these five markers (the del-allele of CHRNA5-rs3841324, the T-allele of CHRNA5-rs615470, the T-allele of CHRNA3-rs6495307, the C-allele of CHRNA3-rs2869546, and the G-allele of CHRNB4-rs11637890) made significantly fewer perseverative responses (P=0.006, 0.003, 0.013, 0.013, and 0.017, respectively) and perseverative errors (P=0.006, 0.004, 0.019, 0.017, and 0.026, respectively; dominant effect; Figure 2). In other words, the major allele of the above five markers was associated with poorer WCST performance in these two domains (recessive effect). Although EAs homozygous for the major allele of the five markers showed less cognitive flexibility, recent tobacco use was found to compensate for the negative effect of these genetic factors and was associated with greater cognitive flexibility in EAs. This finding is depicted in Figure 3. Among EAs homozygous for the major alleles of the above four markers (CHRNA5-rs3841324, CHRNA5-rs615470, CHRNA3-rs6495307, and CHRNA3-rs2869546), current tobacco users (ie, those who had smoked within 2 weeks) performed significantly better in two WCST domains than did past tobacco users (ie, who had not smoked in the preceding 2 weeks; %PR: P=0.038, 0.010, 0.040, and 0.044, respectively; %PE: P=0.048, 0.017, 0.050, and 0.055, respectively). A similar trend for an interactive effect of CHRNB4-rs11637890 genotype C/C and tobacco recency on cognitive flexibility was also observed (%PR: P=0.088; %PE: P=0.135).
Table 4. Effects of CHRNA5–CHRNA3–CHRNB4 Markers and Marker × Tobacco Recency on Cognitive Flexibility in European Americans.
| N |
%PRa |
%PEb |
%N-PEc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (±SEM) | Markerd effect | Marker × TobRCe effect | Mean (±SEM) | Markerd effect | Marker × TobRCe effect | Mean (±SEM) | Markerd effect | Marker × TobRCe effect | ||
| 1: CHRNA5_rs3841324 | ||||||||||
| Ins/Ins | 178 | 19.17 (±1.11) | F(2, 443) | F(2, 443) | 16.69 (±0.87) | F(2, 443) | F(2, 443) | 13.72 (±0.84) | F(2, 443) | F(2, 443) |
| Del/Ins | 199 | 15.21 (±1.11) | =3.80 | =2.44 | 13.64 (±0.86) | =3.73 | =2.16 | 13.73 (±0.83) | =0.06 | =0.58 |
| Del/Del | 76 | 15.11 (±1.67) | P=0.023 | P=0.089 | 13.54 (±1.31) | P=0.025 | P=0.116 | 14.20 (±1.26) | P=0.942 | P=0.563 |
| 2: CHRNA5_rs684513 | ||||||||||
| C/C | 275 | 16.44 (±0.91) | F(2, 445) | F(2, 445) | 14.52 (±0.71) | F(2, 445) | F(2, 445) | 14.08 (±0.70) | F(2, 445) | F(2, 445) |
| C/G | 156 | 16.87 (±1.19) | =0.19 | =1.17 | 14.98 (±0.93) | =0.22 | =1.38 | 12.92 (±0.91) | =0.87 | =1.21 |
| G/G | 24 | 14.84 (±3.17) | P=0.828 | P=0.310 | 13.35 (±2.48) | P=0.801 | P=0.252 | 15.72 (±2.43) | P=0.419 | P=0.300 |
| 3: CHRNA5_rs16969968 | ||||||||||
| Asp/Asp | 208 | 15.38(±1.03) | F(2, 449) | F(2, 449) | 13.76 (±0.82) | F(2, 449) | F(2, 449) | 13.90 (±0.79) | F(2, 449) | F(2, 449) |
| Asp/Asn | 177 | 17.15 (±1.09) | =2.50 | =1.92 | 15.18 (±0.86) | =2.27 | =1.63 | 13.59 (±0.84) | =0.12 | =1.06 |
| Asn/Asn | 74 | 19.91 (±1.80) | P=0.083 | P=0.147 | 17.11 (±1.41) | P=0.104 | P=0.197 | 14.37 (±1.38) | P=0.886 | P=0.348 |
| 4: CHRNA5_rs615470 | ||||||||||
| C/C | 214 | 18.64 (±0.99) | F(2, 448) | F(2, 448) | 16.23 (±0.78) | F(2, 448) | F(2, 448) | 13.82 (±0.78) | F(2, 448) | F(2, 448) |
| C/T | 192 | 14.40 (±1.05) | =4.61 | =3.80 | 13.14 (±0.83) | =4.09 | =3.17 | 14.03 (±0.83) | =0.03 | =0.44 |
| T/T | 52 | 15.03 (±1.97) | P=0.010 | P=0.023 | 13.30 (±1.55) | P=0.017 | P=0.043 | 13.69 (±1.56) | P=0.973 | P=0.642 |
| 5: CHRNA3_rs578776 | ||||||||||
| C/C | 215 | 16.89 (±1.04) | F(2, 448) | F(2, 448) | 14.88 (±0.81) | F(2, 448) | F(2, 448) | 14.13 (±0.78) | F(2, 448) | F(2, 448) |
| C/T | 197 | 16.84 (±1.07) | =0.12 | =0.96 | 14.97 (±0.84) | =0.15 | =1.27 | 13.18 (±0.80) | =0.70 | =0.60 |
| T/T | 46 | 15.62 (±2.37) | P=0.884 | P=0.383 | 13.85 (±1.85) | P=0.856 | P=0.282 | 15.21 (±1.78) | P=0.495 | P=0.551 |
| 6: CHRNA3_rs6495307 | ||||||||||
| C/C | 185 | 18.87 (±1.10) | F(2, 451) | F(2, 451) | 16.39 (±0.86) | F(2, 451) | F(2, 451) | 13.54 (±0.83) | F(2, 451) | F(2, 451) |
| C/T | 199 | 15.39 (±1.07) | =3.10 | =2.36 | 13.92 (±0.84) | =2.85 | =2.18 | 14.34 (±0.81) | =0.35 | =0.14 |
| T/T | 77 | 15.12 (±1.72) | P=0.046 | P=0.096 | 13.33 (±1.34) | P=0.059 | P=0.114 | 13.27 (±1.30) | P=0.703 | P=0.873 |
| 7: CHRNA3_rs1051730 | ||||||||||
| C/C | 205 | 15.35 (±1.06) | F(2, 450) | F(2, 450) | 13.74 (±0.83) | F(2, 450) | F(2, 450) | 13.99 (±0.80) | F(2, 450) | F(2, 450) |
| C/T | 180 | 17.25 (±1.10) | =2.40 | =1.76 | 15.27 (±0.86) | =2.20 | =1.50 | 13.70 (±0.84) | =0.08 | =1.27 |
| T/T | 75 | 19.79 (±1.82) | P=0.092 | P=0.173 | 17.02 (±1.42) | P=0.112 | P=0.223 | 14.32 (±1.38) | P=0.920 | P=0.282 |
| 8: CHRNA3_rs3743078 | ||||||||||
| G/G | 248 | 16.71 (±0.98) | F(2, 452) | F(2, 452) | 14.70 (±0.76) | F(2, 452) | F(2, 452) | 14.06 (±0.74) | F(2, 452) | F(2, 452) |
| C/G | 177 | 16.89 (±1.12) | =0.03 | =1.84 | 15.04 (±0.87) | =0.08 | =2.20 | 13.08 (±0.84) | =0.99 | =0.53 |
| C/C | 37 | 16.22 (±2.55) | P=0.971 | P=0.160 | 14.32 (±1.99) | P=0.926 | P=0.111 | 15.83 (±1.92) | P=0.372 | P=0.591 |
| 9: CHRNA3_rs2869546 | ||||||||||
| T/T | 207 | 18.84 (±1.06) | F(2, 446) | F(2, 446) | 16.40 (±0.83) | F(2, 446) | F(2, 446) | 13.92 (±0.79) | F(2, 446) | F(2, 446) |
| C/T | 190 | 15.37 (±1.10) | =3.09 | =2.09 | 13.81 (±0.86) | =2.90 | =1.87 | 13.78 (±0.82) | =0.01 | =0.595 |
| C/C | 59 | 15.09 (±1.92) | P=0.046 | P=0.125 | 13.44 (±1.50) | P=0.056 | P=0.155 | 13.88 (±1.42) | P=0.992 | P=0.552 |
| 10: CHRNB4_rs11637890 | ||||||||||
| C/C | 192 | 18.52 (±1.06) | F(2, 446) | F(2, 446) | 16.10 (±0.83) | F(2, 446) | F(2, 446) | 13.45 (±0.82) | F(2, 446) | F(2, 446) |
| C/G | 202 | 15.05 (±1.02) | =2.97 | =1.42 | 13.58 (±0.79) | =2.50 | =1.15 | 14.24 (±0.78) | =0.31 | =0.13 |
| G/G | 62 | 15.77 (±2.26) | P=0.050 | P=0.241 | 14.12 (±1.77) | P=0.083 | P=0.316 | 13.22 (±1.73) | P=0.730 | P=0.875 |
Percentage of perseverative responses.
Percentage of perseverative errors.
Percentage of non-perseverative errors.
Main effect of SNPs on cognitive flexibility.
Interactive effect of gene variants and tobacco recency.
Subscripted numbers in the parenthesis under F values are degree of freedom of groups and degree of freedom of errors, respectively.
P-values in bold: statistically significant results.
Table 5. Effects of CHRNA5–CHRNA3–CHRNB4 Markers and Marker × Tobacco Recency on Cognitive Flexibility in African Americans.
| N |
%PRa |
%PEb |
%N-PEc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (±SEM) | Markerd effect | Marker × TobRCe effect | Mean (±SEM) | Markerd effect | Marker × TobRCe effect | Mean (±SEM) | Markerd effect | Marker × TobRCe effect | ||
| 1: CHRNA5-rs3841324 | ||||||||||
| Ins/Ins | 481 | 21.97 (±0.90) | F(2, 749) | F(2, 749) | 19.32 (±0.69) | F(2, 749) | F(2, 749) | 19.58 (±0.61) | F(2, 749) | F(2, 749) |
| Del/Ins | 245 | 20.81 (±1.35) | =0.99 | =0.21 | 18.44 (±1.03) | =1.16 | =0.36 | 16.73 (±0.92) | =3.61 | =3.14 |
| Del/Del | 33 | 26.54 (±3.99) | P=0.371 | P=0.812 | 23.21 (±3.04) | P=0.313 | P=0.696 | 20.69 (±2.70) | P=0.027 | P=0.043 |
| 2: CHRNA5-rs684513 | ||||||||||
| C/C | 509 | 21.52 (±0.88) | F(2, 779) | F(2, 779) | 19.04 (±0.67) | F(2, 779) | F(2, 779) | 18.62 (±0.61) | F(2, 779) | F(2, 779) |
| C/G | 248 | 21.82 (±1.26) | =0.15 | =1.03 | 19.07 (±0.96) | =0.13 | =1.13 | 17.79 (±0.88) | =3.56 | =0.41 |
| G/G | 32 | 19.71 (±3.67) | P=0.862 | P=0.358 | 17.62 (±2.80) | P=0.881 | P=0.323 | 24.74 (±2.54) | P=0.035 | P=0.663 |
| 3: CHRNA5-rs16969968 | ||||||||||
| Asp/Asp | 721 | 21.31 (±0.75) | F(2, 770) | F(2, 770) | 18.80 (±0.57) | F(2, 770) | F(2, 770) | 18.54 (±0.52) | F(2, 770) | F(2, 770) |
| Asp/Asn | 53 | 24.39 (±2.49) | =0.73 | =0.41 | 21.75 (±1.90) | =1.17 | =0.54 | 20.22 (±1.73) | =0.53 | =0.46 |
| Asn/Asn | 6 | 19.99 (±6.61) | P=0.481 | P=0.664 | 17.28 (±5.03) | P=0.312 | P=0.584 | 16.69 (±4.58) | P=0.590 | P=0.631 |
| 4: CHRNA5-rs615470 | ||||||||||
| C/C | 297 | 20.98 (±1.13) | F(2, 769) | F(2, 769) | 18.64 (±0.86) | F(2, 769) | F(2, 769) | 18.24 (±0.78) | F(2, 769) | F(2, 769) |
| C/T | 349 | 22.37 (±1.08) | =0.56 | =0.13 | 19.46 (±0.82) | =0.31 | =0.23 | 18.94 (±0.74) | =0.32 | =0.13 |
| T/T | 133 | 20.56 (±1.84) | P=0.570 | P=0.877 | 18.48 (±1.41) | P=0.736 | P=0.793 | 17.97 (±1.27) | P=0.729 | P=0.881 |
| 5: CHRNA3-rs578776 | ||||||||||
| T/T | 248 | 20.54 (±1.28) | F(2, 755) | F(2, 755) | 18.25 (±0.97) | F(2, 755) | F(2, 755) | 18.55 (±0.88) | F(2, 755) | F(2, 755) |
| C/T | 362 | 22.52 (±1.04) | =1.11 | =0.20 | 19.62 (±0.80) | =0.88 | =0.20 | 18.61 (±0.72) | =0.12 | =0.24 |
| C/C | 155 | 20.11 (±1.66) | P=0.331 | P=0.815 | 18.00 (±1.26) | P=0.413 | P=0.817 | 17.97 (±1.14) | P=0.886 | P=0.784 |
| 6: CHRNA3-rs6495307 | ||||||||||
| C/C | 274 | 20.94 (±1.16) | F(2, 774) | F(2, 774) | 18.62 (±0.88) | F(2, 774) | F(2, 774) | 18.19 (±0.80) | F(2, 774) | F(2, 774) |
| C/T | 368 | 22.53 (±1.03) | =1.01 | =0.03 | 19.63 (±0.79) | =0.69 | =0.01 | 18.81 (±0.72) | =0.30 | =0.06 |
| T/T | 142 | 19.93 (±1.78) | P=0.365 | P=0.972 | 17.97 (±1.36) | P=0.502 | P=0.990 | 17.83 (±1.24) | P=0.739 | P=0.946 |
| 7: CHRNA3_rs1051730 | ||||||||||
| C/C | 648 | 21.13 (±0.79) | F(2, 774) | F(2, 774) | 18.67 (±0.60) | F(2, 774) | F(2, 774) | 18.05 (±0.55) | F(2, 774) | F(2, 774) |
| C/T | 124 | 22.62 (±1.72) | =0.99 | =1.91 | 19.92 (±1.32) | =1.01 | =2.19 | 20.66 (±1.20) | =1.99 | =2.06 |
| T/T | 12 | 27.74 (±5.38) | P=0.370 | P=0.149 | 23.55 (±4.10) | P=0.364 | P=0.112 | 18.79 (±3.73) | P=0.137 | P=0.128 |
| 8: CHRNA3-rs3743078 | ||||||||||
| C/C | 294 | 20.24 (±1.19) | F(2, 775) | F(2, 775) | 18.15 (±0.91) | F(2, 775) | F(2, 775) | 19.72 (±0.82) | F(2, 775) | F(2, 775) |
| C/G | 374 | 22.42 (±1.03) | =0.98 | =0.38 | 19.50 (±0.79) | =0.65 | =0.67 | 17.49 (±0.71) | =2.13 | =2.41 |
| G/G | 117 | 21.10 (±1.79) | P=0.377 | P=0.685 | 18.68 (±1.36) | P=0.524 | P=0.512 | 18.42 (±1.23) | P=0.120 | P=0.090 |
| 9: CHRNA3-rs2869546 | ||||||||||
| T/T | 316 | 22.30 (±1.07) | F(2, 776) | F(2, 776) | 19.62 (±0.81) | F(2, 776) | F(2, 776) | 18.91 (±0.74) | F(2, 776) | F(2, 776) |
| C/T | 348 | 21.30 (±1.11) | =0.74 | =0.10 | 18.67 (±0.85) | =0.69 | =0.07 | 18.14 (±0.77) | =0.27 | =0.13 |
| C/C | 122 | 19.72 (±1.88) | P=0.477 | P=0.903 | 17.85 (±1.44) | P=0.502 | P=0.929 | 18.38 (±1.30) | P=0.765 | P=0.875 |
| 10: CHRNB4-rs11637890 | ||||||||||
| C/C | 575 | 21.41 (±0.83) | F(2, 777) | F(2, 777) | 18.86 (±0.63) | F(2, 777) | F(2, 777) | 18.21 (±0.57) | F(2, 777) | F(2, 777) |
| C/G | 192 | 21.98 (±1.42) | =0.39 | =1.02 | 19.52 (±1.08) | =0.36 | =1.10 | 19.78 (±0.98) | =1.03 | =0.37 |
| G/G | 22 | 24.84 (±3.96) | P=0.675 | P=0.360 | 21.08 (±3.02) | P=0.697 | P=0.334 | 17.62 (±2.72) | P=0.357 | P=0.691 |
Percentage of perseverative responses.
Percentage of perseverative errors.
Percentage of non-perseverative errors.
Main effect of SNPs on cognitive flexibility.
Interactive effect of gene variants and tobacco recency.
Subscripted numbers in the parenthesis under F values are degree of freedom of groups and degree of freedom of errors, respectively.
P-values in bold: statistically significant results.
Figure 2.
Genetic effects of five CHRNA5–CHRNA3–CHRNB4 variants on cognitive flexibility in European Americans (EAs). Subjects homozygous for major alleles of five CHRNA5–CHRNA3–CHRNB4 markers (rs3841324 ins-allele, rs615470 C-allele, rs6495307 C-allele, rs2869546 T-allele, and rs11637890 C-allele) showed significantly more perseverative responses and perseverative errors than those with minor alleles of these five markers. Data shown on the Y-axis are mean values of WCST scores (±SEM). %PR, percentage of perseverative responses; %PE, percentage of perseverative errors; %N-PE, percentage of non-perseverative errors.
Figure 3.
Interactive effects of five CHRNA5–CHRNA3–CHRNB4 SNPs and tobacco recency on cognitive flexibility in European Americans (EAs). Among subjects homozygous for the CHRNA5-rs3841324 major ins-allele, CHRNA5-rs615470 major C-allele, CHRNA3-rs6495307 major C-allele, or CHRNA3-rs2869546 major T-allele, current tobacco users (smoked ⩽2 weeks) made significantly fewer perseverative responses and perseverative errors than did past tobacco users (smoked >2 weeks). Data shown on the Y-axis are mean values of WCST scores (±SEM). %PR, percentage of perseverative responses; %PE, percentage of perseverative errors; %N-PE, percentage of non-perseverative errors.
The functional marker CHRNA5-rs16969968 showed a nonsignificant effect on cognitive flexibility in EAs, but in an opposite direction from the above five markers. Its major allele (Asp) was associated with a trend for better performance in two WCST domains (%PR: P=0.083 (additive model) or P=0.064 (recessive model); P=0.104 (additive model) or P=0.069 (recessive model)). Among EAs homozygous for the rs16969968 major (Asp) allele, current smokers made more perseverative responses and perseverative errors than did past smokers (%PR: 20.08±1.66 vs 14.96±1.84, P=0.066; %PE: 17.12±1.30 vs 13.28±1.44, P=0.088). In AAs (see Table 5), only two CHRNA5 markers (rs3841324 and rs684513) showed a statistical association with non-perseverative errors (rs3841324: P=0.027; rs684513: P=0.035).
In addition, interactive effects of markers in haplotype blocks (three blocks in EAs and two blocks in AAs, see Figure 1) and haplotype × recent tobacco use on cognitive flexibility were analyzed using HAPSTAT. As shown in Table 6, three common haplotypes (Asn-C-C: 35.0%, Asp-T-C: 32.0%, and Asp-C-T: 30.9%) in haplotype block 2 (rs16969968-rs615470-rs578776) had a protective role for cognitive flexibility in EAs, resulting in fewer perseverative responses (P=0.008, 0.0004, and 0.003, respectively) and perseverative errors (P=0.016, 0.001, and 0.007, respectively). However, current smoking reversed the favorable role of the three haplotypes (A-C-C, G-T-C, and G-C-T) in cognitive flexibility, resulting in greater numbers of perseverative responses (P=0.004, 0.0004 and 0.002, respectively) and perseverative errors (P=0.006, 0.001 and 0.004, respectively). Haplotypes in block 1 (rs3841324-rs684513) and block 3 (rs6495307-rs1051730-rs3743078) and their interaction with tobacco recency did not show significant effects on cognitive flexibility (Table 6). In AAs, we did not observe an apparent effect of haplotypes in block 1 (rs3841324-rs684513) and block 2 (rs615470-rs578776; see Figure 1) and their interaction with tobacco recency on cognitive flexibility (data not shown).
Table 6. Influence of CHRNA5–CHRNA3–CHRNB4 Haplotypes and Haplotype × Tobacco Recency on Cognitive Flexibility in European Americans.
|
%PRa |
%PEb |
%N-PEc |
||||
|---|---|---|---|---|---|---|
| Estimated | P-value | Estimated | P-value | Estimated | P-value | |
| Haplotype block 1 (rs3841324-rs684513): | ||||||
| Ins-C (38.9%) | 4.64 | 0.151 | 3.39 | 0.181 | −3.18 | 0.190 |
| Del-C (38.5%) | −4.19 | 0.199 | −3.21 | 0.210 | −1.19 | 0.623 |
| Ins-G (22.3%) | 2.40 | 0.416 | 2.04 | 0.379 | −3.11 | 0.160 |
| Haplotype block 1 × tobacco recency (⩽2 weeks vs >2 weeks): | ||||||
| Ins-C*TobRC | −6.00 | 0.101 | −4.27 | 0.137 | 3.31 | 0.232 |
| Del-C*TobRC | 2.17 | 0.552 | 1.66 | 0.562 | 0.57 | 0.836 |
| Ins-G*TobRC | −4.87 | 0.146 | −3.85 | 0.144 | 3.22 | 0.201 |
| Haplotype block 2 (rs16969968-rs615470-rs578776): | ||||||
| Asn-C-C (35.0%) | −17.15 | 0.008 | −12.30 | 0.016 | −1.56 | 0.779 |
| Asp-T-C (32.0%) | −22.68 | 0.0004 | −16.45 | 0.001 | −1.29 | 0.817 |
| Asp-C-T (30.9%) | −19.10 | 0.003 | −13.70 | 0.007 | −2.14 | 0.704 |
| Haplotype block 2 × tobacco recency (⩽2 weeks vs >2 weeks): | ||||||
| Asn-C-C* TobRC | 21.10 | 0.004 | 15.77 | 0.006 | 4.55 | 0.457 |
| Asp-T-C* TobRC | 25.70 | 0.0004 | 19.20 | 0.001 | 3.90 | 0.525 |
| Asp-C-T* TobRC | 22.37 | 0.002 | 16.69 | 0.004 | 5.65 | 0.359 |
| Haplotype block 3 (rs6495307-rs1051730-rs3743078): | ||||||
| T-C-G (37.3%) | 5.51 | 0.661 | 3.79 | 0.700 | 1.70 | 0.856 |
| C-T-G (34.8%) | 10.61 | 0.404 | 7.66 | 0.442 | 1.42 | 0.882 |
| C-C-C (25.9%) | 8.72 | 0.489 | 6.35 | 0.519 | 0.75 | 0.936 |
| Haplotype block 3 × tobacco recency (⩽2 weeks vs >2 weeks): | ||||||
| T-C-G* TobRC | −7.67 | 0.553 | −5.41 | 0.592 | −4.56 | 0.637 |
| C-T-G* TobRC | −12.05 | 0.357 | −8.68 | 0.396 | −3.86 | 0.693 |
| C-C-C* TobRC | −11.41 | 0.378 | −8.27 | 0.415 | −2.82 | 0.771 |
Percentage of perseverative responses.
Percentage of perseverative errors.
Percentage of non-perseverative errors.
Estimates for the effect of haplotypes and haplotype–smoking interactions on cognitive flexibility.
P-values in bold: statistically significant results.
Haplotypes with frequency>1% are listed.
DISCUSSION
Several lines of evidence suggest that nicotine enhances aspects of cognition through nicotinic receptors and variation in the CHRNA5–CHRNA3–CHRNB4 gene cluster influences the risk for ND. In this study, we further examined the effect of nicotinic receptor gene variation alone and in combination with recent smoking on cognitive flexibility (working memory, attention, set-shifting, and so on). Two major findings were obtained.
First, we confirmed that variants within this gene cluster modulated cognitive flexibility, an effect that was population specific. In EAs, 5 of the 10 genetic markers examined showed an effect on cognitive flexibility. The major alleles of five markers were associated with less cognitive flexibility (ie, more perseverative responses and perseverative errors; Table 4 and Figure 2). The association of multiple markers and cognitive flexibility may be due to the high degree of intercorrelation of markers in the CHRNA5–CHRNA3–CHRNB4 gene region in EAs (see Figure 1). Moreover, three haplotypes (Asn-C-C, Asp-T-C, and Asp-C-T) in haplotype block 2 and their interaction with tobacco recency greatly influenced WCST performance in two domains (perseverative responses and perseverative errors; Table 6). In AAs, only two CHRNA5 markers (rs3841324 and rs684513) were associated with non-perseverative errors (Table 5). As the WCST measurements in homozygous subjects (with genotype rs3841324 del/del or rs684513 G/G) varied considerably, the positive result may be due to type I error. Therefore, variation in this gene cluster may affect cognitive flexibility differentially in individuals of European ancestry compared with those of African ancestry; moreover, the effect of variants in this region on cognitive flexibility was stronger in EAs than in AAs.
Another major finding was that, in EAs, recent smoking offset the genetic effect of CHRNA5–CHRNA3–CHRNB4 variants on cognitive flexibility. When all AA or EA subjects were examined jointly (irrespective of genotype information), no difference in cognitive flexibility was seen between current tobacco users and past tobacco users. The only subgroup of subjects whose cognitive flexibility was improved as a function of recent tobacco use was the subset of EAs who were homozygous for the major allele of five SNPs (rs3841324 and rs615470 in CHRNA5, rs6495307 and rs2869546 in CHRNA3, and rs11637890 in CHRNB4; Figure 3). This finding implies that nicotine can improve working memory, attention, and set-shifting in subjects who might otherwise have cognitive deficits. Nicotine can enhance various aspects of cognitive processing, such as attention and memory by activation of nicotinic receptors (Marchant et al, 2008). However, in AAs, the effect of marker × tobacco recency was not salient. This is consistent with the findings that AAs have a lower prevalence of ND than EAs (Kandel and Chen, 2000). The present study suggests that nicotine modulates the effect of genetic factors on cognitive flexibility in a population-specific way.
The findings from this study may help to understand the high smoking prevalence among patients who suffer from some psychiatric disorders. Mounting evidence suggests that cognitive impairment of executive function is a core symptom of disorders such as schizophrenia and bipolar disorder. For example, patients with schizophrenia made significantly more perseverative responses, indicating a more pronounced and specific deficit in cognitive flexibility (Galderisi et al, 2009), and patients with bipolar disorder have a deficit in their ability to monitor the contents of working memory (Thompson et al, 2007). As activation of nicotinic receptors can enhance working memory and attention, these receptors may be useful therapeutic targets for cognitive dysfunction. Understandably, tobacco use may be viewed as a way of nicotine self-delivery for the treatment of cognitive deficits. Nicotine-patch therapy has been used to reduce smoking and improve cognitive function in patients with schizophrenia (Ziedonis and George, 1997). Functional magnetic resonance imaging studies have also indicated that nicotine-patch therapy improves cognitive function in schizophrenia patients by activating a network of brain regions, including the anterior cingulate cortex and bilateral thalamus (Jacobsen et al, 2004). Moreover, selective agonists for α7 and α4β2 nicotinic receptors have been used to treat neuropsychiatric and neurodegenerative disorders in which cognitive impairment is a key symptom (Cincotta et al, 2008). Nicotinic receptors consisting of subunit peptides encoded by CHRNA5, CHRNA3, and/or CHRNB4 could be a unique target for the treatment of cognitive dysfunction in patients with substance dependence and/or other psychiatric disorders.
Our findings could also help in understanding a potential biological mechanism for the link between nicotinic receptor gene variants and tobacco use. A number of studies have shown an association of ND with genetic variants in the CHRNA5–CHRNA3–CHRNB4 gene cluster (Saccone et al, 2007; Bierut et al, 2008; Weiss et al, 2008). However, the mechanism of the positive association has not been elucidated. Several such variants (including CHRNA5-rs16969968, CHRNA5- rs684513, CHRNA3-rs578776, CHRNA3-rs1051730, and CHRNA3-rs3743078) were included in this study. Our findings that a subgroup of EAs homozygous for the major allele of the five variants in the gene cluster made more perseverative responses and perseverative errors and that tobacco use seemed to enhance cognitive flexibility suggest that remediation of cognitive impairments may be a motivator for smoking in some of these individuals. The genetic moderation of brain (or cognitive) function and its effects on smoking behavior have been supported by other studies. Jacobsen et al (2006) showed that the C957T polymorphism (rs6277) of the dopamine D2 receptor gene (DRD2) moderated the effect of nicotine on working memory performance and cortical processing efficiency. A study by Loughead et al (2009) indicated that smokers with the Val/Val genotype of the catechol-O-methyltransferase gene (COMT) Val158Met polymorphism (rs4680) were more sensitive to an abstinence challenge than carriers of the Met allele. Both our data and those of others suggest that increased susceptibility to ND and smoking relapse may be partly due to certain gene variants that compromise prefrontal neural signaling, leading to alterations in cognitivefunction.
The current study has several strengths. First, to our knowledge, it is the largest study of the genetic effects on cognitive function measured using the WCST. Second, the use of three major domains (perseverative responses, perseverative errors, and non-perseverative errors) of the WCST provides a more comprehensive assessment of cognitive flexibility than analysis that focuses on only one domain. An increase in the number of perseverative errors (resulting from poor working memory) has been associated with frontal lobe dysfunction (Monchi et al, 2001). Moreover, a relatively greater increase in perseverative compared with non-perseverative errors may occur either when impairments in working memory are severe or cognitive inflexibility is present (Hartman et al, 2003). Third, using the percentage of WCST responses or errors can more reliably reflect the difference in cognitive flexibility among individual subjects than using absolute numbers of WCST responses or errors.
This study also has limitations. First, there were no data available for exposure to nicotine on the day of testing. Thus, it is unknown whether nicotine exposure close in time to the WCST has a stronger effect on cognition than more distal nicotine exposure that was within the preceding 2 weeks. Second, although current smokers who are recently abstinent may experience nicotine withdrawal, leading to a deleterious effect on cognitive performance, we were unable to address the presence of nicotine withdrawal symptoms in this study. Third, cognitive flexibility is moderated by a number of different genes (in particular, dopamine-related genes), and the role of the nicotinic receptor gene variants in modulating cognitive function may be clearer when they are examined in the context of interaction with other genes. Animal studies have shown that mice deficient in the dopamine transporter (DAT KO) gene exhibited cognitive deficits, as well as substantial differences from wild-type mice, with respect to nicotinic receptor content and function (Weiss et al, 2007). On the basis of these findings, gene–gene interactions, which were not examined here, may be important determinants of cognitive flexibility. Fourth, as multiple markers in the CHRNA5–CHRNA3–CHRNB4 gene cluster showed effects on cognitive flexibility and their statistical significance cannot withstand conservative multiple testing corrections (at α=0.05/(10*3)=0.002 for 10 markers and three WCST indices by Bonferroni's correction), it is unknown whether they are susceptibility loci for cognitive flexibility or are in LD with a functional variant. However, Bonferroni's correction seems to have been too stringent for this study because markers in the CHRNA5–CHRNA3–CHRNB4 gene cluster region are closely correlated (especially in EAs). If we use the program SNPSpD (single–nucleotide polymorphism spectra decomposition; Nyholt, 2004) to correct for multiple testing, with marker LD information taken into consideration, then the experiment-wide significance threshold required to limit the type I error rate to 5% would be 0.006 for AAs and 0.007 for EAs. Thus, the results from markers rs3841324 and rs615470 are significant. Moreover, haplotype analyses with three markers (rs16969968-rs615470-rs578776) in haplotype block 2 showed that three common haplotypes and their interaction with tobacco recency strongly influenced two domains (perseverative responses and perseverative errors) of WCST performance (Table 6). In addition, the potentially functional variant rs3841324 (in which the del-allele was associated with increased expression of CHRNA5 (Wang et al, 2009)) was associated with cognitive flexibility. Nevertheless, the strongly ND-associated SNPs (ie, the nonsynonymous rs16969968 in CHRNA5 and the 3′ UTR rs1051730 in CHRNA3 (Saccone et al, 2007)) showed only a trend toward an effect on cognitive flexibility when they were examined individually. Thus, the challenge here is to distinguish those that are likely to be functional so as to prioritize them for follow-up studies of their function.
To summarize, the present study showed an important role of variants in the CHRNA5–CHRNA3–CHRNB4 gene cluster in regulating cognitive flexibility. On the basis of these findings, nicotinic acetylcholine receptors could be useful targets for pharmacotherapy of cognitive dysfunction in patients with psychiatric and substance dependence disorders, including those with ND.
Acknowledgments
This work was supported in part by funds from the National Institute on Drug Abuse (R01 DA12849, R01 DA12690, R01 DA021264, K24 DA022288, and K99/R00 DA022891), the National Institute on Alcohol Abuse and Alcoholism (R01 AA11330, P50 AA12870, K08 AA13732, and K24 AA13736), and the National Center for Research Resources (M01 RR06192; University of Connecticut General Clinical Research Center); as well as by the US Department of Veterans Affairs (the National Center for PTSD Research, the VA Medical Research Program and the VA Connecticut–Massachusetts Mental Illness Research, Education and Clinical Center (MIRECC), the VA Research Enhancement Award Program (REAP), and the MERIT Program). It was also partially supported by the Alcoholic Beverage Medical Research Foundation (ABMRF) grant (H Zhang). AnnMarie Lacobelle (the VA Connecticut Healthcare Systems, West Haven, CT, USA) provided excellent technical assistance. David Oslin (University of Pennsylvania Medical Center, Philadelphia, PA, USA), Yari Nunez (APT Foundation, New Haven, CT, USA), and Jessica Bona (the University of Connecticut Health Center, Farmington, CT, USA) assisted in WCST data collection. John Farrell provided critical database management services. We thank the individuals who volunteered to participate in this study and the expert interviewers who phenotyped the participants.
Dr Kranzler has received consulting fees from Ortho-McNeil Pharmaceuticals (Raritan, NJ), H Lundbeck A/S (Copenhagen, Denmark), Forest Pharmaceuticals (St Louis, MO), elbion NV (Leuven, Belgium), Sanofi-Aventis (Bridgewater, NJ), Solvay Pharmaceuticals (Brussels, Belgium), and Alkermes (Cambridge, MA). He has received research support from Ortho-McNeil Pharmaceuticals and Bristol-Myers Squibb Company (New York, NY), and honoraria from Forest Pharmaceuticals and Alkermes. Dr Gelernter reports that he has received compensation for professional services in the previous 3 years from the following entities: Yale University School of Medicine, Veterans Affairs Healthcare System (VA), and the National Institutes of Health (NIAAA, NIDA, and NIMH), and related to academic lectures and editorial functions in various scientific venues (including the ACNP). Dr Kranzler has received compensation for professional services from the National Institutes of Health (NIAAA and NIDA) and for academic lectures and editorial functions in various scientific venues (including the ACNP). Other authors have no conflict of interest to report.
References
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canas J, Quesada JF, Antoli A, Fajardo I. Cognitive flexibility and adaptability to environmental changes in dynamic complex problem-solving tasks. Ergonomics. 2003;46:482–501. doi: 10.1080/0014013031000061640. [DOI] [PubMed] [Google Scholar]
- Cincotta SL, Yorek MS, Moschak TM, Lewis SR, Rodefer JS. Selective nicotinic acetylcholine receptor agonists: potential therapies for neuropsychiatric disorders with cognitive dysfunction. Curr Opin Investig Drugs. 2008;9:47–56. [PubMed] [Google Scholar]
- De La Vega FM, Isaac HI, Scafe CR.2006A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns Pac Symp Biocomput 487–498.PMID: 7094263. [PubMed]
- Ernst M, Heishman SJ, Spurgeon L, London ED. Smoking history and nicotine effects on cognitive performance. Neuropsychopharmacology. 2001;25:313–319. doi: 10.1016/S0893-133X(01)00257-3. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Galderisi S, Davidson M, Kahn RS, Mucci A, Boter H, Gheorghe MD, et al. Correlates of cognitive impairment in first episode schizophrenia: the EUFEST study. Schizophr Res. 2009;115:104–114. doi: 10.1016/j.schres.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Greenwood PM, Fossella JA, Parasuraman R. Specificity of the effect of a nicotinic receptor polymorphism on individual differences in visuospatial attention. J Cogn Neurosci. 2005;17:1611–1620. doi: 10.1162/089892905774597281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve KW, Stickle TR, Love JM, Bianchini KJ, Stanford MS. Latent structure of the Wisconsin Card Sorting Test: a confirmatory factor analytic study. Arch Clin Neuropsychol. 2005;20:355–364. doi: 10.1016/j.acn.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, Saccone NL, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M, Steketee MC, Silva S, Lanning K, Andersson C. Wisconsin Card Sorting Test performance in schizophrenia: the role of working memory. Schizophr Res. 2003;63:201–217. doi: 10.1016/s0920-9964(02)00353-5. [DOI] [PubMed] [Google Scholar]
- Heaton, PAR Staff . Psychological Assessment Resources: Odessa, FL; 1999. Wisconsin Card Sorting Test (Computer Version 3 for Windows Research Edition) [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Jacobsen LK, D'Souza DC, Mencl WE, Pugh KR, Skudlarski P, Krystal JH. Nicotine effects on brain function and functional connectivity in schizophrenia. Biol Psychiatry. 2004;55:850–858. doi: 10.1016/j.biopsych.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Jacobsen LK, Krystal JH, Mencl WE, Westerveld M, Frost SJ, Pugh KR. Effects of smoking and smoking abstinence on cognition in adolescent tobacco smokers. Biol Psychiatry. 2005;57:56–66. doi: 10.1016/j.biopsych.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Jacobsen LK, Pugh KR, Mencl WE, Gelernter J. C957T polymorphism of the dopamine D2 receptor gene modulates the effect of nicotine on working memory performance and cortical processing efficiency. Psychopharmacology (Berl) 2006;188:530–540. doi: 10.1007/s00213-006-0469-1. [DOI] [PubMed] [Google Scholar]
- Kandel DB, Chen K. Extent of smoking and nicotine dependence in the United States: 1991–1993. Nicotine Tob Res. 2000;2:263–274. doi: 10.1080/14622200050147538. [DOI] [PubMed] [Google Scholar]
- Lee J, Laurin N, Crosbie J, Ickowicz A, Pathare T, Malone M, et al. Association study of the nicotinic acetylcholine receptor alpha4 subunit gene, CHRNA4, in attention-deficit hyperactivity disorder. Genes Brain Behav. 2008;7:53–60. doi: 10.1111/j.1601-183X.2007.00325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29:299–312. doi: 10.1002/gepi.20098. [DOI] [PubMed] [Google Scholar]
- Loughead J, Wileyto EP, Valdez JN, Sanborn P, Tang K, Strasser AA, et al. Effect of abstinence challenge on brain function and cognition in smokers differs by COMT genotype. Mol Psychiatry. 2009;14:820–826. doi: 10.1038/mp.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Marchant NL, Trawley S, Rusted JM. Prospective memory or prospective attention: physiological and pharmacological support for an attentional model. Int J Neuropsychopharmacol. 2008;11:401–411. doi: 10.1017/S146114570700819X. [DOI] [PubMed] [Google Scholar]
- McGehee DS. Nicotine and synaptic plasticity in prefrontal cortex. Sci STKE. 2007;2007:e44. doi: 10.1126/stke.3992007pe44. [DOI] [PubMed] [Google Scholar]
- Monchi O, Petrides M, Petre V, Worsley K, Dagher A. Wisconsin Card Sorting revisited: distinct neural circuits participating in different stages of the task identified by event-related functional magnetic resonance imaging. J Neurosci. 2001;21:7733–7741. doi: 10.1523/JNEUROSCI.21-19-07733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Raimondi E, Rubboli F, Moralli D, Chini B, Fornasari D, Tarroni P, et al. Chromosomal localization and physical linkage of the genes encoding the human alpha 3, alpha 5, and beta 4 neuronal nicotinic receptor subunits. Genomics. 1992;12:849–850. doi: 10.1016/0888-7543(92)90324-l. [DOI] [PubMed] [Google Scholar]
- Rezvani AH, Levin ED. Cognitive effects of nicotine. Biol Psychiatry. 2001;49:258–267. doi: 10.1016/s0006-3223(00)01094-5. [DOI] [PubMed] [Google Scholar]
- Robinson AL, Heaton RK, Lehman RA, Stilson DW. The utility of the Wisconsin Card Sorting Test in detecting and localizing frontal lobe lesions. J Consult Clin Psychol. 1980;48:605–614. doi: 10.1037//0022-006x.48.5.605. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, et al. 2010Variation in Nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes NeuropsychopharmacologyMay 19; e-pub ahead of print. PMID: 20485328. [DOI] [PMC free article] [PubMed]
- Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA. High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol. 1999;52:295–299. doi: 10.1136/mp.52.5.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JM, Gray JM, Hughes JH, Watson S, Young AH, Ferrier IN. Impaired working memory monitoring in euthymic bipolar patients. Bipolar Disord. 2007;9:478–489. doi: 10.1111/j.1399-5618.2007.00470.x. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RD, Lobos EA, Sun LW, Neuman RJ. Mutational analysis of the nicotinic acetylcholine receptor alpha 4 subunit gene in attention deficit/hyperactivity disorder: evidence for association of an intronic polymorphism with attention problems. Mol Psychiatry. 2003;8:103–108. doi: 10.1038/sj.mp.4001257. [DOI] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von NA, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Nosten-Bertrand M, McIntosh JM, Giros B, Martres MP. Nicotine improves cognitive deficits of dopamine transporter knockout mice without long-term tolerance. Neuropsychopharmacology. 2007;32:2465–2478. doi: 10.1038/sj.npp.1301385. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Characterization of a likelihood based method and effects of markers informativeness in evaluation of admixture and population group assignment. BMC Genet. 2005a;6:50. doi: 10.1186/1471-2156-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005b;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Poling J, Gruen JR, Gelernter J. Cognitive flexibility is associated with KIBRA variant and modulated by recent tobacco use. Neuropsychopharmacology. 2009;34:2508–2516. doi: 10.1038/npp.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FM, Liang Y, Dani JA. Endogenous nicotinic cholinergic activity regulates dopamine release in the striatum. Nat Neurosci. 2001;4:1224–1229. doi: 10.1038/nn769. [DOI] [PubMed] [Google Scholar]
- Ziedonis DM, George TP. Schizophrenia and nicotine use: report of a pilot smoking cessation program and review of neurobiological and clinical issues. Schizophr Bull. 1997;23:247–254. doi: 10.1093/schbul/23.2.247. [DOI] [PubMed] [Google Scholar]