Abstract
Ubiquitin modification of signal transducing receptors at the plasma membrane is necessary for rapid receptor internalization and downregulation. We have investigated whether ubiquitylation alters a receptor cytoplasmic tail to reveal a previously masked internalization signal, or whether ubiquitin itself carries an internalization signal. Using an α–factor receptor–ubiquitin chimeric protein, we demonstrate that monoubiquitin can mediate internalization of an activated receptor that lacks all cytoplasmic tail sequences. Furthermore, fusion of ubiquitin in-frame to the stable plasma membrane protein Pma1p stimulates endocytosis of this protein. Ubiquitin does not carry a functional tyrosine- or di-leucine-based internalization signal. Instead, the three-dimensional structure of the folded ubiquitin polypeptide carries an internalization signal that consists of two surface patches surrounding the critical residues Phe4 and Ile44. We conclude that ubiquitin functions as a novel regulated internalization signal that can be appended to a plasma membrane protein to trigger downregulation.
Keywords: endocytosis/internalization signal/protein targeting/receptor downregulation/ubiquitin
Introduction
Proteins that undergo regulated internalization must either carry an internalization signal that can be activated, or the protein must be modified with a signal in response to environmental cues. One example of an induced protein modification that regulates internalization is the phosphorylation that occurs on the G protein-coupled β-adrenergic receptor in response to ligand binding. Phosphorylated serines in the receptor cytoplasmic tail provide binding sites for arrestin, a protein that serves as a specialized adaptor to link the activated receptor to endocytic machinery (reviewed in Krupnick and Benovic, 1998).
Ubiquitylation is another modification that can regulate receptor internalization (reviewed in Bonifacino and Weissman, 1998; Hicke, 1999). Ubiquitin is a 76-amino-acid polypeptide that is conjugated to substrate proteins through the formation of an isopeptide bond between the ɛ amino group of lysine residues in target proteins and the C–terminal glycine of ubiquitin. Although ubiquitylation of cytosolic proteins and endoplasmic reticulum membrane proteins generally serves as a recognition tag for destruction by the proteasome, ubiquitylation of plasma membrane proteins can serve to target their entry into the endocytic pathway.
In Saccharomyces cerevisiae, G protein-coupled signaling receptors, as well as permeases and transporters, are ubiquitylated at the cell surface (Hicke, 1999 and references therein). The α–factor (Ste2p) and a–factor (Ste3p) receptors bind mating pheromones and the receptor–pheromone interaction initiates a signal transduction pathway that leads to changes in the yeast cell that are required for mating. Upon ligand binding, the receptors are rapidly internalized and transported to the lysosome-like vacuole, where they are permanently inactivated by degradation (Davis et al., 1993; Schandel and Jenness, 1994). Pheromone binding stimulates phosphorylation and ubiquitylation of Ste2p and Ste3p cytoplasmic tails, and both modifications are required for rapid receptor internalization (Reneke et al., 1988; Hicke and Riezman, 1996; Roth and Davis, 1996). A 9-amino-acid sequence in the Ste2p tail, SINNDAKSS, promotes internalization (Rohrer et al., 1993; Hicke and Riezman, 1996). The serines within this sequence are required for phosphorylation of the receptor; phosphorylation then positively regulates ubiquitylation at the SINNDAKSS lysine (Hicke et al., 1998). Lysine residues in the α–factor receptor tail are critical components of receptor internalization signals because they serve as sites of ubiquitin conjugation (Hicke and Riezman, 1996; Terrell et al., 1998). Unlike recognition and degradation by the proteasome, which generally require the formation of a polyubiquitin chain at least four subunits long, rapid Ste2p internalization occurs after monoubiquitylation of the receptor at a single lysine residue. Furthermore, the fusion of monoubiquitin in-frame to a receptor tail lacking all post-translational ubiquitylation sites is sufficient to promote internalization (Terrell et al., 1998).
Ubiquitin-dependent internalization has also been demonstrated or implicated for cell surface proteins in mammalian cells. A number of proteins that are degraded in the lysosome are ubiquitylated at the cell surface, including the epidermal growth factor (EGF) receptor, the growth hormone receptor and the epithelial Na+ channel (Bonifacino and Weissman, 1998). Ubiquitylation and internalization of several growth factor receptors require the Cbl protooncogene, a ubiquitin protein ligase (Miyake et al., 1998; Joazeiro et al., 1999; Lee et al., 1999; Waterman et al., 1999). Similarly, degradation of the epithelial Na+ channel in the lysosome requires ubiquitylation by the ubiquitin protein ligase Nedd4 (Staub et al., 1997). The growth hormone receptor undergoes ligand-stimulated ubiquitylation and cellular ubiquitylation machinery is also required for its ligand-stimulated internalization. However, in this case ubiquitylation of the receptor itself may not be required (Strous et al., 1996, 1997; Govers et al., 1999).
The mechanism by which ubiquitin directs receptor internalization is unknown. Ubiquitylation of a receptor tail may trigger endocytosis by inducing a structural change in the tail that exposes a previously masked internalization signal. Alternatively, the ubiquitin polypeptide itself may carry internalization information that is appended to an activated receptor. To distinguish between these models we have defined the cis-acting information that is required for ubiquitin-dependent internalization. We have used plasma membrane protein–ubiquitin chimeric proteins to show that receptor tail sequences are not required for internalization. Instead, ubiquitin itself carries the information necessary to promote internalization of cell surface proteins. The internalization signal within ubiquitin is not a linear peptide sequence, but consists of two patches on the three-dimensional surface of the polypeptide in which Phe4 and Ile44 are the crucial residues.
Results
Truncated receptors fused to ubiquitin internalize α–factor rapidly
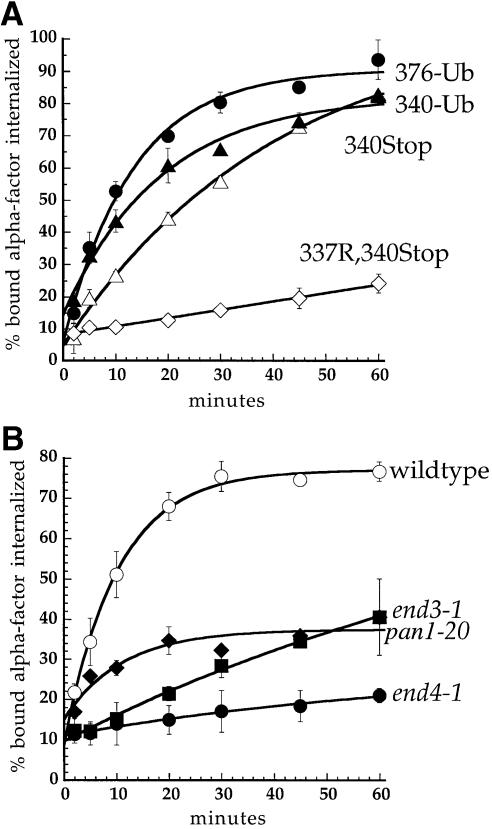
The α–factor receptor carries two strong ubiquitylation sites in its cytoplasmic tail (see Figure 1). These residues, Lys337 and Lys374, can individually mediate rapid rates of ligand-stimulated receptor internalization (M.Houlberg, S.C.Shih and L.Hicke, unpublished data). Receptors truncated following each of these sites, at amino acid 340 or 378, undergo ligand-stimulated internalization that depends on lysines present in the tail (Figure 2A; Terrell et al., 1998). We have previously shown that ubiquitin fused in-frame to Ste2p at amino acid 376 (Ste2p-376–Ub, Figure 1) can rescue internalization of the receptor lacking its ubiquitylation sites (Terrell et al., 1998). To determine whether ubiquitin could also rescue internalization of a more severely truncated receptor, we fused ubiquitin to the receptor at amino acid 340 immediately following the SINNDAK337SS sequence (Ste2p-340–Ub, Figure 1). Ste2p-340–Ub carried a mutation of Lys337 to Arg, to eliminate its only post-translational ubiquitylation site [Lys304 is not ubiquitylated (Hicke and Riezman, 1996)]. Ubiquitin was fused to receptors following a 15-amino-acid Gly/Ser linker that was included to allow independent folding of the receptor tail and ubiquitin domains (Robinson and Sauer, 1996). In addition, Lys48 of ubiquitin was mutated to arginine to remove the primary site of polyubiquitin chain formation, thereby preventing potential degradation of the chimeric protein during transit through the biosynthetic pathway. Chimeric receptors containing a ubiquitin moiety fused either at amino acid 340 or 376 rapidly internalized α–factor (Figure 2A). This internalization was directed by the fused ubiquitin moiety, not by ubiquitin that may become conjugated to one of the seven lysine residues of the fused ubiquitin, because internalization was also rapid when ubiquitin lacking all its lysines is fused to the receptor in the same way (Terrell et al., 1998). Furthermore, fused ubiquitin was not conjugated to another ubiquitin moiety through its C–terminus because mutating or deleting the ubiquitin C–terminal glycines (Gly75 and Gly76) that are important for isopeptide bond formation had no effect on internalization of the chimeric receptor (unpublished data).
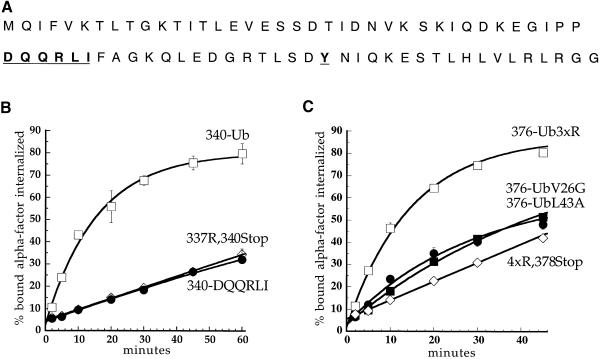
Fig. 1. Cytoplasmic tail variants of Ste2p. The cytoplasmic tail of Ste2p (residues 298–431) is represented schematically. Filled circles, lysine ubiquitylation sites; open circles, lysine→arginine mutations; TMD, transmembrane domain; (g)15, Gly/Ser linker; UB, ubiquitin.
Fig. 2. Ubiquitin fused in-frame to Ste2p at amino acids 340 or 376 directs internalization of Ste2p–Ub chimeric proteins. All curves represent the average of at least three independent experiments and error bars depict the standard deviation. (A) Ste2p and Ste2p–Ub variants were introduced into ste2Δ cells and the resulting strains were assayed for their ability to internalize 35S-labeled pheromone. Cells were harvested and incubated with [35S]α–factor at 4°C, unbound α–factor was removed, and casamino acid medium pre-warmed to 30°C was added to initiate internalization. At various times aliquots of the cells were removed and the ratio of internal to total cell-associated α–factor was measured. Internalization half-times were determined by a simple exponential curve fit using Kaleidagraph software. Ste2p- 376–Ub (LHY558, •; t1/2 = 9.2 min); Ste2p-340–Ub (LHY844, ▴; t1/2 = 12.8 min); Ste2p-340Stop (LHY847, ▵; t1/2 = 28.3 min); Ste2p-337R,340Stop (LHY319, ⋄; t1/2 >200 min). (B) Wild-type (LHY558, ○), end3-1 (LHY1488, ▪), end4-1 (LHY1490, •) or pan1-20 (LHY1580, ♦) strains expressing Ste2p-376–Ub (LHP361) as their sole source of Ste2p were propagated in casamino acid medium and shifted to the non-permissive temperature of 37°C for 20 min. 35S–labeled α–factor was added and the ratio of internalized to total cell-associated α–factor was determined at different time points.
Internalization of α–factor by Ste2p is dependent on a number of proteins identified in genetic screens for endocytosis mutants (Raths et al., 1993; Munn and Riezman, 1994; Munn et al., 1995; Wendland et al., 1996). To ensure that the Ste2p–Ub chimeric proteins internalized α–factor by the same pathway as wild-type Ste2p, we tested whether rapid internalization of pheromone by Ste2p-376–Ub was dependent on the End3, End4 and Pan1 proteins. end3-1, end4-1 and pan1-20 temperature-sensitive mutant strains expressing Ste2p-376–Ub or Ste2p-340–Ub were unable to internalize α–factor rapidly at the non-permissive temperature of 37°C (Figure 2B; K.E.Sloper-Mould and L.Hicke, unpublished data). These data indicate that similar endocytic machinery is required to internalize wild-type Ste2p and the receptor–ubiquitin chimeras. In addition, they demonstrate that End3p, End4p and Pan1p function to promote endocytosis downstream of receptor ubiquitylation.
Ubiquitin carries information sufficient for internalization
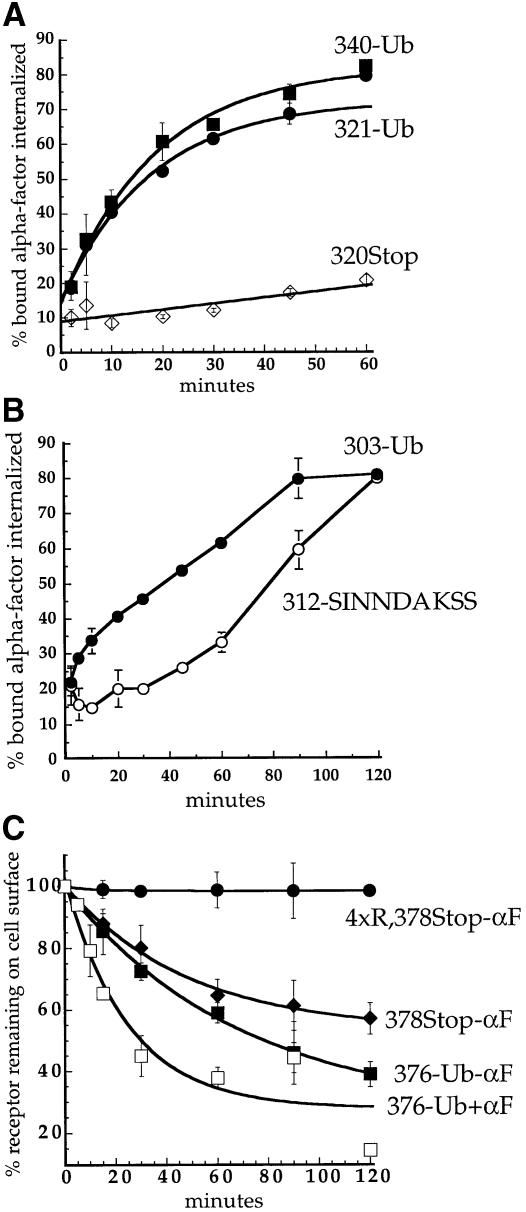
In Ste2p-340–Ub, ubiquitin is fused to the receptor immediately after the SINNDAKSS internalization signal. The SINNDAKSS sequence is the only information necessary for internalization of the truncated receptor Ste2p-345Stop because deletion of residues upstream of SINNDAKSS in this truncated receptor has little or no effect on α–factor internalization (Rohrer et al., 1993). The SINNDAKSS sequence contains a strong ubiquitylation site at Lys337 and three serine residues that are required for phosphorylation of Ste2p-345Stop (Hicke et al., 1998). To determine whether Ste2p phosphorylation is required in addition to ubiquitylation to mediate rapid internalization, we mutated the three SINNDAKSS serines to alanines in Ste2p-340–Ub. Mutating these serines had little effect on internalization of α–factor by the chimeric receptor (S.C.Shih and L.Hicke, unpublished data). To determine whether any other SINNDAKSS residues are required together with ubiquitin to mediate Ste2p internalization, we created a chimeric receptor that carried ubiquitin fused at amino acid 321, upstream of SINNDAKSS (Ste2p-321–Ub, see Figure 1). Ste2p simply truncated at amino acid 320 did not internalize α–factor and carried no sequences important for internalization (Rohrer et al., 1993; Figure 3A). In contrast, Ste2p-321–Ub internalized α–factor as rapidly as Ste2p-340–Ub (Figure 3A). These data demonstrate that the SINNDAKSS sequence and surrounding residues are not required for internalization of a receptor that carries a ubiquitin moiety.
Fig. 3. Ubiquitin is sufficient to internalize Ste2p. (A and B) α–factor internalization assays were performed on cells expressing Ste2p mutants as described for Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. Internalization half-times were determined by an exponential curve fit using Kaleidagraph software, except for Ste2p-312–SINNDAKSS, which required a linear curve fit. (A) Ste2p-340–Ub (LHY844, ▪; t1/2 = 12.8 min); Ste2p-320Stop (LHY852, ⋄; t1/2 >200 min); Ste2p-321–Ub (LHY1194, •; t1/2 = 18.5 min). (B) Ste2p-303–Ub (LHY1212, •; t1/2 = 57.6 min); Ste2p-312–SINNDAKSS (LHY1078, ○; t1/2 = ∼76 min). The fused ubiquitin moiety in Ste2p-303–Ub and the lysine ubiquitin acceptor site in Ste2p-312–SINNDAKSS were approximately the same distance from the plasma membrane in these mutants (see Figure 1). (C) Assays that measure the number of α–factor binding sites present at the cell surface at various times after internalization at 30°C in the presence or absence of pheromone were performed on cells expressing different Ste2p variants (see Materials and methods). Ste2p-4xR,378Stop (LHY319, no α–factor, •); Ste2p-378Stop (LHY18, no α–factor, ♦); Ste2p-376–Ub (LHY558, no α–factor, ▪; + α–factor, □).
Ste2p truncated further upstream at amino acid 312 carries only 14 amino acids of its cytoplasmic tail and is unable to internalize α–factor. Fusing the SINNDAKSS ubiquitylation motif to the severely truncated Ste2p-312Stop receptor can restore internalization (Rohrer et al., 1993). Although internalization of Ste2p-312–SINNDAKSS is relatively slow, it is absolutely dependent on the SINNDAKSS lysine residue, demonstrating that fusion of a post-translation ubiquitylation site just 20 amino acids distal to the transmembrane domain can mediate internalization (Rohrer et al., 1993). To test whether a ubiquitin moiety is sufficient for receptor internalization we fused ubiquitin to Ste2p at amino acid 303. Together with its Gly/Ser linker, this places ubiquitin approximately as far from the last transmembrane domain of the receptor as ubiquitin that is post-translationally conjugated to Ste2p-312–SINNDAKSS, but the Ste2p-303–Ub chimera carries just six amino acids of the receptor tail (see Figure 1). Figure 3B shows that the Ste2p-303–Ub chimera internalized α–factor, indicating that ubiquitin can promote receptor internalization in the absence of almost the entire receptor tail.
Wild-type α–factor receptors are ubiquitylated and internalized at a slow rate in the absence of pheromone (Hicke et al., 1998); however, a truncated receptor that lacks ubiquitylation sites is not constitutively internalized (Figure 3C). We tested whether the fusion of ubiquitin was sufficient to induce constitutive internalization of this truncated receptor. To assay constitutive internalization of a receptor–ubiquitin chimera we measured the clearance of α–factor binding sites from cells expressing Ste2p-376–Ub. In the absence of pheromone, Ste2p-376–Ub was internalized slightly faster than Ste2p-378Stop, which required post-translational ubiquitylation (Figure 3C). This observation indicates that the presence of a ubiquitin moiety fused to a receptor is sufficient to induce receptor endocytosis even in the absence of α–factor. The addition of ligand further stimulated the internalization of Ste2p-376–Ub (Figure 3C), indicating that ubiquitylation is not the only ligand-stimulated event that promotes internalization. The ligand-induced increase in the internalization rate may result from the relief of steric hindrance due to dissociation of the receptor-associated heterotrimeric G protein. Alternatively, ligand stimulation may lead to modifications of components of the endocytic machinery that enhance their activity, as has been demonstrated in mammalian cells (Slepnev et al., 1998; Wilde et al., 1999).
Fusion of ubiquitin can induce the endocytosis of a stable plasma membrane protein
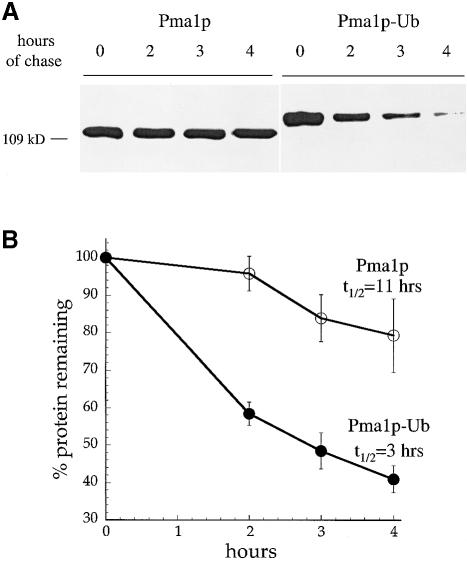
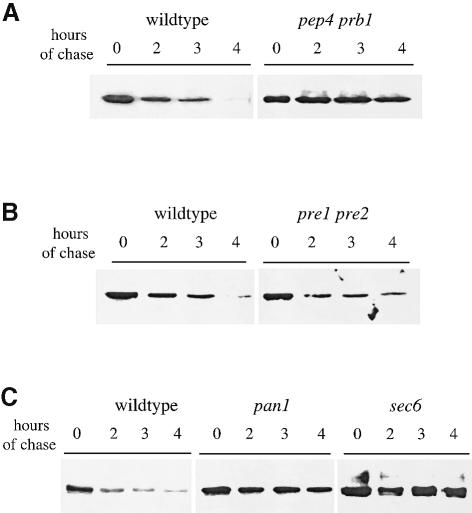
If ubiquitin is a sufficient signal for endocytosis, then it should be able to direct internalization of a heterologous plasma membrane protein. Pma1p is an abundant and stable plasma membrane proton ATPase with a half-life of >11 h (Benito et al., 1991; Figure 4). Previously, it was shown that the fusion of sequences that serve as ubiquitylation motifs from the Ste6 peptide transporter or the a–factor receptor could induce rapid internalization and degradation of Pma1p from the cell surface (Kölling and Losko, 1997; Roth et al., 1998). We fused ubiquitin to the C-terminal amino acid of a hemagglutinin (HA)-epitope-tagged version of Pma1p under control of the glucose-repressible GAL1 promoter. The levels of Pma1p and Pma1p–Ub were analyzed at various times after the repression of new synthesis by incubation of cells in glucose. Figure 4A shows that Pma1p–Ub was degraded more rapidly than Pma1p. Quantification of these data indicated that appending ubiquitin to Pma1p decreased its half-life from 11 to 3 h (Figure 4B). To confirm that Pma1p–Ub was degraded in the vacuole after being internalized from the cell surface, and not by the proteasome in the biosynthetic pathway, we expressed Pma1p and Pma1p–Ub in pep4 prb1 cells that lack active vacuolar hydrolases (Klionsky et al., 1990) and in pre1 pre2 cells that carry defective proteolytic subunits of the proteasome (Heinemeyer et al., 1991, 1993). Figure 5A and B shows that the degradation of HA-Pma1p–Ub depended on active vacuolar hydrolases but not on the proteasome.
Fig. 4. Ubiquitin fused to the C-terminus of Pma1p increases its turnover rate. Strains carrying GAL-HA-PMA1 or GAL-HA-PMA1-UBI plasmids were grown to log phase in casamino acid medium containing 2% galactose and 1% raffinose to induce protein expression from the GAL1 promoter. The cells were harvested, resuspended in glucose-containing medium to inhibit Pma1p or Pma1p–Ub synthesis, and allowed to recover for 20 min at 30°C. An aliquot of cells was removed immediately (time = 0 h) and additional aliquots were removed after 2, 3 and 4 h. Cell lysates were prepared, and proteins were separated by SDS–PAGE on 6% gels and analyzed by immunoblotting with anti-HA antiserum. The immunoblots shown were developed with chemiluminescence and half-times of degrad- ation were calculated from blots of the same lysates developed with chemifluorescence to ensure that the signal was linear. (A) Degradation of Pma1p and Pma1p–Ub expressed in wild-type cells. (B) Degradation of Pma1p and Pma1–Ub turnover shown in (A) was quantified. The curves represent the average of three independent experiments and error bars represent the standard deviations.
Fig. 5. Pma1p–Ub is degraded in the vacuole after internalization from the cell surface. Pma1p and Pma1p–Ub were expressed under control of the GAL1 promoter in different strains. The turnover rate of the Pma1p proteins was measured as described in Figure 4, except that cells were shifted for 20 min to the non-permissive temperature before the first aliquot of cells was withdrawn. (A) Degradation of Pma1p–Ub in pep4 prb1 cells (LHY1479, t1/2 = 15 h) and congenic wild-type cells (LHY1477, t1/2 = 2.8 h) incubated at 37°C. (B) Degradation of Pma1p–Ub in pre1 pre2 cells (LHY1473, t1/2 = 2.9 h) and congenic wild-type cells (LHY1471, t1/2 = 4.2 h) incubated at 38°C. (C) Degradation of Pma1p–Ub in pan1-20 cells (LHY1469; t1/2 = 13.4 h), sec6 cells (LHY1487; t1/2 = 11.5 h) and wild-type cells (LHY1465; t1/2 = 3 h) incubated at 37°C. The immunoblots shown were developed with chemiluminescence and half-times of degradation were calculated from blots of the same lysates developed with chemifluorescence to ensure that the signal was linear.
Since one mutant form of Pma1p, Pma1-7p, can move directly from the Golgi to the vacuole and is degraded without traveling to the cell surface (Chang and Fink, 1995), we asked whether degradation of Pma1p–Ub required transport to and internalization from the plasma membrane. Mutations in the SEC6 gene block transport of proteins from secretory vesicles to the plasma membrane but do not affect the transport of proteins that arrive at the vacuole through intracellular pathways. In particular, the degradation of Pma1-7p is not inhibited in a sec6 mutant (Chang and Fink, 1995). Figure 5C shows that Pma1p–Ub degradation was inhibited in sec6 cells incubated at the non-permissive temperature. A similar result was observed in other mutants, sec1 and sec4 cells, that block the fusion of transport vesicles with the plasma membrane (S.C.Shih and L.Hicke, unpublished data). These data indicate that Pma1p–Ub must reach the cell surface to be degraded, suggesting that degradation occurs after internalization from the plasma membrane and subsequent transport to the vacuole. Furthermore, Pma1p–Ub degradation was blocked in pan1-20 cells (Figure 5C), which are defective in the endocytosis of wild-type Ste2p and Ste2p–Ub but deliver proteins normally from the biosynthetic pathway to the vacuole (Wendland et al., 1996; Figure 2B). Together, these experiments demonstrate that fusing ubiquitin to the C-terminus of Pma1p increased internalization of the protein from the cell surface.
Ubiquitin-dependent internalization is not mediated by a linear peptide signal but requires the three-dimensional structure of the folded ubiquitin polypeptide
Ubiquitin induces internalization of plasma membrane proteins to which it is attached. A simple explanation for this effect would be the presence in the ubiquitin polypeptide of a previously defined internalization signal, an aromatic residue- or di-leucine-based sequence. Transfer of these sequences can induce internalization of heterologous proteins (Collawn et al., 1991; Letourneur and Klausner, 1992). The primary sequence of ubiquitin carries two candidates for these signals: Tyr59, which is exposed on the surface of the molecule (Vijay-Kumar et al., 1985); and DQQRLI, which is similar to the DKQTLL internalization signal that was defined in the CD3 subunit of the T-cell receptor (Letourneur and Klausner, 1992) (Figure 6A). Mutation of Tyr59 to alanine had no effect on the ability of ubiquitin to promote internalization of α–factor by Ste2p-376–Ub (see Figure 7C). A sequence related to the tyrosine-based NPXY signal, NPFXD, has been shown to function as a transferable internalization signal in yeast (Tan et al., 1996). Ubiquitin does not contain an NPFXD sequence although it has two Phe residues on its surface, one of which is important for endocytosis (see below).
Fig. 6. Ubiquitin internalization information is not a tyrosine- or di-leucine-based signal but is carried in the three-dimensional structure of the polypeptide. (A) Sequence of ubiquitin. Shown in bold and underlined are residues that may be part of well-characterized plasma membrane protein internalization motifs. (B and C) α–factor internalization assays were performed on cells expressing different Ste2p and Ste2p–Ub mutants (see Figure 1). Cells expressing these receptors were grown to log phase in SD minimal medium and assayed as described in Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. (B) Ste2p-340–Ub (LHY713, □); Ste2p-337R,340Stop (LHY848, ⋄); Ste2p-340-DQQRLI (LHY1518, •). (C) Ste2p-376–Ub3xR (LHY1127, □); Ste2p-4xR,378Stop (LHY319, ⋄); Ste2p-376–UbV26G (LHY1157, ▪); Ste2p-376–UbL43A (LHY1162, •).
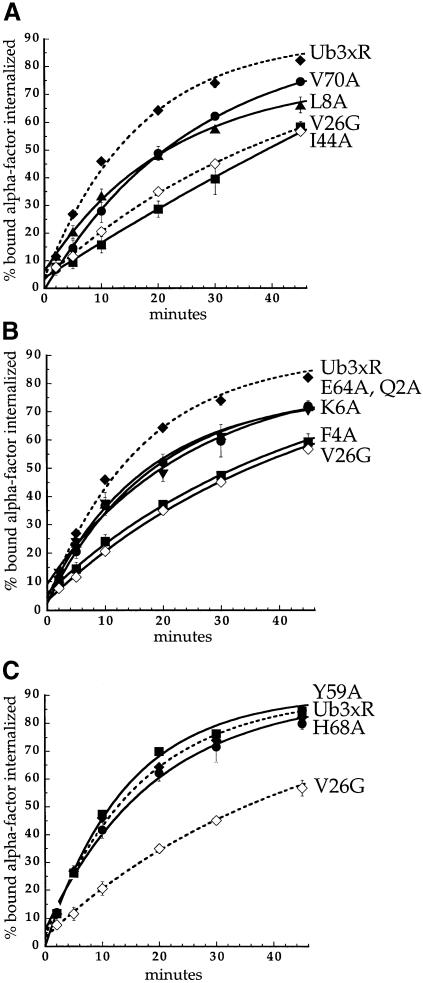
Fig. 7. Mutations in ubiquitin surface residues that inhibit Ste2p–Ub internalization. α–factor internalization assays were performed on cells expressing different Ste2p–Ub mutant receptors. The Ste2p–Ub chimeras carried mutations of ubiquitin lysines 29, 48 and 63 to arginine. Cells expressing these receptors were grown to log phase in SD minimal medium and assayed as described in Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. (A) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbI44A (LHY1186, ▪); Ste2p-376–UbL8A (LHY1340, ▴); Ste2p-376–UbV70A (LHY1338, •). (B) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbF4A (LHY1516, ▪); Ste2p-376–UbQ2A (LHY1386, ▴); Ste2p-376–UbE64A (LHY1596, •); Ste2p-376–UbK6A (LHY1676, ▾). (C) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbY59A (LHY1153, ▪); Ste2p-376–UbH68A (LHY1381, •).
The potential di-leucine motif in ubiquitin actually has the sequence LI, not LL, but LI has been shown to substitute for LL (Letourneur and Klausner, 1992; Pieters et al., 1993; Odorizzi, 1994; Dietrich et al., 1997). The leucine in DQQRLI, Leu43, is a hydrophobic residue buried in the interior of the folded ubiquitin structure (Vijay-Kumar et al., 1985) and therefore mutation of this residue to alanine is likely to disrupt proper folding of the polypeptide (see below). Because critical residues in this putative signal are buried within the folded polypeptide, it is unlikely that DQQRLI is recognized by the endocytic machinery. Nevertheless, to rule out the possibility that DQQRLI is a di-leucine signal in ubiquitin, we tested whether DQQRLI could restore internalization of a version of Ste2p that was truncated at amino acid 340 and lacked its single ubiquitylation site (see Figure 1). It has been shown for the CD3 di-leucine signal that the transfer of six amino acids, DKQTLL, is sufficient to induce the internalization of a heterologous protein (Letourneur and Klausner, 1992). However, DQQRLI did not rescue internalization of pheromone by the truncated receptor, although fusion of the entire ubiquitin polypeptide did (Figure 6B). Thus, DQQRLI does not act as a di-leucine internalization signal, and ubiquitin does not carry one of the classic internalization signals that have been defined previously.
To determine whether ubiquitin carries a linear internalization signal, or whether the three-dimensional folded structure of the ubiquitin polypeptide is necessary for promoting receptor internalization, we made mutations in Ste2p-376–Ub that were expected to disrupt folding of the ubiquitin moiety. Figure 6C shows the rapid internalization of Ste2p-376–Ub3xR, in which Lys29, Lys48 and Lys63 of ubiquitin have been mutated to Arg to prevent all polyubiquitin chain formation (Arnason and Ellison, 1994). The truncated receptor that lacks all post-translational ubiquitylation sites and was not fused to ubiquitin (Ste2p-4xR,378Stop) was internalized 4.3-fold more slowly (Table I). This represents the ubiquitin-independent background level of internalization for this experiment. Mutation of ubiquitin Val26, a buried residue in the three-dimensional ubiquitin structure, to Gly severely destabilizes ubiquitin by retarding folding kinetics (Khorasanizadeh et al., 1996). In addition, UbV26G can not serve as a sole source of ubiquitin for a yeast cell (unpublished data), consistent with an important role for this amino acid in the structure of the protein. The mutation of Val26 to Gly in Ste2p-376–Ub3xR also inhibits internalization of α–factor by the chimeric protein (Figure 6C). To analyze the effect of mutating another buried hydrophobic residue that is likely to be required for protein stability we mutated Leu43 to alanine. Ubiquitin with the mutation Leu43 to Ala also can not serve as the sole source of ubiquitin in the cell (K.E.Sloper-Mould and L.Hicke, unpublished data) and the mutation abolishes the internalization of α–factor by Ste2p-376–Ub3xR (Figure 6C). The effect of mutating Val26 and Leu43 within the ubiquitin hydrophobic core indicates that a properly folded ubiquitin polypeptide is required to promote internalization.
Table I. Effect of surface ubiquitin mutations on endocytosis.
| Mutation | Internalization half-time (min) | Relative inhibition of internalization |
|---|---|---|
| 376-Ub3xR | 11.4 | 1 |
| 4xR,378Stop | 49.5 | 4.3 |
| V26G | 31.3 | 2.7 |
| L43A | 34.7 | 3.0 |
| F4A | 30.2 | 2.6 |
| I44A | 39.4 | 3.4 |
| Q2A | 17.8 | 1.6 |
| K6A | 19.4 | 1.7 |
| L8A | 21.0 | 1.8 |
| E64A | 17.9 | 1.6 |
| V70A | 17.9 | 1.6 |
| Y59A | 9.8 | 0.9 |
| H68A | 12.7 | 1.1 |
The ubiquitin internalization signal is two surface patches bridged by a lysine residue
To define the internalization signal carried in the three-dimensional ubiquitin structure we performed scanning alanine mutagenesis of all the surface residues in the globular domain of ubiquitin. We mutated ubiquitin amino acids individually or in clusters of 2–4 residues in the Ste2p-376–Ub3xR chimeric protein. Most of these mutations had little or no effect on the internalization of α–factor by the chimera (for example see Figure 7C). However, the mutation of two amino acids markedly decreased the internalization rate of Ste2p-376–Ub3xR. The Ile44A and Phe4A mutations decreased the internalization rate 3.4- and 2.6-fold respectively, effects similar to those observed by disrupting the folded structure of ubiquitin (Figure 7A and B; Table I). We defined mutations that internalized α–factor 1.5- to 2-fold more slowly than Ste2p-376–Ub3xR to have a partial defect in α–factor internalization. The mutations to alanine of five amino acids that lie near Phe4 or Ile44 have the most severe defects of mutations in this class. These mutations, Q2A, K6A, L8A, E64A and V70A, decreased internalization of Ste2p–Ub ∼2-fold (Figure 7A and B; Table I). The mutations of other ubiquitin surface residues to alanine in the globular domain resulted in a <1.5-fold decrease in α–factor internalization rates (Figure 7C; K.E.Sloper-Mould and L.Hicke, unpublished data).
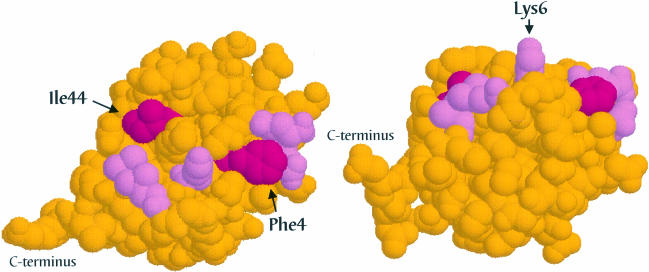
We mapped the amino acids that are required for internalization of Ste2p–Ub on the surface of the ubiquitin polypeptide (Figure 8). We observed that the residues important for internalization are localized to two surface patches that are bridged by Lys6. One of these patches consists of the hydrophobic residues Ile44, Leu8 and Val70. The other patch, Phe4, Gln2 and Glu64, is located on the same side of the ubiquitin molecule but is not contiguous with the first patch. The two patches are bridged by a protruding side chain belonging to Lys6. Mutation of Lys6 has a partial effect on the function of ubiquitin in internalization, whereas mutation of neighboring His68 has little if any effect (Figure 7C). Thus, two surface residues in the globular domain of ubiquitin are critical for its function as an internalization signal. These residues, Ile44 and Phe4, are found on distinct surfaces of the ubiquitin polypeptide and are part of patches formed with nearby residues that play a significant but lesser role in internalization.
Fig. 8. Internalization information carried by ubiquitin is present in two surface patches bridged by a lysine residue. The three-dimensional structure of ubiquitin (Vijay-Kumar et al., 1985) is presented at two different angles in images constructed with RasMol molecular visualization software (Sayle and Milner-White, 1995). Residues essential for internalization are highlighted in magenta; surrounding residues that function in endocytosis but play a lesser role are shown in pink.
Discussion
At least two models exist to explain the mechanism by which ubiquitin triggers internalization of plasma membrane proteins. It has been suggested that ubiquitylation might promote the unfolding of linked proteins (Pickart, 1998); therefore, ubiquitin may expose a concealed internalization signal in endocytic cargo by inducing a structural change in the protein. Alternatively, ubiquitin itself may contain an internalization signal that is appended to a protein. Here we show that ubiquitin carries an endocytosis signal in its amino acid sequence that is sufficient to promote internalization of plasma membrane proteins. This signal is not one of the previously defined linear peptide internalization signals, but consists of patches on the surface of the folded ubiquitin polypeptide.
We have demonstrated that the fusion of ubiquitin in-frame to receptors that lack ubiquitin sites restores the ability of the receptors to be internalized. Endocytosis of the Ste2p–Ub chimeric proteins follows a pathway similar to that taken by wild-type Ste2p because internalization of Ste2p or Ste2p-376–Ub requires the same cellular machinery, including the Pan1, End3 and End4 proteins. The previously defined SINNDAKSS internalization signal is not required for endocytosis of a receptor chimera that carries a ubiquitin moiety. Therefore, SINNDAKSS serves as a strong ubiquitylation motif but does not in itself carry additional information important for receptor internalization. These results suggest that serine phosphorylation is necessary for internalization solely to promote ubiquitylation, although we have not formally demonstrated that the Ste2p–Ub proteins are not phosphorylated in the Gly/Ser linker or on the ubiquitin moiety.
Other ubiquitylation motifs that promote endocytosis have been defined in the cytoplasmic domains of the a–factor receptor, Ste3p, and a peptide transporter, Ste6p (Kölling and Losko, 1997; Roth et al., 1998). A PEST-like sequence that is required for ubiquitylation of the uracil permease has also been identified (Marchal et al., 1998). Like the SINNDAKSS motif, these sequences are rich in serines, threonines and negatively charged amino acids, and include lysine residues. We propose that these motifs may all promote endocytosis by simply serving as sites for interaction with the ubiquitylation machinery, allowing a ubiquitin internalization signal to be appended.
Ubiquitin is able to promote internalization when fused to a severely truncated receptor that carries only six amino acids of its cytoplasmic tail (Ste2p-303–Ub), suggesting that ubiquitin carries information sufficient for internalization. The SINNDAKSS sequence is also able to promote internalization when fused to a receptor that lacks all internalization information (Ste2p-312–SINNDAKSS) (Rohrer et al., 1993). Interestingly, the Ste2p-303–Ub chimera (t1/2 = 57.6 min) is internalized more rapidly than Ste2p-312–SINNDAKSS (t1/2 = 76 min), as is Ste2p-340–Ub (t1/2 = 12.8 min) compared with Ste2p-340Stop (t1/2 = 28.3 min). These observations suggest that ubiquitylation is rate limiting for internalization, at least for truncated versions of the receptor. More evidence that ubiquitin is sufficient for internalization comes from the observation that the fusion of ubiquitin is enough to induce the internalization of a truncated receptor that lacks ubiquitin sites even in the absence of pheromone.
Ubiquitin is not only necessary and sufficient for Ste2p internalization, but can serve as a transferable internalization signal. It was shown previously that large sequences carrying ubiquitylation motifs from Ste3p and Ste6p could be added to the stable plasma membrane ATPase to promote rapid degradation of this protein (Kölling and Losko, 1997; Roth et al., 1998). We have now shown that fusion of only ubiquitin itself to the C-terminus of Pma1p significantly increased turnover of this protein. The increase in degradation induced by ubiquitin was due to increased internalization and subsequent degradation of this protein in the vacuole. Degradation of Pma1p–Ub was prevented in a mutant that blocks the fusion of secretory vesicles with the plasma membrane, in mutants that can not endocytose, and in a mutant that lacks active vacuolar hydrolases. In contrast, mutations that inhibit proteasome activity did not affect Pma1p–Ub degradation, indicating that the proteasome is not involved. Thus, Ste2p–Ub and Pma1p–Ub travel the same endocytic pathway as ubiquitylated wild-type Ste2p and are ultimately degraded in the vacuole. Ubiquitin by itself can function as an internalization signal.
Our results do not rule out the possibility that the optimal function of ubiquitin in promoting internalization is context dependent. On the contrary, we found that the distance of ubiquitin from the plasma membrane influences how efficiently it triggers endocytosis. The internalization of Ste2p-303–Ub (t1/2 = 57.6 min) is less efficient than internalization of Ste2p-321–Ub (t1/2 = 18.5 min) or Ste2p-340–Ub (t1/2 = 12.8 min). Perhaps ubiquitin is more accessible to interacting proteins when it is further from the membrane. The orientation of ubiquitin with respect to the membrane and other endocytic cargo may also affect its efficacy in promoting internalization. Finally, some ubiquitylated plasma membrane proteins such as Ste3p, Gap1p and the EGF receptor also carry Tyr/Phe-based or di-leucine internalization sequences (Chang et al., 1993; Hein and André, 1997; Roth et al., 1998). How these internalization signals function together with ubiquitin remains to be determined.
What is the internalization information carried by ubiquitin? Ubiquitin has one tyrosine residue and a DQQRLI sequence that may act as traditional tyrosine- or di-leucine-based signals. However, we have shown that neither of these established linear signals function to promote endocytosis. Instead, we find that the ubiquitin signal requires three-dimensional structure, since destabilizing the ubiquitin folded structure inactivates the signal. The ubiquitin structure consists of a single globular domain made up of a five-stranded β-sheet and an α-helix. Protruding from this domain is a flexible 4-amino-acid C–terminal tail (Vijay-Kumar et al., 1985). The structure has a dense hydrophobic core and several surface hydrophobic patches. We have found that two patches on the surface of the ubiquitin globular domain carry information necessary for endocytosis of a receptor–ubiquitin chimera; we have not yet determined the role of the ubiquitin tail in endocytosis. Two surface residues, Ile44 and Phe4, were absolutely required for internalization because their mutation to alanine affected Ste2p–Ub internalization as severely as mutations that destabilized the ubiquitin structure. Scanning alanine mutagenesis of other ubiquitin surface residues revealed that the majority of surface amino acids play little or no role in internalization. In most of the ubiquitin protein, even the simultaneous mutation of 3–4 consecutive amino acids had little effect on internalization. Although most of the ubiquitin surface residues are not involved in endocytosis, they do play a significant role in other cellular functions because most mutants could not serve as a sole source of ubiquitin for a yeast cell (K.E.Sloper-Mould and L.Hicke, unpublished data).
Ile44 is part of a hydrophobic surface patch that includes Leu8 and Val70. The residues in this patch are important for the degradation of ubiquitylated proteins by the proteasome (Beal et al., 1996). Leu8 and Val70 play a role in endocytosis, although they are not as important as Ile44. Pickart and colleagues have demonstrated that mutations in these three hydrophobic residues result in mutant ubiquitin molecules that are able to conjugate relatively efficiently to substrate proteins, indicating that the mutant proteins can be recognized by the ubiquitylation machinery and have a reasonably stable structure (Beal et al., 1996). Therefore, the effect of the I44A, L8A and V70A mutations on endocytosis is unlikely to be due to unstable structure of the mutant proteins. Phe4 is the critical residue in another patch on the surface of ubiquitin important for endocytosis. This patch also includes Gln2 and Glu64. It is possible that mutations in Phe4 destabilize the ubiquitin protein; however, the side chain of this residue is completely exposed on the ubiquitin surface (see Figure 8). Furthermore, mutation of Phe4 to Tyr, a conservative substitution that is not expected to disrupt structure, has as severe an effect on endocytosis as mutation to Ala (K.E.Sloper-Mould and L.Hicke, unpublished data). Therefore, we believe that mutation of Phe4 might abrogate endocytosis not by disturbing ubiquitin structure, but by disrupting a protein–protein interaction that promotes ubiquitin-dependent internalization.
In mammalian cells, tyrosine- and di-leucine-based internalization signals promote endocytosis by interacting with adaptors (AP complexes) and clathrin. Yeast AP complexes play little or no role in Ste2p internalization (Huang et al., 1999). In addition, internalization of the EGF receptor, which is ubiquitylated, does not require the AP2 adaptor protein (Nesterov et al., 1999). Therefore, ubiquitin may promote internalization by linking internalized proteins to the endocytic machinery via a specialized type of adaptor. Such a mechanism promotes the internalization of the β-adrenergic receptor. Upon activation, this receptor becomes phosphorylated on serine residues; phosphorylated serines then provide binding sites for the protein arrestin, a specialized adaptor that links activated receptor to the clathrin-based endocytic machinery (Goodman et al., 1996). Our finding that monoubiquitin, in the absence of receptor tail sequences, is sufficient for internalization suggests that an adaptor protein that recognizes monoubiquitin would need to be sequestered away from the free ubiquitin in the cell that might compete with ubiquitylated plasma membrane proteins for binding. This might be accomplished by localization of the adaptor to specific regions of the plasma membrane. Alternatively, an adaptor that recognizes ubiquitin may require a multivalent ubiquitin interaction that occurs non-covalently upon multimerization of ubiquitylated receptors. Perhaps one of the patches important for internalization promotes clustering of endocytic cargo proteins and the other provides a binding site for interacting proteins. Alternatively, the Ile44 and Phe4 patches, bridged by Lys6, could provide binding sites for one or more plasma membrane proteins that would interact with endocytic cargo to promote efficient internalization.
Materials and methods
Strains, plasmids, media and reagents
Yeast strains are listed in Table II. The plasmids containing STE2 variants were transformed into ste2Δ strains by single-step gene transplacement at the ura3 locus. All mutant Ste2 proteins were able to restore mating and α–factor binding to the ste2Δ parental strain. The expression of Ste2p mutant proteins was assessed by immunoblotting using anti-Ste2p polyclonal antibodies. Each mutant Ste2p migrated at the expected molecular weight. Two individual transformants of each mutant were assayed for their ability to internalize α–factor, and in each case both transformants demonstrated similar internalization kinetics.
Table II. Yeast strains.
| Strain | Genotype |
|---|---|
| LHY10 | MATa ura3 ste2::LEU2 leu2 his3 trp1 bar1-1 |
| LHY18 | ura3::ste2-378Stop[URA3], same as LHY10 |
| LHY319 | ura3::ste2-337R,352R,358R,374R,378Stop[URA3], same as LHY10 |
| LHY558 | ura3::ste2-337R,352R,358R,374R,376-UBIK48R[URA3], same as LHY10 |
| LHY713 | ura3::ste2-337R340-UBIK48R[URA3], same as LHY10 |
| LHY844 | ura3::ste2-337R,340-UBIK48R[URA3], same as LHY10 |
| LHY847 | ura3::ste2-340Stop[URA3], same as LHY10 |
| LHY848 | ura3::ste2-337R,340Stop[URA3], same as LHY10 |
| LHY852 | ura3::ste2-320Stop[URA3], same as LHY10 |
| LHY1078 | ura3::ste2-312SINNDAKSS[URA3], same as LHY10 |
| LHY1127 | ura3::ste2-337R,352R,358R,374R376-Ub29R,48R,63R,[URA3], same as LHY10 |
| LHY1153 | ura3::ste2-337R,352R,358R,374R376-Ub29R,48R,Y59A,63R[URA3], same as LHY10 |
| LHY1157 | ura3::ste2-337R,352R,358R,374R376-UbV26G,29R,48R,63R[URA3], same as LHY10 |
| LHY1162 | ura3::ste2-337R,352R,358R,374R-Ub29R,L43A,48R,63R[URA3], same as LHY10 |
| LHY1186 | ura3::ste2-337R,352R,358R,374R-Ub29R,I44A,48R,63R[URA3], same as LHY10 |
| LHY1194 | ura3::ste2-321-UBIK48R[URA3], same as LHY10 |
| LHY1212 | ura3::ste2-303-UBIK48R[URA3], same as LHY10 |
| LHY1338 | ura3::ste2-337R,352R,358R,374R-Ub29R,48R,63R,V70A[URA3], same as LHY10 |
| LHY1340 | ura3::ste2-337R,352R,358R,374R-UbL8A,29R,48R,63R[URA3], same as LHY10 |
| LHY1381 | ura3::ste2-337R,352R,358R,374R-Ub29R,48R,63R,H68A[URA3], same as LHY10 |
| LHY1386 | ura3::ste2-337R,352R,358R,374R-UbQ2A,29R,48R,63R[URA3], same as LHY10 |
| LHY1464 | pGAL-HA-PMA1[URA3] MATa ura3 leu2 his4 lys2 bar1-1 |
| LHY1465 | pGAL-HA-PMA1-UBIK48R[URA3] MATa ura3 leu2 his4 lys2 bar1-1 |
| LHY1469 | pGAL-HA-PMA1-UBIK48R[URA3] MATα pan1-20 ura3 leu2 trp1 lys2 his3 bar1-1 |
| LHY1471 | pGAL-HA-PMA1-UBIK48R[URA3] MATa ura3 leu2-3,112 his3-11,15 |
| LHY1474 | pGAL-HA-PMA1-UBIK48R[URA3] MATa pre1-1 pre2-2 ura3 leu2-3,112 his3-11,15 |
| LHY1477 | pGAL-HA-PMA1-UBIK48R[URA3] MATa ura3-1 leu2-3,112 his3-11 ade2-1 can1-100 |
| LHY1479 | pGAL-HA-PMA1-UBIK48R[URA3] MATa pep4::HIS3 prb1::LEU2 ura3-1 leu2-3,122 his3-11 ade2-1 can1-100 |
| LHY1487 | pGAL-HA-PMA1-UBIK48R[URA3] MATa sec6 his4 leu2 lys2 bar1 |
| LHY1488 | ura3::ste2-337R,352R,358R,374R,376-UBIK48R [URA3] MATa ste2::LEU2 end4-1 ura3 trp1 leu2 bar1 his3 and/or his4 |
| LHY1490 | ura3::ste2-337R,352R,358R,374R,376-UBIK48R[URA3] MATa ste2::LEU2 end3-1 ura3 leu2 his4 ade2 bar1 |
| LHY1516 | ura3::ste2-337R,352R,358R,374R-UbF4A,29R,48R,63R[URA3], same as LHY10 |
| LHY1518 | ura3::ste2-337R,340-DQQRLI[URA3], same as LHY10 |
| LHY1580 | ura3::ste2-337R,352R,358R,374R,376-UBIK48R[URA3] MATa ste2::LEU2 pan1-20 ura3-52 his3-Δ200 trp1Δ901 leu2-3,112 lys2-801 suc2-Δ9 bar1::LYS2 |
| LHY1596 | ura3::ste2-337R,352R,358R,374R-Ub29R,48R,63R,E64A[URA3], same as LHY10 |
| LHY1676 | ura3::ste2-337R,352R,358R,374R-UbK6A,29R,48R,63R[URA3], same as LHY10 |
All mutations in the STE2 sequence were constructed by site-directed mutagenesis using the two-step polymerase chain reaction (PCR) method (Higuchi et al., 1988). DNA sequences of the mutagenized PCR fragments and in-frame ligation junctions were confirmed by automated sequencing. To introduce an in-frame ubiquitin fusion, a unique BamHI site was created at different positions of STE2. This was followed by an in-frame ligation of a PCR-amplified DNA fragment encoding a glycine-rich linker (GGGSGGGTGGGSGGG; Robinson and Sauer, 1996) and UbK48R flanked by BamHI restriction sites. The BamHI site introduced at the codon for amino acid 320 resulted in three amino acid changes: F319L, Y320D and G321P. These residues were mutated back to the original sequence by site-directed mutagenesis resulting in LHP583. To fuse ubiquitin at amino acid 303, a unique NheI site was introduced at amino acid 302 followed by in-frame ligation of DNA encoding glycine-linked UbK48R as described above (LHP611).
Plasmid LHP719 was constructed by amplification of a PCR fragment from pJR3 (Rohrer et al., 1993) using a primer that introduced a BamHI site at the codon for amino acid 340, followed by the sequence encoding DQQRLI-Stop. This PCR product was ligated into a plasmid carrying a BamHI site introduced into STE2 at amino acid 340.
Plasmid LHP609 containing GAL1-HA-PMA1-UBI was derived from pRK315, which contains a unique BamHI site behind the last codon of GAL1-HA-PMA1 (Kölling and Losko, 1997). The ∼250 bp BamHI fragment containing glycine-linked ubiquitin from LHP412 was cloned into the BamHI site of pRK315 and the sequence of the PMA1-UBI junction was confirmed.
All strains were propagated in synthetic minimal (SD) medium (Sherman, 1991), casamino acid medium lacking uracil (0.67% yeast nitrogen base without amino acids, 2% glucose, 0.5% vitamin assay casamino acids from Difco, supplemented with 50 mg/l adenine, histidine, tryptophan and methionine) or rich (YPUAD) medium (2% bacto peptone, 1% yeast extract, 2% glucose). The purification of 35S-labeled α–factor has been described previously (Singer and Riezman, 1990; Dulic et al., 1991). Ste2p polyclonal antibody was purified as described (Hicke and Riezman, 1996). The HA 12CA5 monoclonal antibody was a kind gift from Dr Robert A.Lamb (Northwestern University).
Ubiquitin mutagenesis
All Ste2p-376–Ub ubiquitin mutants were constructed by site-directed mutagenesis of template plasmid LHP587 using specific primers designed to introduce the required mutations (Q2A, F4A, K6A, L8A, V26G, L43A, I44A, Y59A, E64A, H68A or V70A). LHP587 (Ste2p-376–Ub3xR) was constructed in two steps. Initially, the KpnI sites in LHP361 were deleted and the BamHI site at the 3′ end of UBI was mutated to KpnI. Using the resulting plasmid (LHP372) as a template, Lys29 and Lys63 in ubiquitin were mutated to arginines with mutagenic primers and a QuickChange™ Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All mutations were confirmed by automated sequencing.
Receptor clearance and α–factor internalization assays
All internalization assays were performed as described previously (Dulic et al., 1991; Terrell et al., 1998). For each assay, the growth and assay conditions are indicated in the figure legend. Internalization half-times were calculated based on an exponential curve-fit performed with KaleidaGraph software (Synergy Software, Reading, PA).
Receptor clearance assays were performed as described (Hicke et al., 1998). Briefly, cells were treated with 10 μg/μl cycloheximide for 20 min at 30°C to inhibit new protein synthesis. At various times aliquots of cells were transferred into buffer containing 10 mM NaN3 and 10 mM NaF. To measure ligand-stimulated receptor clearance, 1 × 10–6 M synthetic α–factor was added prior to removing the 0 min time point aliquot. Synthetic α–factor was then dissociated from poisoned cells by shaking the cells for 3 h at 30°C. The number of receptors that remained on the cell surface at each time point was determined by incubating cells with 35S-labeled α–factor at a saturating concentration.
Cell lysates and immunoblots
To express Pma1p or Pma1p–Ub from the GAL1 promoter, cells were grown to log phase at the appropriate temperature in casamino acid medium containing 2% galactose and 1% raffinose. Cells were harvested and resuspended to 5 × 106 cells/ml in casamino acid medium containing 2% glucose to inhibit Pma1p or Pma1p–Ub synthesis. After cells were allowed to recover at 30°C, or were shifted to the restrictive temperature for 20 min, a 10 ml aliquot was removed from the culture and resuspended in 20 mM (final concentration) NaF/NaN3 and placed on ice. This represented the 0 h chase time point. After 2, 3 and 4 h, 10 ml aliquots were removed and treated as described above.
The cells were collected by centrifugation at 4°C and washed once in 1 ml TEPI, 5 mM NEM (Terrell et al., 1998) in 2.2 ml microcentrifuge tubes. Cells were resuspended in 100 μl TEPI, ∼0.2 g of glass beads were added, and cells were lysed by vortexing for 4 min at maximum speed. The cell lysate was diluted with 100 μl of urea, 5% SDS buffer (Terrell et al., 1998) and heated to 37°C for 10 min. The diluted lysate was removed and transferred to a 1.5 ml tube, and centrifuged at 4°C for 10 min at 20 800 g to remove cell debris. Bromophenol Blue (0.0002%) and β-mercaptoethanol (2%) were added to the cell lysate and proteins were resolved by 6% SDS–PAGE and transferred to Immobilon membranes (Millipore Corp., Bedford, MA). Membranes were blocked with 5% non-fat dried milk, 0.1% NP-40, 50 mM Tris–HCl pH 7.5, 0.15 M NaCl, followed by incubation with 12CA5 monoclonal HA antibody at 1:5000 dilution. Goat anti-mouse immunoglobulin conjugated with horseradish peroxidase (Sigma, St Louis, MO) was used at 1:5000 dilution and the immunoblot was developed with SuperSignal reagents (Pierce, Rockford, IL). To quantify the degradation of Pma1p and Pma1p–Ub, goat anti-mouse IgG + IgM alkaline phosphatase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as secondary antibody and visualized using ECF substrate (Amersham Pharmacia Biotech, Piscataway, NJ). The intensity of each protein signal was determined using Molecular Dynamics Storm 860 ImageQuant 1.11 software (Sunnyvale, CA). Degradation half-times were determined from a linear curve fit using Microsoft Excel software (Microsoft Corporation).
Acknowledgments
Acknowledgements
GAL-HA-PMA1 plasmids were generously provided by Ralf Kölling (Heinrich-Heine-Universität, Düsseldorf) and James Haber (Brandeis University). Monoclonal antiserum against the HA epitope (12CA5) was a gift from Robert Lamb (Northwestern University). Howard Riezman kindly provided numerous plasmids and strains. We thank Rebecca Dunn, Rick Gaber and Andreas Matouschek for helpful discussion. The manuscript was improved by comments from Amy deHart, Rebecca Dunn, Dan Linzer, Andreas Matouschek and Joshua Schnell. S.C.S. was supported by NIH Training Grant T32GM08061. The Burroughs Wellcome Fund, the Searle Scholars Program and the National Institutes of Health (R01 DK53257) supported this research.
References
- Arnason T. and Ellison, M.J. (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol., 14, 7876–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal R., Deveraux, Q., Xia, G., Rechsteiner, M. and Pickart, C. (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl Acad. Sci. USA, 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B., Moreno, E. and Lagunas, R. (1991) Half-life of the plasma membrane ATPase and its activating system in resting yeast cells. Biochim. Biophys. Acta, 1063, 265–268. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman, A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. and Fink, G.R. (1995) Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol., 128, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-P., et al. (1993) Ligand-induced internalization of the epidermal growth factor receptor is mediated by multiple endocytic codes analogous to the tyrosine motif found in constitutively internalized receptors. J. Biol. Chem., 268, 19312–19320. [PubMed] [Google Scholar]
- Collawn F.J., Kuhn, L.A., Liu, L.-F.S., Trainer, J.A. and Trowbridge, I.S. (1991) Transplanted LDL and mannose-6-phosphate receptor internalization signals promote high-efficiency endocytosis of the transferrin receptor. EMBO J., 10, 3247–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N.G., Horecka,J.L. and Sprague,G.F. (1993) Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol., 122, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J., Kastrup, J., Nielsen, B., Odum, N. and Geisler, C. (1997) Regulation and function of the CD3-γ DxxxLL motif: a binding site for adaptor protein-1 and adaptor protein-2 in vitro. J. Cell Biol., 138, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V., Egerton, M., Elguindi, I., Raths, S., Singer, B. and Riezman, H. (1991) Yeast endocytosis assays. Methods Enzymol., 194, 697–710. [DOI] [PubMed] [Google Scholar]
- Goodman O.B. Jr Krupnick, J.G., Santini, F., Gurevich, V.V., Penn, R.B., Gagnon, A.W., Keen, J.H. and Benovic, J.L. (1996) β-Arrestin acts as a clathrin adaptor in endocytosis of the β2-adrenergic receptor. Nature, 383, 447–450. [DOI] [PubMed] [Google Scholar]
- Govers R., ten Broeke, T., van Kerkhof, P., Schwartz, A.L. and Strous, G.J. (1999) Identification of a novel conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J., 18, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein C. and André, B. (1997) A C-terminal di-leucine motif and nearby sequences are required for NH4+-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol. Microbiol., 24, 607–616. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W., Kleinschmidt, J.A., Saidowsky, J., Escher, C. and Wolf, D.H. (1991) Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J., 10, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Gruhler, A., Möhrle, V., Mahé, Y. and Wolf, D.H. (1993) PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem., 268, 5115–5120. [PubMed] [Google Scholar]
- Hicke L. (1999) Gettin' down with ubiquitin: turning off cell surface receptors, transporters and channels. Trends Cell Biol., 9, 107–112. [DOI] [PubMed] [Google Scholar]
- Hicke L. and Riezman, H. (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell, 84, 277–287. [DOI] [PubMed] [Google Scholar]
- Hicke L., Zanolari, B. and Riezman, H. (1998) Cytoplasmic tail phosphorylation of the α–factor receptor is required for its ubiquitination and internalization. J. Cell Biol., 141, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel, B. and Saiki, R.K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res., 16, 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.M., D'Hondt, K., Riezman, H. and Lemmon, S.K. (1999) Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J., 18, 3897–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A., Wing, S.S., Huang, H., Leverson, J.D., Hunter, T. and Liu, Y.C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin–protein ligase. Science, 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S., Peters, I.D. and Roder, H. (1996) Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nature Struct. Biol., 3, 193–205. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Herman, P.K. and Emr, S.D. (1990) The fungal vacuole: composition, function and biogenesis. Microbiol. Rev., 54, 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R. and Losko, S. (1997) The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J., 16, 2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnick J. and Benovic, J.L. (1998) The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol., 38, 289–319. [DOI] [PubMed] [Google Scholar]
- Lee P.S., Wang, Y., Dominguez, M.G., Yeung, Y.G., Murphy, M.A., Bowtell, D.D. and Stanley, E.R. (1999) The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis and attenuates macrophage proliferation. EMBO J., 18, 3616–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F. and Klausner, R.D. (1992) A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell, 69, 1143–1157. [DOI] [PubMed] [Google Scholar]
- Marchal C., Haguenauer-Tsapis, R. and Urban-Grimal, D. (1998) A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol., 18, 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S., Lupher, M.L., Jr, Druker, B. and Band, H. (1998) The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor α. Proc. Natl Acad. Sci. USA, 95, 7927–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A.L. and Riezman, H. (1994) Endocytosis is required for the growth of vacuolar H+-ATPase-defective yeast: identification of six new END genes. J. Cell Biol., 127, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A.L., Stevenson,B.J., Geli,M.I. and Riezman,H. (1995) end5, end6 and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell, 6, 1721–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov A., Carter, R.E., Sorkina, T., Gill, G.N. and Sorkin, A. (1999) Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant μ2 subunit and its effects on endocytosis. EMBO J., 18, 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi C.G., Trowbridge, I.S., Xue, L., Hopkins, C.R., Davis, C.D. and Collawn, J.F. (1994) Sorting signals in the MHC class II invariant chain cytoplasmic tail and transmembrane region determine trafficking to an endocytic processing compartment. J. Cell Biol., 126, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (1998) Polyubiquitin chains. In Peters,J.M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY, pp. 19–63. [Google Scholar]
- Pieters J., Bakke, O. and Dobberstein, B. (1993) The MHC class II-associated invariant chain contains two endosomal targeting signals within its cytoplasmic tail. J. Cell Sci., 106, 831–846. [DOI] [PubMed] [Google Scholar]
- Raths S., Rohrer, J., Crausaz, F. and Riezman, H. (1993) end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol., 120, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneke J.E., Blumer, K.J., Courchesne, W.E. and Thorner, J. (1988) The carboxy-terminal segment of the yeast α–factor receptor is a regulatory domain. Cell, 55, 221–234. [DOI] [PubMed] [Google Scholar]
- Robinson C.R. and Sauer, R.T. (1996) Covalent attachment of Arc repressor subunits by a peptide linker enhances affinity for operator DNA. Biochemistry, 35, 109–116. [DOI] [PubMed] [Google Scholar]
- Rohrer J., Bénédetti, H., Zanolari, B. and Riezman, H. (1993) Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled α-pheromone receptor in yeast. Mol. Biol. Cell, 4, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A.F. and Davis, N.G. (1996) Ubiquitination of the yeast a–factor receptor. J. Cell Biol., 134, 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A.F., Sullivan, D.M. and Davis, N.G. (1998) A large PEST-like sequence directs the ubiquitination, endocytosis and vacuolar degradation of the yeast a–factor receptor. J. Cell Biol., 142, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle R.A., Milner-White, E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- Schandel K.A. and Jenness, D.D. (1994) Direct evidence for ligand-induced internalization of the yeast α–factor pheromone receptor. Mol. Cell. Biol., 14, 7245–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Singer B. and Riezman, H. (1990) Detection of an intermediate compartment involved in transport of α–factor from the plasma membrane to the vacuole in yeast. J. Cell Biol., 110, 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev V.I., Ochoa, G.-C., Butler, M.H., Grabs, D., De Camilli, P. (1998) Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science, 281, 821–824. [DOI] [PubMed] [Google Scholar]
- Staub O., Gautschi, I., Ishikawa, T., Breitschopf, K., Ciechanover, A., Schild, L. and Rotin, D. (1997) Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J., 16, 6325–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous G., van Kerkhof, P., Govers, R., Ciechanover, A. and Schwartz, A.L. (1996) The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J., 15, 3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Strous G., van Kerkhof, P., Govers, R., Rotwein, P. and Schwartz, A. (1997) Growth hormone-induced signal transduction depends on an intact ubiquitin system. J. Biol. Chem., 272, 40–43. [DOI] [PubMed] [Google Scholar]
- Tan P.K., Howard, J.P. and Payne, G.S. (1996) The sequence NPFXD defines a new class of endocytosis signal in Saccharomyces cerevisiae. J. Cell Biol., 135, 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J., Shih, S., Dunn, R. and Hicke, L. (1998) A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell, 1, 193–202. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg, C.E., Wilkinson, K.D. and Cook, W.J. (1985) Three-dimensional structure of ubiquitin at 2.8 Å resolution. Proc. Natl Acad. Sci. USA, 82, 3582–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman H., Levkowitz, G., Alroy, I. and Yarden, Y. (1999) The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem., 274, 22151–22154. [DOI] [PubMed] [Google Scholar]
- Wendland B., McCaffery, J.M., Xiao, Q. and Emr, S.D. (1996) A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol., 135, 1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Beattie, E.C., Lem, L., Riethof, D.A., Liu, S.H., Mobley, W.C., Soriano, P. and Brodsky, F.M. (1999) EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell, 96, 677–687. [DOI] [PubMed] [Google Scholar]