Abstract
The current study focuses on the molecular mechanisms responsible for actin assembly on a defined membrane surface: the phagosome. Mature phagosomes were surrounded by filamentous actin in vivo in two different cell types. Fluorescence microscopy was used to study in vitro actin nucleation/polymerization (assembly) on the surface of phagosomes isolated from J774 mouse macrophages. In order to prevent non–specific actin polymerization during the assay, fluorescent G–actin was mixed with thymosin β4. The cytoplasmic side of phagosomes induced de novo assembly and barbed end growth of actin filaments. This activity varied cyclically with the maturation state of phagosomes, both in vivo and in vitro. Peripheral membrane proteins are crucial components of this actin assembly machinery, and we demonstrate a role for ezrin and/or moesin in this process. We propose that this actin assembly process facilitates phagosome/endosome aggregation prior to membrane fusion.
Keywords: actin assembly/ezrin–radixin–moesin family/latex beads/phagocytosis
Introduction
Although it has been recognized for many years that de novo actin assembly in cells occurs mainly on the cytoplasmic surface of membranes (Tilney, 1976), this process has remained elusive. Identification of the mechanistic details of this crucial cell function has been hampered both by the complexity and rapidity of its actions, and by the lack of eukaryotic membrane model systems that are suitable for both in vivo and in vitro analyses. Rapid polymerization of actin is widely thought to be responsible for the generation of ‘driving’ forces allowing the plasma membrane to be somehow ‘pushed’ in structures such as microvilli, lamellipod or pseudopod extensions (Condeelis et al., 1988; Small et al., 1995; Mitchison and Cramer, 1996). In all membrane systems examined so far, actin assembles on membranes in a manner that is quite different from the better understood microtubules. The latter are nucleated within defined structures such as the perinuclear microtubule organizing center, and grow by addition of tubulin monomers to the end away from the nucleator. In contrast, actin monomers are inserted ‘at’ the membrane where the fast growing ‘barbed’ or plus ends of the actin filaments are invariably located (Tilney, 1976; Tilney and Portnoy, 1989; Hartwig, 1992; Small et al., 1995; Mitchison and Cramer, 1996; Carlier, 1998).
Eukaryotic cells express a large number of actin-binding proteins (ABPs); their activities not only allow a network of cross-linked actin filaments to be formed for mechanical rigidity, but also regulate the rapid turnover of actin filaments necessary for many cell motility processes (Pollard and Cooper, 1986; Vandekerckhove and Vancompernolle, 1992; Carlier, 1998). Other ABPs, such as thymosin β4 (Tβ4), function by sequestering a large pool of monomeric G–actin. This peptide is found especially concentrated (up to 0.5 mM) in macrophages, fibroblasts, neutrophils and platelets (Gondo et al., 1987; Cassimeris et al., 1992; Weber et al., 1992). Upon cell activation, Tβ4 can release ATP G–actin when needed for rapid actin filament assembly (Cassimeris et al., 1992; Weber et al., 1992; Pantaloni and Carlier, 1993). For the main part of the present study, we took advantage of chemically synthesized Tβ4 (Echner and Voelter, 1988) as a tool to buffer free G–actin to a level in between the critical concentrations for monomer addition at the two ends of the actin filament (Weber et al., 1992). In this way, the in vitro growth of actin is restricted to the faster growing, barbed ends of the filaments, as is the case in vivo (Tilney and Portnoy, 1989; Hartwig, 1992; Small et al., 1995; Mitchison and Cramer, 1996).
Among the large number of ABPs described so far, many are capable of inducing or facilitating actin assembly in vitro in the absence of membranes. It has been extremely difficult, however, to assess the possible functions of these proteins in actin assembly as it occurs on a eukaryotic membrane surface under physiological conditions. An increasingly important role for both actin-based motility of the intracellular bacteria Listeria and actin assembly at the leading edge of eukaryotic cells has recently been given to the actin-related protein complex ARP2/3 (Welch et al., 1998; Mullins and Pollard, 1999; Svitkina and Borisy, 1999). ABPs that can bind actin and phosphoinositides on the membrane simultaneously, such as talin, vinculin and the ezrin/radixin/moesin (ERM) proteins, also represent particularly interesting candidates for involvement in actin nucleation on membranes (Niggli et al., 1995; Gilmore and Burridge, 1996; Bretscher, 1999; Mangeat et al., 1999). It has recently been proposed that membrane-bound dimers of talin could nucleate actin in vivo (Isenberg and Goldmann, 1998). The ERM proteins, which belong to the same superfamily as talin, are thought to function as mechanical linkers between the actin cytoskeleton and the plasma membrane (Bretscher, 1989; Hanzel et al., 1991; Algrain et al., 1993; Berryman et al., 1995). The ERM proteins share ∼75% overall amino acid identity and localize to identical subcellular domains in many cell types, raising the possibility that they have at least partially overlapping functions (Doi et al., 1999; reviewed in Tsukita and Yonemura, 1997; Bretscher, 1999; Mangeat et al., 1999).
The first step of phagocytosis is known to be an actin-dependent process. Particles inducing phagocytosis such as erythrocytes, or bacteria covered with ligands such as immunoglobulins induce rapid and massive actin assembly via signal transduction in different cell types, allowing the phagosome enclosing the particle to be formed (Reaven and Axline, 1973; Greenberg et al., 1991; Allen and Aderem, 1995; Caron and Hall, 1998; reviewed in Greenberg and Silverstein, 1993). After the uptake process, most of the F–actin is believed to be lost from the phagosomes; this is a dogma in the field. However, the actin cytoskeleton must also interact with mature phagosomes, since the transport of latex bead phagosomes within macrophages, as well as their ability to fuse in vivo and in vitro with endocytic organelles, can be inhibited by cytochalasin D (cyto. D) and latrunculin A (Toyohara and Inaba, 1989; A.Jahraus, M.Egeberg, A.Habermann, B.Hinner, E.Sackmann, A.Pralle, H.Faulstich, H.Defacque and G.Griffiths, submitted; A.Al-Haddad, M.A.Shonn, B.Redlich, A.Blocker, J.K.Burkhardt, H.Yu, J.A.Hammer III, D.G.Weiss, W.Steffen, G.Griffiths and S.Kuznetsov, submitted). Moreover, actin is found around mature phagosomes in some cell types (Stockem et al., 1983; Kersken et al., 1986; Toyohara and Inaba, 1989), as well as on isolated phagosomes (Desjardins et al., 1994a; Rezabek et al., 1997). Furthermore, many ABPs such as α–actinin, moesin (Desjardins et al., 1994a), MARKS (myristoylated, alanine-rich, C–kinase substrate) (Allen and Aderem, 1995), annexins (Desjardins et al., 1994a; Diakonova et al., 1997), a 30 kDa actin bundling protein (Rezabek et al., 1997) and several myosins (Swanson et al., 1999) are also associated with fully formed phagosomes in different cell types, although the precise roles of these proteins remain unclear.
In this paper, we introduce phagosomes enclosing latex beads as a novel eukaryotic membrane model for analyzing actin assembly on a defined eukaryotic membrane surface. Inert, 1 μm non-degradable beads can be internalized by mouse J774 macrophage cells, leading to the de novo assembly of phagosomes that subsequently mature and show significant and continual biochemical changes at least until 24 h after their internalization (Desjardins et al., 1994a). These easily purified organelles have been used to analyze microtubule binding and bi-directional motility, as well as membrane fusion; all of these functions vary significantly as the phagosome ages, with each of these processes showing a rather unique kinetic pattern (Desjardins et al., 1994b; Blocker et al., 1996, 1997; Griffiths, 1996; Claus et al., 1998; Jahraus et al., 1998). Here we show that in both J774 and NBT-2 cell lines, filamentous actin can be seen in close proximity to many, but not all phagosomes. In J774 cells, this activity was significantly lower at the 12 h phagosome stage. In vitro assays were then set up to monitor actin nucleation/polymerization (referred to for simplicity as ‘assembly’) on purified phagosomes, and led to the finding that these organelles can assemble actin de novo on their cytoplasmic surface. As in vivo, their capacity to assemble actin oscillates with a low point at 12 h. Evidence is also provided that the ERM proteins ezrin and/or moesin are essential for this membrane-bound actin assembly process.
Results
Relationship between actin cytoskeleton and phagosomes in cells
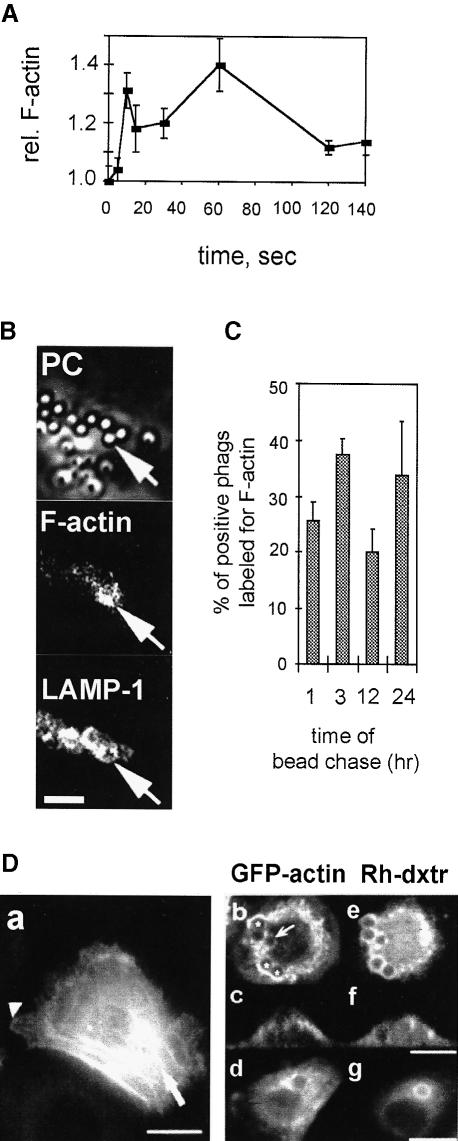
We first asked whether naked latex beads added to J774 cells could induce actin assembly, as has been described for the process of receptor-mediated phagocytosis (Greenberg et al., 1991; Greenberg and Silverstein, 1993; Allen and Aderem, 1996). For this, F–actin content was quantified at different times after bead addition on cells using a standard rhodamine–phalloidin (Rh–Ph) fluorimetric assay. Two major peaks of actin assembly could be seen, a first between 5 and 10 s (129 ± 9% of control values, n = 3), and a second between 60 and 90 s (130 ± 10% of control values, n = 3) (Figure 1A). These changes in F–actin upon phagocytosis of naked latex beads correspond to the formation of pseudopods surrounding the particle to be internalized (Reaven and Axline, 1973; our unpublished results using high resolution cryo-scanning electron microscopy).
Fig. 1. Actin–phagosome interactions at early and later stages of phagocytosis. (A) Latex bead internalization in J774 cells induces a rapid F–actin polymerization/depolymerization cycle. Relative F–actin content of J774 cells following stimulation with latex beads at 0 s. Results are the mean ± SD of one typical experiment performed in triplicate. (B) F–actin labeling on fusogenic phagosomes. J774 cells were pulsed with latex beads for 1 h and chased for 3 h. Indirect immunofluorescence labeling of LAMP–1, Rh–Ph staining (F–actin) and phase-contrast (PC) images of the same x–y optical section. Arrows show phagosomes labeled for both LAMP–1 and F–actin. Bar, 5 μm. (C) Quantitation of phagosomes positive for F–actin per cell. J774 cells were pulsed with latex beads for 1 h and chased for the indicated times. After immunostaining, the percentage of phagosomes positive for both LAMP–1 and F–actin was determined. Results are the mean ± SD of one typical experiment. (D) GFP–actin can localize around fusogenic phagosomes in fibroblasts. (a) NBT-2 cells differentiated by EGF treatment showing GFP–actin localized in stress fibers (arrow) or the leading edge of the cell (arrowhead). (b–g) Differentiated NBT-2 cells that have been pulsed with rhodamine–dextran for 5 min and subsequently internalized 3 μm fibronectin-coated latex beads for 1 h. x–y (b, d, e, g) and x–z (c, f) optical sections showing GFP–actin (b–d) and rhodamine–dextran (Rh-dxtr) (e–g) in cells. (b) Arrow, rhodamine–dextran-positive phagosome weakly labeled for GFP–actin; *, more strongly labeled phagosomes. Bars, 10 μm.
We next searched for the presence of F–actin around fully formed phagosomes in J774 cells. For this, J774 cells were incubated with latex beads for different pulse and chase times, fixed and labeled for their F–actin content with Rh–Ph. The glycosylated lysosome-associated membrane proteins LAMP–1 and -2 are known to be transferred to phagosomes from late endosomes and lysosomes (Desjardins et al., 1994b; Allen and Aderem, 1995). Therefore, in order to visualize phagosomes at stages in which they are fusogenic (Jahraus et al., 1998) and exclude non-internalized beads from the analysis, LAMP–1 was also labeled by indirect immunofluorescence labeling and visualized together with F–actin. In a first set of experiments, cells were pulsed and chased for 5 min with latex beads. High F–actin labeling surrounded all latex beads that were in the process of being internalized (and which were negative for LAMP–1) (data not shown), consistent with all previous data (Greenberg and Silverstein, 1993; Allen and Aderem, 1995; Caron and Hall, 1998). We next pulsed the cells with latex beads for 1 h followed by 3 h of chase.
After this, the phagosomes were positive for LAMP–1 and some of them had significant F–actin labeling (Figure 1B). Different sets of cells were then pulsed with latex beads for 1 h and chased for various times prior to double immunofluorescence labeling. The percentage of LAMP–1-positive phagosomes that were positive for F–actin was determined by direct counting. After a 1 h chase, an average of 25.5% of phagosomes were labeled for actin, and this parameter rose to 37.5% after 3 h. Intriguingly, only 18% of phagosomes were labeled at the 12 h time point with a return to a high value (33%) after a 24 h chase (Figure 1C).
NBT-2 cells have previously been stably transfected with a green fluorescent protein (GFP)–actin construct (Westphal et al., 1997; Choidas et al., 1998). EGF treatment for 24 h allowed these cells to be differentiated into fibroblasts (Valles et al., 1990). After differentiation, GFP–actin was concentrated into stress fibers as well as at the leading edge of the cells, indicating that it incorporates normally into F–actin (Figure 1D) (Choidas et al., 1998). We then pre-labeled early endosomes prior to bead uptake with rhodamine–dextran for 5 min, before adding 3 μm fibronectin-coated latex beads for 1 h followed by 1 h of chase. The beads were coated with fibronectin because those differentiated cells, as for other fibroblastic cells, could undergo phagocytosis mediated by fibronectin receptors (Rabinovitch, 1995). Labeling of phagosomes with rhodamine–dextran was taken as a measure of fusion with late endocytic organelles. Many fusogenic phagosomes were clearly surrounded by GFP–actin structures (Figure 1D). The labeling was not homogeneous since some phagosomes were negative (data not shown), or weakly positive (Figure 1Db). Phagosome-associated actin did not seem to be related to the cell cortex but rather surrounded some phagosomes (Figure 1Dc), and consisted in some cases of actin patches in the perinuclear region (Figure 1Db and d). Thus, in both J774 and NBT–2 cells, F–actin was found closely associated with a significant fraction of mature phagosomes.
Phagosomes induce actin assembly in vitro
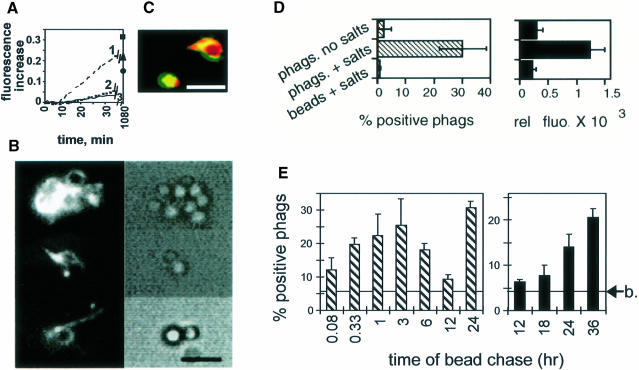
An in vitro assay was next set up to test whether the cytoplasmic surface of purified phagosomes from J774 cells could induce actin assembly. Actin is prevented from spontaneous polymerization in eukaryotic cells by proteins such as Tβ4 (Gondo et al., 1987; Cassimeris et al., 1992; Weber et al., 1992). Therefore, as a general strategy throughout this study, we buffered monomeric G–actin with a 3–fold excess of chemically synthesized Tβ4. Pyrene-actin is known to have an increased fluorescence when it incorporates into F–actin, which allows the kinetics of actin polymerization to be measured easily by spectrofluorimetry (Kouyama and Mihashii, 1981). Under our in vitro conditions, but in the absence of phagosomes, Tβ4 inhibited the initial rate of spontaneous pyrene-actin polymerization by >80% (Figure 2A), as expected from earlier studies performed with purified Tβ4 (Weber et al., 1992; Pantaloni and Carlier, 1993).
Fig. 2. Phagosomes polymerize actin from a pool of G–actin–Tβ4. (A) In vitro inhibition of spontaneous actin polymerization by Tβ4. Polymerization of 2 μM pyrene-G–actin, measured by spectrofluorimetry, in the presence of 6 μM (curve 3), 2 μM (curve 2) or 0 μM Tβ4 (curve 1). The fluorescence increase at steady state (1080 min) for each condition is also indicated: ▪, 0 μM Tβ4; ▴, 2 μM Tβ4; •, 6 μM Tβ4. Fluorescence increase was expressed in arbitrary units. Results are representative of several experiments. (B) Typical fields of phagosomal-induced actin polymerization. Phagosomes incubated with rhodamine–actin (left panel) observed by fluorescence microscopy; phase-contrast microscopy visualizes the latex beads (right panel). Bar, 3 μm. (C) Co-localization of polymerized actin and phagosomal membranes. Phagosomes labeled with R18 were repurified by flotation and assayed for fluorescein–actin assembly activity. Membrane, red; actin, green; overlap, yellow. Bar, 2 μm. (D) Correlation between the average fluorescence intensity and the number of positive phagosomes by fluorescence microscopy. Phagosomes (phags) or fish-skin gelatin-coated beads (beads) in non-polymerizing (no salts) or polymerizing (+ salts) conditions were observed by fluorescence microscopy. Individual phagosomes having actin patches as in (B) were expressed as a percentage (left panel). The average fluorescence intensity of at least 20 individual phagosomes (from five different fields) was measured using the NIH image software (right panel). (E) The ability of phagosomes to assemble actin is dependent on their maturation state. Phagosomes formed after internalization of avidin beads in cells were purified after different pulse and chase times, and assayed for their ability to polymerize actin by fluorescence microscopy. Time of bead pulse: 5 and 20 min for the two first time points, respectively, and 1 h for the other points. Arrow, background of fluorescence obtained for non-internalized latex beads. Left, average ± SD of three separate experiments. Right, typical data ± SD of one representative experiment.
A mixture of rhodamine-labeled G–actin–Tβ4 was then added to purified phagosomes (prepared after a 1 h pulse followed by a 1 h chase of fish-skin gelatin-coupled latex beads into cells: ‘2 h phagosomes’). Using fluorescence microscopy, rhodamine–actin dots and bundles of actin attached to phagosomes were clearly visible after 15 min of incubation (Figure 2B). In a few cases a partial or even complete halo of actin could be seen around phagosomes (Figure 2C). When phagosomes were first pre-labeled with the fluorescent lipidic probe R18 (octadecyl Rhodamine B chloride) and then used in combination with fluorescein–G–actin (instead of rhodamine–G–actin), polymerized actin and labeled phagosomal membranes co-localized (Figure 2C). The percentage of individual phagosomes with rhodamine–actin structures was determined, and in an average of three experiments 30% (± 8%) of phagosomes were found to be positive. In contrast, non-internalized beads were not labeled significantly (0.9 ± 0.3% of positive beads). There was a good correlation between the average intensity of the fluorescence signal and the percentage of positive phagosomes counted (Figure 2D).
The phagosome actin assembly process was also quantified by fluorescence-activated cell sorting (FACS) analysis in bulk solution. After incubation of phagosomes with fluorescein–G–actin and Tβ4, ∼12% of the phagosomes showed a significant signal for fluorescein–actin, whereas non-internalized beads had no significant signal under the same conditions. This technique was therefore useful for quantifying actin assembly on phagosomes, although it was less sensitive than fluorescence microscopy. In the absence of Tβ4 and with 2 μM fluorescein–G–actin, 48% of individual phagosomes were positive by FACS analysis, although background signals on non-internalized beads also increased considerably (data not shown). No significant signal was seen by incubating phagosomes with fluorescein–F–actin (stabilized by phalloidin) instead of G–actin (data not shown). Collectively, these results indicate that isolated phagosomes can induce actin assembly even when the bulk of G–actin is sequestered by Tβ4, and do not appear to bind significantly to preformed actin filaments.
Actin assembly varies with phagosome age
We next examined the ability of phagosomes to assemble actin in vitro relative to their maturation state in the cells. For this, latex beads coupled to avidin were internalized into macrophages for different times. Each phagosome preparation was tested for its integrity and its actin assembly activity by fluorescence microscopy. As assessed by the accessibility of fluorescein–biotin to avidin beads within phagosomes, the integrity of each phagosome preparation was similar (average of 82 ± 6.7% intact phagosomes for the different time points) (not shown). However, actin assembly activity changed significantly with phagosome age in a manner remarkably consistent with their in vivo activity (Figure 1C). Phagosome-induced actin polymerization increased until 3 h, was the lowest at the 12 h time point, but unexpectedly increased continuously at later time points (18–36 h phagosomes) (Figure 2E). Since 2 h phagosomes exhibit a significant ability to assemble actin and also many other biological activities (Blocker et al., 1996; Jahraus et al., 1998), we used this time point in the following experiments as a model for an ‘active’ phagosome stage.
Actin polymerizes preferentially from barbed ends close to the membrane
Assuming a value of the equilibrium dissociation constant of 1.7 μM for the G–actin–Tβ4 complex and using the appropriate formulae (Pantaloni and Carlier, 1993), the calculated concentration of free G–actin in our assay is 0.5 μM. This concentration should therefore be close to, or below, the critical concentration above which G–actin can grow at the pointed ends (0.5–1 μM) (Pollard and Cooper, 1986). We tested this prediction by taking advantage of the use of cyto. D to block barbed ends (Cooper et al., 1987). Latex beads conjugated to polylysine were used as a control. On such beads, filament growth occurs by initial binding of actin nuclei to the bead surface, allowing the filaments to grow out via their barbed ends; these beads therefore expose many free barbed ends (Brown and Spudich, 1981). Co-incubation of either phagosomes (Figure 3A) or polylysine beads (see Brown and Spudich, 1981) with 100 nM cyto. D during the assay in both cases blocked their ability to assemble actin, consistent with barbed end growth. This argues against a significant role for pointed end growth of filaments on phagosomes under our in vitro assembly conditions.
Fig. 3. Molecular characterization of the phagosomal-induced actin polymerization. (A) Polymerization is inhibited by low cyto. D concentrations. Two hour phagosomes were incubated in polymerization conditions, without (control) or with 100 nM cyto. D. The number of positive phagosomes was determined by fluorescence microscopy. The errors reported are the SD from three separate experiments. (B) Endogenous actin filaments are not required for the phagosomal-induced actin assembly. F–actin beads or 2 h phagosomes were pre-incubated in buffer F for 10 min at 25°C, with (hatched bars) or without (black bars) 2 μM cyto. D, and purified by flotation in PHEM buffer with protease inhibitors. Polymerization of actin was determined in the absence (F–actin beads) or the presence (2 h phagosomes) of Tβ4 using the flow cytometry assay. The errors reported are the SD from three separate experiments. (C) Labeling of assembled filaments with myosin S–1 visualized by electron microscopy, showing the presence of actin bundles emanating from the membrane fragments attached to a latex bead (arrowhead) (left). Right panel, higher magnification of insert; arrowheads symbolize the direction of polarity. Bars, 0.5 μm. (D) Actin assembly on the surface of phagosomes. Polylysine beads incubated first with fluorescein–G–actin (Actin 1) and subsequently with rhodamine–G–actin (Actin 2) are shown in the upper panel. Two hour phagosomes incubated first with rhodamine–G–actin–Tβ4 (Actin 1) and subsequently with fluorescein–G–actin–12 μM Tβ4 (Actin 2) are shown in the lower panel. Optical sections of different representative fields are shown. Fluorescein–actin (green), rhodamine–actin (red) and latex beads (visualized by phase contrast, here in dark blue) images are overlapped. Bar, 5 μm.
Actin is bound to purified phagosomes, as shown by two-dimensional gel electrophoresis and Western blot analyses (Desjardins et al., 1994a; our unpublished data). Therefore, phagosome-induced actin polymerization might be due to barbed end elongation of pre-existing filaments that remain attached to the phagosomal membrane after isolation. If so, then pre-incubating phagosomes with cyto. D (to block exposed barbed ends), followed by a rapid repurification, might be expected to block the subsequent growth of actin. As a control, we used polylysine beads that had polymerized unlabeled F–actin; pre-treatment of these beads with 2 μM cyto. D (followed by re-isolation) efficiently inhibited subsequent growth of fluorescein–G–actin. However, in the case of phagosomes, such pre-incubation had little effect on subsequent actin assembly activity, as determined by FACS analysis (Figure 3B). Evidently, the phagosomal membrane assembles actin filaments de novo rather than on pre-existing barbed end templates.
The polarity of the (in vitro formed) filaments with respect to the phagosomal membrane was next examined by decoration of actin filaments with myosin subfragment S–1. The visualization by electron microscopy of the S–1 arrowhead decoration allows the pointed end of each filament and thus its polarity to be defined (Tilney, 1976). However, this approach was made especially difficult by the large size of the latex beads (1 μm). In order to facilitate the visualization of membranes and actin, the phagosomes were therefore broken by osmotic shock. Since ezrin is localized to the cytoplasmic surface of phagosomes (see below), we labeled these broken phagosomes on electron microscope grids using anti-ezrin and immunogold. The great majority of the phagosomal membranes were labeled, implying that the cytoplasmic surface of membranes was still accessible for actin assembly and S–1 decoration (data not shown). In separate experiments, actin assembly on broken phagosomes and S–1 decoration were performed. The majority of filaments (∼70%) whose polarity was clearly identified had their barbed ends orientated towards the phagosomal membrane (Figure 3C).
As an alternative approach to assess actin polarity, 2 h phagosomes were incubated with 2 μM rhodamine–G–actin (red) in the presence of Tβ4 (6 μM), and then chased with 4 μM fluorescein–G–actin (green) (in the presence of 12 μM Tβ4). At the end of the experiment, fluorescein–actin was located close to the phagosomes, consistent with actin growth at the membrane (Figure 3D). As a control, we performed the same kind of experiment with polylysine beads (in the absence of Tβ4), which assemble actin by addition of monomers distal to the beads (Brown and Spudich, 1981). For technical reasons, the best results were obtained when fluorescein–actin was used first and then ‘chased’ with rhodamine–actin. In this case, rhodamine–actin was found away from the bead surface on the polylysine beads (Figure 3D), consistent with the addition of monomers at the distal ends of the filaments.
Peripheral membrane proteins are essential for actin assembly
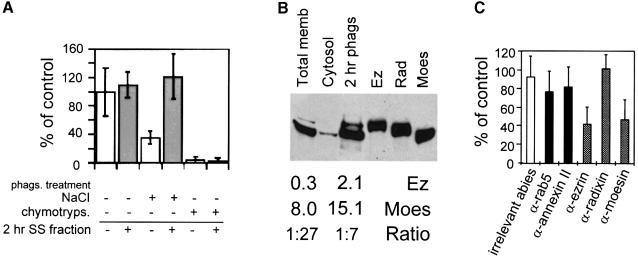
As determined by fluorescence microscopy, pre-treatment of 2 h phagosomes with chymotrypsin abolished their ability to assemble actin (Figure 4A). Pre-treatment with 1.3 M NaCl (Figure 4A) or with unbuffered carbonate (data not shown) led to averages of 62 and 57% inhibition, respectively. Addition of the salt-stripped (SS) fraction obtained from 2 h phagosomes fully reconstituted actin nucleation on SS phagosomes, but not on chymotrypsin-treated phagosomes (Figure 4A). These results suggest that actin assembly on phagosomal membranes depends on peripheral membrane proteins.
Fig. 4. Ezrin and moesin are two abundant phagosomal proteins necessary for actin assembly. (A) Actin assembly requires peripheral and integral membrane proteins. Two hour phagosomes were treated with control buffer, 1.3 M NaCl or 30 μg/ml chymotrypsin prior to gradient isolation. Where indicated, the phagosomes were then pre-incubated with an equal volume of SS fraction (30 ng/ml of proteins) from 2 h phagosomes (2 h SS fraction) (gray bars). (B) Distribution of ERMs in J774 cells. Western blot analysis of ERM proteins in J774 total membrane extracts, cytosol and 2 h phagosomes was performed as described in Materials and methods by using a monoclonal anti-ERM antibody. Ez, Moes and Rad, 1 μg of recombinant ezrin, moesin and radixin, respectively. A relatively long exposure time of the autoradiogram is shown here. Shorter exposure times allowed quantitation of the band intensity. The relative values (bottom) are typical signals (arbitrary units) obtained, as well as ezrin:moesin ratio. (C) Effect of anti-ABP antibodies on in vitro actin assembly. Two hour phagosomes were pre-incubated in PBS alone, or with antibodies (dilutions: α-ezrin, 1:200; α-moesin, 1:20; α-radixin, 1:10; α-rab5, α-annexin II; irrelevant antibodies, 1:200). In (A) and (C), after pre-incubation the phagosomes were repurified by a sucrose gradient, and actin assembly activity was checked by fluorescence microscopy. Results show the percentage of positive phagosomes for each sample relative to control phagosomes. The errors reported are the SD from three separate experiments.
Effect of antibodies against phagosomal membrane proteins
The possible involvement of some membrane proteins (especially ABPs) was then examined. Previous work had shown that phagosomal membranes contain several ABPs, among them annexin II (Desjardins et al., 1994a; Diakonova et al., 1997) and moesin, which is particularly abundant (Desjardins et al., 1994a). We first examined the presence of moesin and its homolog ezrin on isolated phagosomes by Western blot analysis. An ∼2–fold enrichment of moesin and a 4–fold enrichment of ezrin in phagosomes over total membranes were found (Figure 4B).
Owing to the low amounts of material recovered, our initial attempts to deplete ezrin from the SS fraction of 2 h phagosomes in order to add it back on SS phagosomes were not successful (our unpublished observations). In order to look for a role of ezrin or moesin in actin assembly, we analyzed the effect of antibodies specific for these proteins. Each antibody was pre-incubated with 2 h phagosomes, and after flotation of phagosomes, actin assembly was assayed by fluorescence microscopy. On average, 60 and 55% of actin assembly was inhibited by the anti-ezrin or the anti-moesin antibodies, respectively (Figure 4C). As an additional control, we pre-absorbed the anti-ezrin antibody on recombinant ezrin before adding it on phagosomes. For an ezrin:antibody ratio of 1:1, the inhibitory effect of anti-ezrin fell to 16%, and was completely suppressed for a 5:1 ratio (not shown). Antibodies specific for rab 5, which is a phagosomal membrane protein (Jahraus et al., 1998) not known to play any role in actin polymerization, as well as antibodies specific for annexin II and the ERM protein radixin (which was not detected on phagosomes) (Figure 4B), had no significant effects (Figure 4C).
Involvement of ezrin and moesin in actin assembly
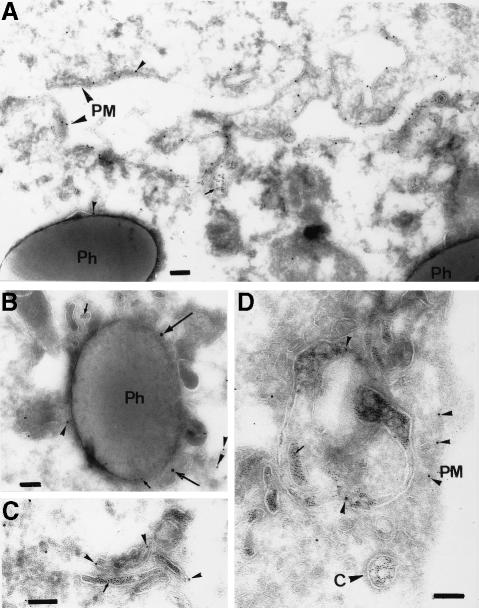
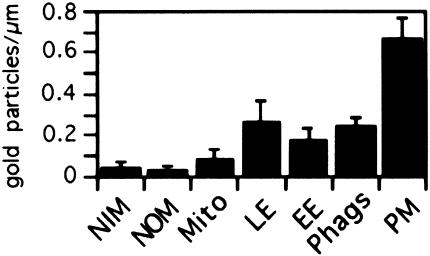
Immunogold localization. Ezrin is an abundant protein on phagosomes, at levels that are similar at all maturation time points, as determined by Western blot analysis (A.Jahraus and G.Griffiths, unpublished data). We therefore selected this protein for a more detailed investigation, and began by studying its intracellular distribution by electron microscopy. For this, J774 cells were pre-labeled with latex beads, as well as two distinguishable gold markers to identify early (5 nm) and late (16 nm) endosomes. The thawed cryosections of these cells were labeled with an affinity-purified anti-ezrin antibody and protein A–gold (10 nm) (Figures 5 and 6). As expected from earlier studies, the highest labeling was observed over the plasma membrane, and especially in areas that probably correspond to membrane ruffles and active extensions. Phagosomes, as well as both early and late endosomes, but not other membranes, were labeled significantly. With respect to our in vitro data, this experiment is important in showing that a significant concentration of ezrin is present around phagosomes in cells.
Fig. 5. Ezrin immunogold labeling on J774 cryosections. J774 cells that had internalized 16 nm gold (large arrows) for 1 h followed by an overnight chase (late endocytic organelles), latex beads for 1 h pulse and 1 h chase (Ph) and 5 nm gold (small arrows) for 5 min (early endosomes). The sections were labeled with anti-ezrin and protein A–gold (10 nm arrowheads). (A) Two adjacent cells with higher than average labeling for ezrin on their plasma membranes (PM), which probably correspond to ruffling membrane domains. One gold particle for ezrin on the phagosomal membrane is also shown. (B) An ezrin-labeled phagosome is shown that is also labeled for both 5 and 15 nm gold, implying that this phagosome had fused with both early and late endocytic structures. (C and D) Labeling for ezrin in early endosomes identifiable by the 5 nm gold. Labeling for ezrin is also seen on the plasma membrane in (D) (PM); a coated pit (indicated by the letter C) labeled with 5 nm gold is unlabeled for ezrin. Bars, 100 nm.
Fig. 6. Quantitation of ezrin immunogold labeling. Quantitation was expressed as the mean density of gold labeling per linear micrometer of membrane. LE, late endosomes; EE, early endosomes; PM, plasma membrane; Phags, phagosomes. The signal detected for mitochondria (Mito), nuclear outer membrane (NOM) and nuclear inner membrane (NIM) corresponds to the background signal (irrelevant antibody) (not shown). The error bars show SEMs.
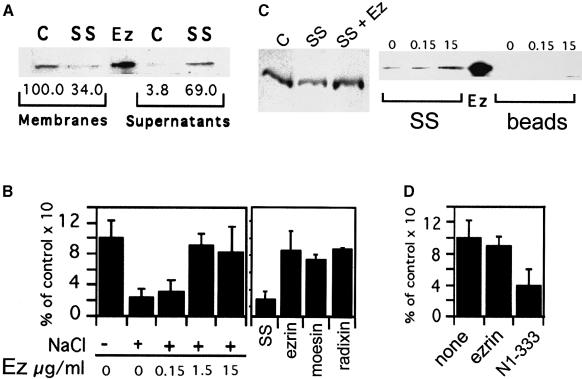
Biochemical analysis. As shown by Western blot analysis, salt-stripping of 2 h phagosomes resulted in the removal of ∼67% of ezrin from the phagosomal membrane (Figure 7A), which correlates to a 62% inhibition of actin assembly (Figure 4A). Remarkably, back-addition of recombinant ezrin to 2 h SS phagosomes fully reconstituted actin assembly in a concentration-dependent manner (Figure 7B, left panel). The morphology of the rhodamine–actin structures visualized by light microscopy on these phagosomes was indistinguishable from that observed on mock phagosomes, and the formation of these structures was inhibited by adding 100 nM cyto. D to the assay (86.3% inhibition) (not shown). The amount of recombinant ezrin that bound to SS phagosomes corresponded approximately to endogenous ezrin concentration on untreated phagosomes, as seen by Western blot analysis (Figure 7C, left panel). Reconstitution of actin assembly on SS phagosomes was dependent on ezrin–membrane interactions, since (i) pre-incubation of non-internalized latex beads with 15 μg/ml ezrin did not allow them to assemble rhodamine–G–actin (not shown), and (ii) recombinant ezrin bound to SS phagosomes but not to membrane-free latex beads (Figure 7C, right panel).
Fig. 7. ERM proteins can mediate actin assembly on phagosomes. (A) Removal of ezrin from phagosomes by high salt treatment. Two hour phagosomes (Membranes) with (SS) or without (C) 1.3 M NaCl treatment were re-purified. The proteins removed by this treatment were concentrated (Supernatants). Western blot analysis was performed using the rabbit polyclonal anti-ezrin antibody. Ez, 60 ng recombinant ezrin. Autoradiograms were scanned and data quantified; the values (bottom) represent the percentage of signal obtained for each condition relative to control, untreated phagosomes. (B) Recombinant ezrin, radixin or moesin restores actin nucleation on SS phagosomes. Left panel, 2 h phagosomes SS or not (+ or – NaCl) were incubated with different concentrations of recombinant ezrin. After pre-incubation the phagosomes were re-purified and actin assembly activity was checked by fluorescence microscopy. Right panel, SS phagosomes were pre-incubated with buffer (SS) or with 5 μg/ml recombinant ezrin, moesin or radixin and re-isolated. (C) Binding of recombinant ezrin to SS phagosomes. Right panel, equal amounts of 2 h SS phagosomes (SS) or fish-skin gelatin beads (beads) were incubated with different concentrations of recombinant ezrin. Left panel, equal amounts of 2 h control phagosomes (C), SS phagosomes (SS), as well as SS phagosomes pre-incubated with 10 μg/ml recombinant ezrin (SS + Ez). Phagosomes were all re-purified by flotation and analyzed by Western blotting with the polyclonal anti-ezrin rabbit antibody. Ez, 100 ng recombinant ezrin. (D) Blocking of endogenous ezrin on phagosomes inhibits their actin assembly activity. Two hour phagosomes were pre-incubated in buffer D alone (none), 15 μg/ml full-length ezrin (ezrin) or 15 μg/ml N-terminal fragment 1–333 (N1–333). In (B) and (D), after re-isolation of phagosomes, actin assembly was assayed by fluorescence microscopy. Results show the percentage of positive phagosomes for each sample relative to control phagosomes. The errors reported are the SD from three separate experiments.
Since all ERMs are known to have homologous domains responsible for membrane attachment (Reczek and Bretscher, 1998; Yonemura et al., 1998) and display functional homologies in vivo (Doi et al., 1999), we postulated that recombinant moesin and radixin should also reconstitute actin assembly on SS phagosomes. In fact, when 5 μg/ml of any recombinant ERM was pre-incubated with SS phagosomes, actin assembly was fully restored (Figure 7B, right panel).
We also tried to block endogenous ezrin functions by the following approach. Interactions between full-length ezrin and its N-terminal fragment N1–333 (amino acids 1–333 of ezrin) have previously been shown to prevent binding of F–actin to ezrin (Roy et al., 1997). This fragment did bind to phagosomes, as seen by Western blot analysis (data not shown). In the fluorescence assay, pre-incubation of phagosomes with the N1–333 fragment inhibited actin assembly by ∼60%, while pre-incubation with full-length ezrin had no effect (Figure 7D).
Discussion
Latex beads have been used for over 30 years as a model for the phagocytic process (Sbarra and Karnovsky, 1959; Singer et al., 1969). These inert particles, which cannot be evicted by cells, serve as a convenient model for the uptake of non-degradable particulate material that can often be medically relevant, as in the case of asbestos or carbon particles arising from cigarette smoking. We show here that latex beads induce a rapid and transient F–actin assembly when added to J774 macrophages, with kinetics that resemble those seen in other systems (Carson et al., 1986; Condeelis and Hall, 1991; Greenberg et al., 1991; Bengtsson et al., 1993). This result suggests that these particles, although lacking any biologically specific surface structure, can switch on the actin assembly machinery in J774 macrophages. Although the nature of the molecules that bind to the latex beads is not known, it is evident that some receptors must be activated. That receptors such as the Fc or complement receptors can activate macrophages to induce actin assembly is now clearly established (Allen and Aderem, 1996; Caron and Hall, 1998).
The phagosome system we introduce here represents the first defined intracellular membrane organelle that is capable of assembling actin in vitro. Other membrane fractions, such as plasma membrane (Shariff and Luna, 1990), eukaryotic cell lysates (Eddy et al., 1997; Zigmond et al., 1997) or crude membrane vesicles (Ma et al., 1998) and bacteria (Cossart and Lecuit, 1998), have been shown to nucleate actin in vitro, either from purified monomeric actin alone or from cytosol. In order to follow actin nucleation under defined conditions that resemble physiological conditions in our study, G–actin was buffered with Tβ4, which regulates the levels of free G–actin in many eukaryotic cells, including macrophages and neutrophils (Gondo et al., 1987; Cassimeris et al., 1992; Weber et al., 1992). The Tβ4 and actin concentrations used in the assay should give ∼0.5 μM free G–actin throughout the course of the analysis, and should prevent the elongation of actin filaments at pointed ends (Pollard and Cooper, 1986). Such a situation is not possible with pure actin, in which monomer concentration always decreases progressively as polymerization proceeds.
Several lines of evidence argue that the actin assembly seen on the surface of phagosomes is physiologically relevant. Actin was assembled in vitro via insertion of monomers at the barbed ends of filaments on the phagosomal membrane, with the filaments growing mostly perpendicularly away from the membrane surface, as in all other cell systems investigated; as has been observed in other systems, these filaments often appeared in bundles (Tilney, 1976; Tilney and Portnoy, 1989; Hartwig, 1992; Small et al., 1995; Mitchison and Cramer, 1996). Since actin is detected biochemically on isolated phagosomes (Desjardins et al., 1994b), actin assembly on phagosomal membranes might result from the growth of pre-existing filaments still attached to isolated phagosomes. However, these purified organelles could not be significantly labeled with fluorescent phalloidin (H.Defacque, unpublished observations), leaving open the molecular organization of this membrane-bound actin pool (see also Carraway and Carraway, 1989). Moreover, pre-incubation of phagosomes or F–actin beads (which have abundant free barbed ends; Brown and Spudich, 1981) with cyto. D followed by a re-isolation of both fractions led to a subsequent inhibition of new actin growth on the F–actin beads but not on phagosomes. The simplest interpretation of these data is that phagosomes do not polymerize actin from pre-existing, free barbed end actin filaments, but rather assemble actin de novo.
Phagosomes are highly dynamic vesicles that are formed de novo by the cell and subsequently mature (Griffiths, 1996). This dynamic behavior is further illuminated here by the fact that actin assembly activity of phagosomes changed drastically and significantly as they aged in the cells. Unexpectedly, the youngest phagosomes (5 min of pulse and chase) had a relatively low in vitro activity. This argues against the notion that what is measured at later time points reflects residual actin nucleation sites on phagosomes that had been involved in the uptake process. The actin assembly capacity of phagosomes peaked at 2–4 h of chase, then decreased drastically at 12 h before an unexpected and unexplained recovery that occurred over the next 24 h. It is intriguing that this pattern of actin assembly on phagosomes mirrors that obtained for phagosomal proteins phosphorylated in vitro by endogenous phagosomal kinases (both activities being high for the 2–4 and 24 h time points, and low for the 12 h time point) (Emans et al., 1996). However, both the kinases and the substrates involved in these processes are yet to be identified.
Ezrin or moesin (which cannot yet be functionally distinguished) appear to be essential for actin assembly by phagosomes. Both moesin (Desjardins et al., 1994a; this study) and ezrin are enriched on phagosomal membranes (Figure 4B). Actin assembly could be blocked by pre-incubation with an anti-ezrin antibody (previously shown to block binding of G- but not F–actin to the N–terminal part of ezrin) (Roy et al., 1997) or an N1–333 fragment of ezrin (which binds to the C–terminal part of ezrin) (reviewed in Mangeat et al., 1999). This fragment and a shorter one (1–310) have been shown to block in vitro ezrin–F–actin interactions (Roy et al., 1997), and to inhibit ezrin functions in cells (Crepaldi et al., 1997). An antibody against moesin also partially inhibited actin assembly by phagosomes. In contrast, an affinity-purified antibody against the closely related ERM protein, radixin, which is not detectable in J774 cells, or against annexin II or rab 5, which are present on phagosomes (Desjardins et al., 1994a; Diakonova et al., 1997), had no effect. Finally, the addition of recombinant ERM proteins could allow phagosomes incapacitated by salt-stripping to regain their full capacity to nucleate actin. Although radixin seems to be absent in both J774 macrophages (our present data) and peripheral blood monocytes (Shcherbina et al., 1999), the ability of recombinant radixin to reconstitute actin assembly in our system presumably reflects high functional homologies of the ERM proteins (Mackay et al., 1997; Doi et al., 1999; reviewed in Tsukita and Yonemura, 1997; Bretscher, 1999; Mangeat et al., 1999).
Collectively, our data argue that ezrin/moesin represents a minimal machinery that needs to be recruited by phagosomes from the cytoplasm in order for actin assembly to proceed. However, these data leave open the question of how these molecules bind to the phagosomal membrane. Three possibilities are: (i) binding to a membrane receptor such as CD44 (which can be detected on phagosomes in J774 cells; H.Defacque, unpublished data), ICAM–1 or -2 (Hirao et al., 1996; Heiska et al., 1998; Yonemura et al., 1998), or to a peripheral membrane protein such as EBP50 (Reczek and Bretscher, 1998); (ii) binding by self-association to pre-existing ezrin or moesin already bound to phagosomes (see below); or (iii) binding to phosphoinositides, especially PIP2 (Niggli et al., 1995; Hirao et al., 1996; Heiska et al., 1998). In separate experiments, we have found that PIP2 can be detected on phagosomes, and also seems to be involved in phagosomal actin assembly. Moreover, an ezrin mutant defective in PIP2 binding was also defective in rescuing the ability of salt-stripped phagosomes to assemble actin; this ezrin mutant also bound less to phagosomes than did wild-type ezrin (H.Defacque, E,Bos, C.Barret, C.Roy, P.Mangeat and G.Griffiths, submitted).
ERM proteins are members of a growing family of homologous peripheral membrane proteins that includes the red blood cell protein band 4.1, merlin/schwanonin and talin (Tsukita and Yonemura, 1997; Bretscher, 1999; Mangeat et al., 1999). In their inactive state they are soluble, due to their head-to-tail interaction (Berryman et al., 1995), and unphosphorylated (Chen et al., 1995). Upon trans-membrane signaling, ERM protein phosphorylation (on serine/threonine or tyrosine residues, depending on the cell system) correlates with their membrane association (Bretscher, 1989; Hanzel et al., 1991; Chen et al., 1995) and is thought to facilitate their association with actin (Simons et al., 1998; Nakamura et al., 1999). Following ERM activation, the N–terminal domain also becomes free to bind Rho-GDI, which can deliver the GDP form of the rho family proteins (Takahashi et al., 1997). Downstream effectors of these GTPases, such as Rho-kinase, can phosphorylate the ERM proteins, raising the possibility of a cycle and amplification of the functions of these proteins (Matsui et al., 1998; Oshiro et al., 1998; Shaw et al., 1998). The role of phosphorylation of ezrin and moesin in our system remains to be investigated.
The ERM proteins are now widely considered as mechanical linkers between the membrane and actin filaments (Bretscher, 1989; Hanzel et al., 1991; Algrain et al., 1993; Berryman et al., 1995). Based on our data, we propose that these molecules may also play a more general role in de novo actin assembly on membrane surfaces, as first proposed by Vaheri et al. (1997). A number of observations would be consistent with this view. First, these proteins are generally enriched in sites where actin assembly occurs, such as the leading edge of motile cells (reviewed in Bretscher, 1999; Mangeat et al., 1999). Secondly, both ezrin (Yao et al., 1996; C.Roy and P.Mangeat, unpublished results) and talin (Isenberg and Niggli, 1998) can accelerate pyrene-actin assembly in vitro. Finally, Mackay et al. (1997) showed that any recombinant ERM protein could reconstitute actin assembly in permeabilized cells, in response to the GTPases Rho and Rac. Our in vitro actin assembly system was not affected by GTP, GTP–γS, C3 transferase or by Rho-GDI (H.Defacque, unpublished results), which suggests that these conditions for actin assembly reconstitute a machinery that operates independently of, and presumably ‘downstream’ of these regulatory molecules.
What could be the function(s) of the de novo assembled actin on phagosomal membranes? As for exocytosis (Muallem et al., 1995) and receptor-mediated endocytosis (Lamaze et al., 1997), we have recently found that in vitro phagosome fusion with endocytic organelles could be variously stimulated or inhibited by reagents that affect the actin cytoskeleton (A.Jahraus, M.Egeberg, A.Habermann, B.Hinner, E.Sackmann, A.Pralle, H.Faulstich, H.Defacque and G.Grifiths, submitted). Light- and electron-microscopy observations, in conjunction with biochemical and rheological studies, argue that most of the actin that polymerizes in our in vitro fusion assay is nucleated by membranes. Actin grows from the surface of both phagosomes and endosomes as large bundles to which other organelles seem to bind and move. We therefore postulate that membrane-induced assembly of actin on phagosomal (and endosomal) membranes provides tracks that help other organelles to move, presumably via myosin motors, towards these actin nucleating sources. A consequence of such a model is that actin assembled by membranes would facilitate the ability of membrane organelles to aggregate before they fuse.
Materials and methods
Reagents
One- or three-micrometer latex beads were obtained from Seradyn (Indianapolis, IN) or Polyscience (Warrington, PA), respectively. Actin purified from rabbit muscle was labeled with fluorescein maleimide or with pyrenyl iodoacetamide according to established protocols (Kouyama and Mihashii, 1981; Pardee et al., 1982). Muscle G–actin labeled with 5-carboxytetramethyl rhodamine was obtained from Cytoskeleton Company (Denver, CO). Where indicated, G–actin was buffered with Tβ4 (synthesized and purified using the Fmoc technique; Echner and Voelter, 1988) by incubating a 3–fold molar excess of Tβ4 (6 μM) to G–actin (2 μM) in buffer G [10 mM Tris pH 8, 0.1 mM ATP, 0.1 mM dithiothreitol (DTT), 0.1 mM CaCl2] for 20 min at 4°C. The mixture of protease inhibitors used was 1 μg/ml pepstatin A, 0.5 μg/ml leupeptin, 4 μg/ml aprotinin (Sigma, St Louis, MO). Purification of human recombinant ezrin (full-length and N1–333), radixin and moesin was performed as described previously (Roy et al., 1997). Rabbit anti-ezrin antibodies were previously characterized and affinity purified (Andreoli et al., 1994; Roy et al., 1997). Affinity-purified rabbit polyclonal antibodies against radixin and moesin were produced similarly from recombinant human radixin and moesin (C.Roy and P.Mangeat, unpublished results). Mouse monoclonal anti-ERM was from Transduction Laboratories (Lexington, KY). LAMP–1 was detected using a rat monoclonal antibody (mAb 1D4B; Developmental Studies Hybridoma Bank, University of Iowa, IA).
J774 cell culture, phagosome purification and treatments
J774A.1 mouse macrophages were maintained in Dulbecco's modified Eagle's medium (Sigma, St Louis, MO) supplemented with 10% fetal calf serum (Gibco-BRL, Eggenstein, Germany), 2 mM glutamine, 100 IU/ml penicillin and 100 mg/ml streptomycin (all from Seromed, Berlin, Germany) (complete medium) at 37°C in a CO2 atmosphere. Phagosomes were formed in cells by internalization of 1 μm beads covalently coupled with fish-skin gelatin (Sigma, St Louis, MO) unless specified otherwise, and isolated using the high buoyant density of latex as described previously (Jahraus et al., 1998). Briefly, a post-nuclear supernatant from J774 macrophages (obtained by homogenization of cells in 3 mM imidazole pH 7.4) was brought to 35% sucrose, and 25 and 10% sucrose layers (both in 3 mM imidazole pH 7.4) were added on the top. Centrifugation was performed in a swinging bucket rotor (SW40; Beckman Instruments, Palo Alto, CA) at 100 000 g for 1 h at 4°C. In some experiments, a total cellular membrane fraction was obtained by collecting the interface of the 35 and 25% sucrose solutions (Desjardins et al., 1994b). The phagosome band was collected from the interface of the 10 and 25% sucrose solutions and further purified by a second sucrose gradient in a swinging bucket rotor (SW60) (Jahraus et al., 1998). J774 cytosol was prepared from spinner cultures as described previously (Jahraus et al., 1998).
Treatments of phagosomes with 30 μg/ml chymotrypsin or with 0.1 M unbuffered Na2CO3 were performed as described previously (Burkhardt et al., 1995; Blocker et al., 1997). After incubation, phagosomes were re-purified using a small-scale version of a sucrose gradient (Jahraus et al., 1998) and stored in liquid nitrogen. For salt-stripping of phagosomes by 1.3 M NaCl, one part of phagosomes was mixed with one part of 53% sucrose/3 mM imidazole pH 7.4, protease inhibitors, 1 mM DTT, with or without 2.6 M NaCl. After incubation for 45 min at 4°C, 0.3% fish-skin gelatin and 62% sucrose/3 mM imidazole pH 7.4 (final sucrose concentration 35%) were added to the phagosomes. The samples were transferred in 11 × 60 mm Ultraclear Beckman tubes and overlaid with 25% sucrose/3 mM imidazole pH 7.4 and 10% sucrose/3 mM imidazole pH 7.4/protease inhibitors/1 mM DTT homogenization buffer (HB). They were centrifuged in an SW60 rotor at 100 000 g for 20 min at 4°C. The 25%/HB interphase was recovered and phagosomes were aliquoted and rapidly frozen in liquid nitrogen. In order to recover the proteins extracted by 1.3 M NaCl, after flotation of the phagosomes the 35% sucrose fraction was desalted and concentrated in a Nanosep concentrator (3 kDa cut-off) (Pall Filtron, Northborough, MA) in 20 mM Na2HPO4 pH 7.2 with protease inhibitors following the manufacturer's instructions and used immediately. Protein concentration was determined with a Bio-Rad Protein assay (Bio-Rad, Munich, Germany).
For R18 staining, R18 (Molecular Probes, Eugene, OR) was added to the purified phagosomes to a final concentration of 0.1 μM for 30 min at 4°C in HB (without DTT). Phagosomes were separated from free R18 using a discontinuous sucrose gradient (14, 25 and 40.6% sucrose/3 mM imidazole pH 7.4/protease inhibitors/1 mM DTT) by centrifugation in an SW60 rotor at 120 000 g for 1 h at 4°C. They were then incubated with fluorescein–G–actin–Tβ4, and actin assembly was followed immediately by fluorescence microscopy.
For phagosome pre-incubation, phagosomes were incubated with a recombinant protein in buffer D (10 mM HEPES pH 7.5, 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, protease inhibitors) for 15 min at 25°C and were repurified by a sucrose gradient as for the salt-stripping experiments. Pre-incubation with antibodies was performed similarly, in phosphate-buffered saline (PBS) instead of buffer D (without DTT). Anti-ezrin antibody was pre-absorbed on recombinant ezrin by incubating the antibody (0.19 μM) with 0, 0.19 or 0.95 μM ezrin in PBS for 2 h at 25°C before adding it on 2 h phagosomes at a 1:100 final dilution; actin assembly was then performed as described in Figure 4.
Quantitative measurement of F–actin in J774 cells
F–actin was quantified by a Rh–Ph binding assay originally developed by Howard and Oresajo (1985). J774 cells were plated on glass coverslips and incubated with 1 μm naked latex beads (1:300 diluted) in internalization medium (MEM, 10 mM HEPES, 5 mM D-glucose pH 7.4) on ice for 15 min. Phagocytosis was triggered by replacement of cold medium with medium pre-warmed at 37°C. After 5, 10, 15, 30, 60, 120 and 140 s, coverslips were fixed with a 3.7% formaldehyde solution for 20 min and permeabilized with 0.1% Triton X–100 (TX-100) for 5 min in CSK buffer [10 mM PIPES, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF) pH 6.8]. After washing several times in PBS, coverslips were stained with 0.33 μM Rh–Ph (Molecular Probes) for 30 min, washed in PBS and Rh–Ph was extracted with 2 ml of MeOH for 1 h in the dark. Non-saturable binding of Rh–Ph was determined in parallel samples by incubating samples in Rh–Ph plus a 100–fold excess (33 μM) of unlabeled phalloidin. Rhodamine fluorescence (excitation wavelength 550 nm; emission wavelength 580 nm) was measured in an Aminco-Bowman Series 2 Luminescence Spectrometer (SLM-Aminco Inc., Northampton, MA). The number of cells in five sampled microscope fields arranged diagonally across each coverslip was counted after methanol extraction and was used to determine the relative amounts of Rh–Ph per cell in each sample. The relative F–actin content was given as the amount of Rh–Ph per cell in a phagocytosis-triggered sample divided by the amount of Rh–Ph per cell in a control sample (coverslips incubated on ice) (Condeelis and Hall, 1991).
Quantitative analysis of F–actin-positive phagosomes in J774 cells
J774 cells grown on Lab-tek chamber slides (Nunc, Naperville, IL) were incubated at 37°C with pre-warmed 1 μm latex beads (1:300 diluted) in complete medium. After 1 h, the cells were washed three times in PBS at 4°C, chased with pre-warmed medium for various times at 37°C and washed again three times with PBS at 4°C. The cells were then washed with CB (10 mM MES pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 5 mM glucose), fixed with 3% paraformaldehyde/CB for 10 min, and permeabilized with 0.2% TX–100 in PBS for 2 min at room temperature. Staining of F–actin with 0.5 μM Rh–Ph and labeling of LAMP–1 with the rat monoclonal anti-LAMP 1 antibody (1:100 diluted) were performed as described previously (Desjardins et al., 1994b; Diakonova et al., 1997). The glass slides were mounted with ProLong antifade reagent (Molecular Probes) according to the manufacturer's protocol.
Immunofluorescence images were collected using a Leica (Bensheim, Germany) TCSNT inverted confocal laser scanning microscope equipped with an immersion oil UV PLAPO 63× 1.32 lens. Settings allowed simultaneous co-localization of fluorescein-labeled phagosomes and Rh–Ph-stained cells. Images were simultaneously recorded from the same optical x–y or x–z sections. They were stored on erasable optical disks, and processed and printed using NIH Image and Adobe Photoshop software. To quantitate the percentage of phagosomes that were positive for both actin and LAMP–1 per cell, random x–y sections of at least 10 cells were analyzed using Adobe Photoshop software. To rule out the possibility that phagosome-associated actin corresponded to cortical actin, we considered x–y sections in the middle part of cells and selected phagosomes that were in the perinuclear region. For each optical section, the fluorescein image (which visualizes LAMP–1) was overlapped with the phase-contrast image (which visualizes latex beads). Phagosomes were then identified as individual latex beads that were positive for LAMP–1. Of these phagosomes, those with rims or patches of F–actin (as seen by overlapping rhodamine and fluorescein images) were counted, and the percentage of phagosomes positive for F–actin per cell was calculated.
NBT-2 cell culture
NBT-2 cells (Tchao, 1982) were stably transfected with a pEGFP-C2 vector including the GFP sequence linked to the Dictyostelium actin-15 sequence (GFP–actin), as described previously (Choidas et al., 1998). The cells were maintained in complete medium supplemented with 1 mg/ml geneticin (Gibco-BRL, Eggenstein, Germany) at 37°C in a CO2 atmosphere. They were differentiated by incubating them with 100 ng/ml mouse EGF (Boehringer Mannheim, Mannheim, Germany) for 24 h. These cells failed to take up naked or fish-skin gelatin-coated beads (our unpublished observations) and thus fibronectin-mediated phagocytosis had to be performed. For this, cells that were allowed to spread to fibronectin-coated coverslips were incubated for 5 min at 37°C with pre-warmed 0.5 mg/ml rhodamine–dextran (average mol. wt 70 000; Sigma, St Louis, MO) in complete medium. They were then washed four times with 0.5% bovine serum albumin (BSA)/PBS at 4°C. Three-micrometer latex beads (0.1% final concentration) were mixed with 3.6 μg/ml fibronectin (Sigma) and incubated for 10 min at 37°C. The beads were sonicated in a water bath and reconstituted (0.1% final concentration) in complete medium. Fibronectin-coated beads were added to the cells and pulsed for 1 h at 37°C. During this period, the rhodamine–dextran will chase into late endocytic structures. The cells were then washed several times with PBS at 4°C, chased with pre-warmed complete medium for 1 h at 37°C and washed again with PBS at 4°C. After 15 min fixation at room temperature with 0.5% paraformaldehyde (PFA)/PBS, the coverslips were mounted and observed under the confocal microscope as described above.
Pyrene–actin assays
The increase of fluorescence upon polymerization of actin (10% pyrenyl-labeled) was followed in buffer G at 25°C for 10 min (Kouyama and Mihashii, 1981) immediately after addition of salts (50 mM KCl, 1 mM MgCl2) in an Aminco-Bowman Series 2 Luminescence Spectrometer. Excitation–emission wavelengths were 365–407 nm.
In vitro actin assembly assay: fluorescence microscopy
Since latex beads interfered with the fluorescence emitted by pyrene-actin, as an alternative approach to follow actin assembly on phagosomes we used fluorescence microscopy. Glass slides (Proper Ltd, UK) and 22 × 22 mm coverslips (Gold Seal, Clay Adams) were coated with 0.5% fish-skin gelatin/H2O and air dried prior to the experiment. A constant number of phagosomes (as determined by bead counts) (Blocker et al., 1997) was incubated between a slide/coverslip in buffer P (20 mM HEPES pH 7.0, 50 mM KCl, 4 mM MgCl2, 0.2 mM CaCl2, 0.1 mM ATP, 0.03% fish-skin gelatin, protease inhibitors) with 2 μM rhodamine–G–actin–6 μM Tβ4 and an antifade reagent (Blocker et al., 1997) at room temperature for 15 min. The samples were then observed using a Zeiss Axioscope microscope and a 63× Plan-Apo 1.4 lens (Zeiss, Oberkochen, Germany). Actin patches or filaments bound to individual phagosomes were observed by eye, and in each experiment at least five different fields arranged diagonally across each coverslip (∼15 phagosomes per field) were sampled for the percentage of labeled phagosomes.
To determine the site of addition of new actin monomers to pre-existing filaments, 2 h phagosomes were incubated with rhodamine–G–actin–Tβ4 for 15 min, diluted 1:10 with 4 μM fluorescein–G–actin–12 μM Tβ4 and then incubated for another 20 min before they were observed under a Zeiss LSM 510 confocal microscope with a 63× oil planapochromat lens (NA 1.40; Carl Zeiss, Inc.). As a control, polylysine beads were incubated with 3 μM fluorescein–G–actin for 3 min, diluted 1:3 with 15 μM rhodamine–G–actin and then incubated for a further 10 min before they were observed at the confocal microscope. For these experiments excitation lines from two lasers were used (488 and 543 nm) and emission was monitored using the respective filters (505–530 and 560 long pass). Samples were line scanned (typically 4.48 μs/pixel, ∼30 s/frame) and power settings were minimized to avoid photobleaching. Each picture represents one optical slice of 1 μm (pinhole size 140 μm).
In vitro actin assembly: FACS analysis
Briefly, latex beads or purified phagosomes were incubated in buffer P with 2 μM fluorescein–G–actin (complexed with 6 μM Tβ4 for phagosomes) for 30 min at 37°C (polymerizing conditions). For each sample, a negative control was made by incubating the same reagents in the absence of 50 mM KCl and 1 mM MgCl2 (non-polymerizing conditions). The samples were then gently fixed in the same tube with 1% PFA/PBS, and 10 000 events were acquired at the FACS (Beckton Dickinson, San Jose, CA). For each preparation, histograms of fluorescence relative to the number of events (corresponding to individual phagosomes) were obtained. The relative percentage of positive beads or phagosomes was determined by defining the percentage of individual beads (or phagosomes) in polymerizing conditions having a fluorescence intensity higher than those of individual beads (or phagosomes) in non-polymerizing conditions. The errors reported are the SD from at least three separate experiments. F–actin beads were prepared by incubating polylysine beads (Brown and Spudich, 1981) with 10 μM G–actin in buffer F, and flotation on a sucrose gradient for 20 min at 100 000 g in PHEM buffer (60 mM PIPES, 25 mM HEPES, 2 mM MgCl2, 10 mM EGTA, 1 mM ATP).
Integrity (latency) of phagosome preparations
J774 cells were fed with 1 μm latex beads coupled with avidin as described previously, and phagosomes were purified after different pulse and chase times as indicated, following our previously described protocol (Jahraus et al., 1998). For each phagosome preparation, a constant number of phagosomes (as determined by bead counts) (Blocker et al., 1997) was gently mixed with 12 μg/ml fluorescein–biotin (Molecular Probes) in PBS, 0.03% fish-skin gelatin and protease inhibitors. After incubation for 5 min at room temperature, samples were immediately observed under a Zeiss Axioscope microscope with a 63× Plan-Apo 1.4 lens. A minimum number of three fields arranged diagonally across each coverslip (minimum number of 50 phagosomes counted) was considered. For each field, fluorescein-positive individual phagosomes and the total number of individual phagosomes were counted by eye, and the percentage of intact phagosomes was sampled. Pre-treatment of phagosomes with 2% TX–100 before fluorescein–biotin incubation resulted in the labeling of ∼100% of phagosomes. The errors reported are the SD from at least two separate experiments.
Electron microscopy
Since 1 μm latex beads have a high electron density, in order to see the membranes clearly, 2 h phagosomes were first broken by osmotic shock in buffer G and protease inhibitors, and then transferred onto freshly glow discharged Formvar-carbon grids for 5 min. S–1 was prepared from rabbit skeletal-muscle myosin, purified according to Margossian and Lowey (1982), and digested as described by Weeds and Taylor (1975). For S–1 labeling, the phagosomes coated on the grids were incubated with 2 μM G–actin–6 μM Tβ4 in buffer F (50 mM Tris pH 8, 0.1 mM ATP, 50 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.1 mM CaCl2) and protease inhibitors for 10 min, washed with S–1 buffer (10 mM imidazole, 1 mM EGTA, 20 mM KCl, 0.1 mM MgSO4) and incubated with 1 mg/ml S–1 in S–1 buffer for 1 min, before negative staining with 1% uranyl acetate. The actin filament polarity on membranes was scored from negatives. For this, 50–100 filaments that clearly emanated from the membranes were considered in each experiment; the percentage of those that unequivocally had their barbed ends towards the membrane was then estimated (average of two independent determinations).
Quantitation of immunogold labeling of ezrin in J774 cells using cryosections was performed with the polyclonal anti-ezrin rabbit antibody (1:100 diluted) and protein A–gold (10 nm). Protocols for the internalization of early and late endocytic (BSA–5 nm and 16 nm gold conjugates, respectively) and phagocytic (1 μm naked latex beads) markers in cells, cryosectioning and immunogold labeling have been described elsewhere (Griffiths, 1993; Diakonova et al., 1997). For quantifying the density of label per micrometer of membrane, ezrin-labeled cryosections were systematically sampled by moving the grid in a defined direction in the electron microscope and imaged on a negative. The density of label was determined by randomly placing a double lattice test line system on the micrographs and relating the number of gold particles per micrometer of membrane, as described previously (Griffiths, 1993). For this analysis we presumed a resolution of 20 nm and only gold particles within this distance from a membrane profile of interest were scored.
Western blot quantitation of ERM proteins
Electrophoresis of cell extracts and Western blotting procedures were performed as described previously (Roy et al., 1997). Briefly, 50 μg of proteins were loaded per lane. After migration and transfer of proteins on membranes, a Coomassie Blue staining of transferred proteins was performed before immunorevelation to confirm that the same amounts of proteins were loaded (data not shown). Membranes were then probed with a monoclonal anti-ERM antibody as described previously (Roy et al., 1997). Autoradiograms were scanned and data quantified using the ImageQuant software. As a standard, known amounts of ezrin were run on separate gels.
Acknowledgments
Acknowledgements
We are grateful for the excellent technical assistance of Lena Johansen and Ann Atzberger. We also thank Drs A.Blocker, A.Jahraus, A.Hyman, R.Tournebize and M.Way for their help and critical comments on the manuscript. The experiment shown in Figure 3B (18–36 h) was performed by Evelyne Bos. We thank Drs M.Zerial (EMBL, Heidelberg, Germany) for the polyclonal anti-rab 5 antibody and V.Gerke (University of Münster, Germany) for the monoclonal anti-annexin II antibody. This work was supported in part by funds from the European Community and the Association pour la Recherche contre le Cancer (to P.M.), and by a network grant from the Human Frontier Science Program Organization (to G.G.).
References
- Algrain M., Turunen, O., Vaheri, A., Louvard, D. and Arpin, M. (1993) Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane–cytoskeletal linker. J. Cell Biol., 120, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen L.H. and Aderem, A. (1995) A role for MARCKS, the α isozyme of protein kinase C and myosin I in zymosan phagocytosis by macrophages. J. Exp. Med., 182, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoli C., Martin, M., Leborgne, R., Reggio, H. and Mangeat, P. (1994) Ezrin has properties to self-associate at the plasma membrane. J. Cell Sci., 107, 2509–2521. [DOI] [PubMed] [Google Scholar]
- Bengtsson T., Jaconi, M.E.E., Gustafson, M., Magnusson, K.E., Theler, J., Lew, D.P. and Stendahl, O. (1993) Actin dynamics in human neutrophils during adhesion and phagocytosis is controlled by changes in intracellular free calcium. Eur. J. Cell Biol., 62, 49–58. [PubMed] [Google Scholar]
- Berryman M., Gary, R. and Bretscher, A. (1995) Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J. Cell Biol., 131, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A., Severin, A., Habermann, A., Hyman, A.A., Griffiths, G. and Burkhardt, J.K. (1996) Microtubule-associated protein-dependent binding of phagosomes to microtubules. J. Biol. Chem., 271, 3803–3811. [DOI] [PubMed] [Google Scholar]
- Blocker A., Severin, F.F., Burkhardt, J.K., Bingham, J.B., Yu, H., Olivo, J.C., Schroer, T.A., Hyman, A.A. and Griffiths, G. (1997) Molecular requirements for bi-directional movement of phagosomes along microtubules. J. Cell Biol., 137, 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1989) Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J. Cell Biol., 108, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1999) Regulation of cortical structure by the ezrin–radixin–moesin protein family. Curr. Opin. Cell Biol., 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Brown S.S. and Spudich, J.A. (1981) Mechanism of action of cytochalasin: evidence that it binds to actin filament ends. J. Cell Biol., 88, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J., Huber, L.A., Dieplinger, H., Blocker, A., Griffiths, G. and Desjardins, M. (1995) Gaining insight into a complex organelle, the phagosome, using two-dimensional gel electrophoresis. Electrophoresis, 16, 2249–2257. [DOI] [PubMed] [Google Scholar]
- Carlier M.F. (1998) Control of actin dynamics. Curr. Opin. Cell Biol., 10, 45–51. [DOI] [PubMed] [Google Scholar]
- Caron E. and Hall, A. (1998) Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science, 282, 1717–1721. [DOI] [PubMed] [Google Scholar]
- Carraway K.L. and Carraway, C.A. (1989) Membrane–cytoskeleton interactions in animal cells. Biochim. Biophys. Acta, 988, 147–171. [DOI] [PubMed] [Google Scholar]
- Carson M., Weber, A. and Zigmond, S.H. (1986) An actin-nucleating activity in polymorphonuclear leukocytes is modulated by chemotactic peptides. J. Cell Biol., 103, 2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Safer, D., Nachmias, V.T. and Zigmond, S.H. (1992) Thymosin β4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. J. Cell Biol., 119, 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cohn, J.A. and Mandel, L.J. (1995) Dephosphorylation of ezrin as an early event in renal microvillar breakdown and anoxic injury. Proc. Natl Acad. Sci. USA, 92, 7495–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choidas A., Jungbluth, A., Sechi, A., Murphy, J., Ullrich, A. and Marriott, G. (1998) The suitability and application of a GFP–actin fusion protein for long-term imaging of the organization and dynamics of the cytoskeleton in mammalian cells. Eur. J. Cell Biol., 77, 81–90. [DOI] [PubMed] [Google Scholar]
- Claus V., Jahraus, A., Tjelle, T., Berg, T., Kirsche, H., Faulstich, H. and Griffiths, G. (1998) Lysosomal enzyme trafficking between phagosomes, endosomes and lysosomes in J774 macrophages. J. Biol. Chem., 273, 9842–9851. [DOI] [PubMed] [Google Scholar]
- Condeelis J. and Hall, A. (1991) Measurement of actin polymerization and cross-linking in agonist-stimulated cells. Methods Enzymol., 196, 486–493. [DOI] [PubMed] [Google Scholar]
- Condeelis J., Hall, A., Bresnick, A., Warren, V., Hock, R., Bennett, H. and Ogihara, S. (1988) Actin polymerization and pseudopod extension during amoeboid chemotaxis. Cell Motil. Cytoskel., 10, 77–90. [DOI] [PubMed] [Google Scholar]
- Cooper J.A. (1987) Effects of cytochalasin and phalloidin on actin. J. Cell Biol., 105, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. and Lecuit, M. (1998) Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J., 17, 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepaldi T., Gautreau, A., Comoglio, P.M., Louvard, D. and Arpin, M. (1997) Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J. Cell Biol., 138, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M., Celis, J.E., Van Meer, G., Dieplinger, H., Jarhaus, A., Griffiths, G. and Huber, L. (1994a) Molecular characterization of phagosomes. J. Biol. Chem., 269, 32194–32200. [PubMed] [Google Scholar]
- Desjardins M., Huber, L., Parton, R.G. and Griffiths, G. (1994b) Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol., 124, 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakonova M., Gerke, V., Ernst, J., Liautard, J.P., van der Vusse, G. and Griffiths, G. (1997) Localization of five annexins in J774 macrophages and on isolated phagosomes. J. Cell Sci., 110, 1199–1213. [DOI] [PubMed] [Google Scholar]
- Doi Y., Itoh, M., Yonemura, S., Ishihara, S., Takano, H., Noda, T. and Tsukita, S. (1999) Normal development of mice and unimpaired cell adhesion/cell motility/actin-based cytoskeleton without compensatory up-regulation of ezrin or radixin in moesin gene knockout. J. Biol. Chem., 274, 2315–2321. [DOI] [PubMed] [Google Scholar]
- Echner H. and Voelter,W. (1988) Eine neue synthese von thymosin α1. Liebigs Ann. Chem., 1095–1097. [Google Scholar]
- Eddy R.J. and Condeelis, J.S. (1997) Capping protein terminates but does not initiate chemoattractant-induced actin assembly in Dictyostelium.J. Cell Biol., 139, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emans E., Nzala, N.N. and Desjardins, M. (1996) Protein phosphorylation during phagosome maturation. FEBS Lett., 1, 37–42. [DOI] [PubMed] [Google Scholar]
- Gilmore A.P. and Burridge, K. (1996) Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4,5-bisphosphate. Nature, 381, 531–535. [DOI] [PubMed] [Google Scholar]
- Gondo H., Kudo, J., White, J.W., Barr, C., Selvanayagam, P. and Saunders, G.F. (1987) Differential expression of the human thymosin-β4 gene in lymphocytes, macrophages and granulocytes. J. Immunol., 139, 3840–3848. [PubMed] [Google Scholar]
- Greenberg S. and Silverstein,S.C. (1993) Phagocytosis. In Paul,W.E. (ed.), Fundamental Immunology. Raven Press, New York, NY, pp. 941–964. [Google Scholar]
- Greenberg S., El Khoury, J., Di Virgilio, F., Kaplan, E.M. and Silverstein, S.C. (1991) Ca2+-independent F-actin assembly and disassembly during Fc receptor-mediated phagocytosis in mouse macrophages. J. Cell Biol., 113, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G. (1993) Cryo and replica techniques for immunolabeling. In Fine Structure Immunochemistry. Springer Verlag, Berlin, Germany, pp. 137–191. [Google Scholar]
- Griffiths G. (1996) On vesicles and membrane compartments. Protoplasma, 195, 37–58. [Google Scholar]
- Hanzel D., Reggio, H., Bretscher, A., Forte, J.G. and Mangeat, P. (1991) The secretion-stimulated 80K phosphoprotein of parietal cells is ezrin and has properties of a membrane cytoskeletal linker in the induced apical microvilli. EMBO J., 10, 2363–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig J.H. (1992) Mechanisms of actin rearrangements mediating platelet activation. J. Cell Biol., 118, 1421–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiska L., Alfthan, K., Gronholm, M., Vilja, P., Vaheri, A. and Carpen, O. (1998) Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM–1 and ICAM-2). Regulation by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem., 273, 21893–21900. [DOI] [PubMed] [Google Scholar]
- Hirao M., Sato, N., Kondo, T., Yonemura, S., Monden, M., Sasaki, T., Takai, Y., Tsukita, S. and Tsukita, S. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane interaction: possible involvement of phosphatidylinositol turnover and rho-dependent signaling pathway. J. Cell Biol., 135, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard T.H. and Oresajo, C.O. (1985) The kinetics of chemotactic peptide-induced change in F-actin content, F-actin distribution and the shape of neutrophils. J. Cell Biol., 101, 1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg G. and Goldmann, W.H. (1998) Peptide-specific antibodies localize the major lipid binding sites of talin dimers to oppositely arranged N-terminal 47 kDa subdomains. FEBS Lett., 426, 165–170. [DOI] [PubMed] [Google Scholar]
- Isenberg G. and Niggli, V. (1998) Interaction of cytoskeletal proteins with membrane lipids. Int. Rev. Cytol., 178, 73–125. [DOI] [PubMed] [Google Scholar]
- Jahraus A., Tjelle, T.E., Berg, T., Habermann, A., Storrie, B., Ullrich, O. and Griffiths, G. (1998) In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J. Biol. Chem., 273, 30379–30390. [DOI] [PubMed] [Google Scholar]
- Kersken H., Vilmart-Seuwen, J., Momayezi, M. and Plattner, H. (1986) Filamentous actin in Paramecium cells: mapping by phalloidin affinity labeling in vivo and in vitro.J. Histochem. Cytochem., 34, 443–454. [DOI] [PubMed] [Google Scholar]
- Kouyama T. and Mihashii, K. (1981) Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labeled F-actin. Eur. J. Biochem., 114, 33–38. [PubMed] [Google Scholar]
- Lamaze C., Fujimoto, L.M., Yin, H.L. and Schmid, S.L. (1997) The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J. Biol. Chem., 272, 20332–20335. [DOI] [PubMed] [Google Scholar]
- Ma L., Cantley, L.C., Janmey, P.A. and Kirschner, M.W. (1998) Corequirement of specific phosphoinositides and small GTP-binding protein cdc42 in inducing actin assembly in Xenopus egg extracts. J. Cell Biol., 140, 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D.J., Esch, F., Furthmayr, H. and Hall, A. (1997) Rho- and rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J. Cell Biol., 138, 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat P., Roy, C. and Martin, M. (1999) ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol., 9, 187–192. [DOI] [PubMed] [Google Scholar]
- Margossian S.S. and Lowey, S. (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol., 85, 55–71. [DOI] [PubMed] [Google Scholar]
- Matsui T., Maeda, M., Doi, Y., Yonemura, S., Amano, M., Kaibuchi, K., Tsukita, S. and Tsukita, S. (1998) Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol., 140, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J. and Cramer, L.P. (1996) Actin-based cell motility and cell locomotion. Cell, 84, 371–379. [DOI] [PubMed] [Google Scholar]
- Muallem S., Kwiatkowska, K., Xu, X. and Yin, H.L. (1995) Actin filament disassembly is a sufficient final trigger for exocytosis in nonexcitable cells. J. Cell Biol., 128, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D. and Pollard, T.D. (1999) Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol., 9, 244–249. [DOI] [PubMed] [Google Scholar]
- Nakamura F., Huang, L., Pestonjamasp, K., Luna, E.J. and Furthmayr, H. (1999) Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell, 10, 2669–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V., Andreoli, C., Roy, C. and Mangeat, P. (1995) Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett., 376, 172–176. [DOI] [PubMed] [Google Scholar]
- Oshiro N., Fukata, Y. and Kaibuchi, K. (1998) Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J. Biol. Chem., 273, 34663–34666. [DOI] [PubMed] [Google Scholar]
- Pantaloni D. and Carlier, M.F. (1993) How profilin promotes actin filament assembly in the presence of thymosin β4. Cell, 75, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Pardee J.D., Simpson, P.A., Stryer, L. and Spudich, J.A. (1982) Actin filaments undergo limited subunit exchange in physiological salt conditions. J. Cell Biol., 94, 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. and Cooper, J. (1986) Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem., 55, 987–1035. [DOI] [PubMed] [Google Scholar]
- Rabinovitch M. (1995) Professional and non-professional phagocytes: an introduction. Trends Cell Biol., 5, 85–87. [DOI] [PubMed] [Google Scholar]
- Reaven E.P. and Axline, S.G. (1973) Subplasmalemmal microfilaments and microtubules in resting and phagocytizing cultivated macrophages. J. Cell Biol., 59, 12–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D. and Bretscher, A. (1998) The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem., 273, 18452–18458. [DOI] [PubMed] [Google Scholar]
- Rezabek B.L., Rodriguez-Paris, J.M., Cardelli, J.A. and Chia, C.P. (1997) Phagosomal proteins of Dictyostelium discoideum.J. Eukar. Microbiol., 44, 284–292. [DOI] [PubMed] [Google Scholar]
- Roy C., Martin, M. and Mangeat, P. (1997) A dual involvement of the amino-terminal domain of ezrin in F- and G-actin binding. J. Biol. Chem., 272, 20088–20095. [DOI] [PubMed] [Google Scholar]
- Sbarra A.J. and Karnovsky, M.L. (1959) The biological basis of phagocytosis. I. Metabolic changes during the ingestion of particles by polymorphonuclear leukocytes. J. Biol. Chem., 234, 1355–1362. [PubMed] [Google Scholar]
- Shariff A. and Luna, E.J. (1990) Dictyostelium discoideum plasma membranes contain an actin-nucleating activity that requires ponticulin, an integral membrane glycoprotein. J. Cell Biol., 110, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.J., Henry, M., Solomon, F. and Jacks, T. (1998) RhoA-dependent phosphorylation and relocalization of ERM proteins into apical membrane/actin protrusions in fibroblasts. Mol. Biol. Cell, 9, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbina A., Bretscher, A., Kenney, D.M., Remold-O'Donnell, E. (1999) Moesin, the major ERM protein of lymphocytes and platelets, differs from ezrin in its insensitivity to calpain. FEBS Lett., 443, 31–36. [DOI] [PubMed] [Google Scholar]
- Simons P.C., Pietromonaco, S.F., Reczek, D., Bretscher, A. and Elias, L. (1998) C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton. Biochem. Biophys. Res. Commun., 253, 561–565. [DOI] [PubMed] [Google Scholar]
- Singer J.M., Adlersberg, L., Hoenig, E.M., Ende, E. and Tchorsch, Y. (1969) Radiolabeled latex particles in the investigation of phagocytosis in vivo: clearance curves and histological observations. J. Reticuloendothel. Soc., 6, 561–589. [PubMed] [Google Scholar]
- Small J.V., Herzog, M. and Anderson, K. (1995) Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol., 129, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockem W., Hoffman, H.U. and Gruber, B. (1983) Dynamics of the cytoskeleton in Amoeba proteus. I. Redistribution of microinjected fluorescein-labeled actin during locomotion, immobilization and phagocytosis. Cell Tissue Res., 232, 79–96. [DOI] [PubMed] [Google Scholar]
- Svitkina T.M. and Borisy, G.G. (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol., 145, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J.A., Johnson, M.T., Beningo, K., Post, P., Mooseker, M. and Araki, N. (1999) A contractile activity that closes phagosomes in macrophages. J. Cell Sci., 112, 307–316. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Sasak, T., Mammoto, A., Takaishi, K., Kameyama, T., Tsukita, S. and Takai, Y. (1997) Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem., 272, 23371–23375. [DOI] [PubMed] [Google Scholar]
- Tchao R. (1982) Novel forms of epithelial cell motility on collagen and on glass surfaces. Cell Motil., 4, 333–341. [DOI] [PubMed] [Google Scholar]
- Tilney L.G. (1976) Actin: its association with membranes and the regulation of its polymerization. In Brinkley,B.R. and Porter,R.K. (eds), International Cell Biology. Rockefeller University Press, Boston, MA, pp. 388–402. [Google Scholar]
- Tilney L.G. and Portnoy, D.A. (1989) Actin filaments and the growth, movement and spread of the intracellular bacterial parasite, Listeria monocytogenes.J. Cell Biol., 109, 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyohara A. and Inaba, K. (1989) Transport of phagosomes in mouse peritoneal macrophages. J. Cell Sci., 94, 143–153. [DOI] [PubMed] [Google Scholar]
- Tsukita S. and Yonemura, S. (1997) ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr. Opin. Cell Biol., 9, 70–75. [DOI] [PubMed] [Google Scholar]
- Vaheri A., Carpen, O., Heiska, L., Helander, T.S., Jaaskelainen, J., Majander-Nordenswan, P., Sainio, M., Timonen, T. and Turunen, O. (1997) The ezrin protein family: membrane–cytoskeleton interactions and disease associations. Curr. Opin. Cell Biol., 9, 659–666. [DOI] [PubMed] [Google Scholar]
- Valles A.M., Boyer, B., Badet, J., Ticker, G.C., Barritault, D. and Thiery, J.P. (1990) Acidic fibroblast growth factor is a modulator of epithelial plasticity in a rat bladder carcinoma cell line. Proc. Natl Acad. Sci. USA, 87, 1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J. and Vancompernolle, K. (1992) Structural relationships of actin-binding proteins. Curr. Opin. Cell Biol., 4, 36–42. [DOI] [PubMed] [Google Scholar]
- Weber A., Nachmias, V.T., Pennise, C.R., Pring, M. and Safer, D. (1992) Interaction of thymosin β4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry, 31, 6179–6185. [DOI] [PubMed] [Google Scholar]
- Weeds A.G. and Taylor, R.S. (1975) Separation of subfragment–1 isoenzymes from rabbit skeletal muscle myosin. Nature, 257, 54–55. [DOI] [PubMed] [Google Scholar]
- Welch M.D., Rosenblatt, J., Skoble, J., Portnoy, D.A. and Mitchison, T.J. (1998) Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science, 281, 105–108. [DOI] [PubMed] [Google Scholar]
- Westphal M., Jungbluth, A., Heidecker, M., Mühlbauer, C., Heizer, C., Schwartz, J.M., Marriott, G. and Gerisch, G. (1997) Microfilament dynamics during cell movement and chemotaxis monitored using a GFP–actin fusion protein. Curr. Biol., 7, 176–183. [DOI] [PubMed] [Google Scholar]
- Yao X., Cheng, L. and Forte, J.G. (1996) Biochemical characterization of ezrin–actin interaction. J. Biol. Chem., 271, 7224–7229. [DOI] [PubMed] [Google Scholar]
- Yonemura S., Hirao, M., Doi, Y., Takahashi, N., Kondo, T. and Tsukita, S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43 and ICAM-2. J. Cell Biol., 140, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H., Joyce, M., Borleis, J., Bokoch, G.M. and Devreotes, P.N. (1997) Regulation of actin polymerization in cell-free systems by GTPγS and cdc42. J. Cell Biol., 138, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]