Abstract
The mechanisms by which peroxisomal membrane proteins (PMPs) are targeted to and inserted into membranes are unknown, as are the required components. We show that among a collection of 16 Saccharomyces cerevisiae peroxisome biogenesis (pex) mutants, two mutants, pex3Δ and pex19Δ, completely lack detectable peroxisomal membrane structures and mislocalize their PMPs to the cytosol where they are rapidly degraded. The other pexΔ mutants contain membrane structures that are properly inherited during vegetative growth and that house multiple PMPs. Even Pex15p requires Pex3p and Pex19p for localization to peroxisomal membranes. This PMP was previously hypothesized to travel via the endoplasmic reticulum (ER) to peroxisomes. We provide evidence that ER-accumulated Pex15p is not a sorting intermediate on its way to peroxisomes. Our results show that Pex3p and Pex19p are required for the proper localization of all PMPs tested, including Pex15p, whereas the other Pex proteins might only be required for targeting/import of matrix proteins.
Keywords: peroxisomal membrane protein/protein degradation/protein targeting/yeast
Introduction
The assembly and maintenance of peroxisomes depend on a variety of processes including targeting and import of peroxisomal matrix and membrane proteins, recruitment of lipids, and proliferation and inheritance of the organelle. The isolation and characterization of mutants that are defective in peroxisome formation (pex) have provided us with a large number of proteins (peroxins) that play an essential role in these processes (Distel et al., 1996). To date, 22 peroxins have been identified, most of which are conserved among different eukaryotic organisms (an updated list can be viewed on the following web site: http://www.mips.biochem.mpg.de/proj/yeast/reviews/pex_table.html). Five of these peroxins, Pex5p, Pex7p, Pex13p, Pex14p and Pex17p, are involved in peroxisomal matrix protein import (for reviews, see Erdmann et al., 1997; Subramani, 1998; Hettema et al., 1999; Tabak et al., 1999). Pex5p and Pex7p are soluble receptors (Marzioch et al., 1994; Dodt and Gould, 1996; Elgersma et al., 1996, 1998; Rehling et al., 1996) that recognize and bind newly synthesized proteins containing a type I or type II peroxisomal targeting signal (PTS), respectively. Pex5p and Pex7p are thought to function as cycling receptors that pick up their cargo in the cytosol, deliver it to docking proteins at the peroxisomal membrane and then shuttle back into the cytosol for a next round of targeting. Protein–protein interaction studies have shown that the membrane-located docking complex is at least comprised of one integral membrane protein, Pex13p (Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996; Girzalsky et al., 1999), and two membrane-associated proteins, Pex14p (Albertini et al., 1997; Brocard et al., 1997; Fransen et al., 1998; Schliebs et al., 1999) and Pex17p (Huhse et al., 1998). Whether these proteins are also part of the translocation machinery or whether the translocation machinery consists of another set of peroxins remains to be elucidated. Nothing is known about the actual membrane translocation process except that proteins can go in retaining a (partially) folded conformation (Glover et al., 1994; McNew and Goodman, 1994; Walton et al., 1995; Häusler et al., 1996). The function of the other peroxins in peroxisome biogenesis remains to be resolved.
Growth and maintenance of the peroxisomal compartment require, besides import of matrix proteins, the specific targeting and insertion of membrane proteins. Peroxisomal membrane proteins (PMPs) are synthesized on free polysomes (Fujiki et al., 1984; Suzuki et al., 1987), and two of them (PMP70 and PMP22) have been shown to be post-translationally inserted into the peroxisomal membrane (Diestelkötter and Just, 1993; Imanaka et al., 1996). These observations have led to the view that PMPs, like matrix proteins, are imported directly from the cytosol (Lazarow and Fujiki, 1985). On the other hand, one PMP (PMP50) was reported to be synthesized on membrane-bound polyribosomes (Bodnar and Rachubinski, 1991); however, the gene encoding PMP50 has not been identified and further studies on its targeting are lacking. Furthermore, under certain conditions PMPs have been found to be associated with the endoplasmic reticulum (ER). These include overexpression of Pex15p in Saccharomyces cerevisiae (Elgersma et al., 1997) and truncation of Pex3p in Hansenula polymorpha (Baerends et al., 1996). Whether these PMPs under physiological conditions follow an ER pathway remains to be analysed. Nevertheless, the import of PMPs probably requires a unique set of proteins because it is not dependent on the components of the pathway for matrix protein import. Consistent with this notion is the identification of a PTS for PMPs (mPTS) that bears no resemblance to either PTS1 or PTS2 (Baerends et al., 1996; Dyer et al., 1996; Wiemer et al., 1996; Elgersma et al., 1997). Two yeast peroxins, Pex3p and Pex19p, have been suggested to be required for peroxisomal membrane formation (Höhfeld et al., 1991; Baerends et al., 1996; Wiemer et al., 1996; Götte et al., 1998). This suggestion is largely based on the observation that pex3 and pex19 mutants lack detectable peroxisomal membrane structures. However, these mutants have not been well characterized biochemically; in particular the fates of PMPs in these mutants are largely unknown. Morphological and biochemical studies in man have identified one additional gene, PEX16, which has been suggested to be required for peroxisomal membrane formation (South and Gould, 1999). Remarkably, the S.cerevisiae genome seems to lack a PEX16 homologue.
Here we describe the localization of PMPs in 16 yeast pexΔ mutants. We show that 14 pexΔ mutants harbour PMP-containing peroxisomal membranes, similar to those originally described in patients suffering from peroxisome biogenesis disorders (Santos et al., 1988). However, two mutants, pex3Δ and pex19Δ, are shown here to be defective in peroxisomal membrane formation. Specifically, these mutants lack detectable peroxisomal membranes and mislocalize their PMPs to the cytosol. We provide evidence that in pex3Δ and pex19Δ cells the mislocalized PMPs, but not the mislocalized matrix proteins, are unstable and rapidly degraded. These results suggest that Pex3p and Pex19p play an essential role either in the assembly or in the maintenance of the peroxisomal membrane. We also found that the peroxisomal localization and the stability of endogenous Pex15p depend on Pex3p and Pex19p. However, overexpressed Pex15p is found in the ER independently of the presence of these proteins. These results suggest that the observed ER localization of Pex15p is probably not due to trapping of the protein on its way to peroxisomes but is caused by mistargeting of the protein resulting from its overexpression. Our results stress that experiments in which PMPs are overexpressed should be interpreted with great care.
Results
pex3Δ and pex19Δ cells lack Pex11p-containing membrane structures
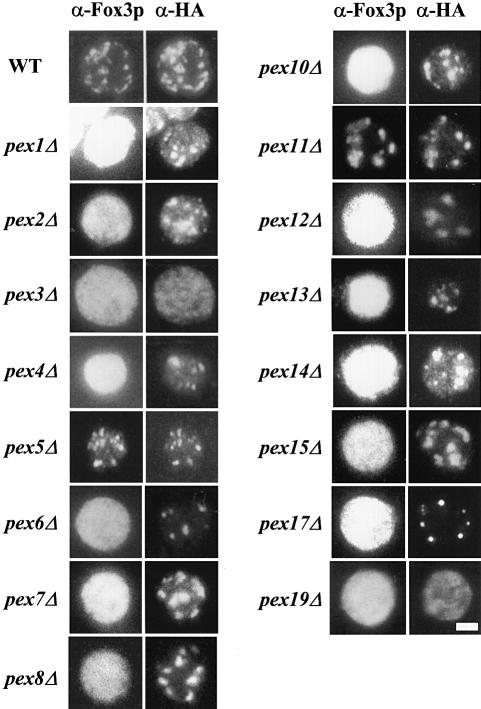
pex mutants in yeast are characterized by the mislocalization of peroxisomal matrix proteins to the cytosol and the absence of (normal) peroxisomes. Little is known, however, about the localization of PMPs in these pex mutants. To identify pex mutants that are defective in localization of PMPs we expressed an HA-tagged version of the peroxisomal membrane marker Pex11p in 16 different yeast strains each containing a deletion of a particular PEX gene. The HA-tagged version of Pex11p was expressed under the control of its own promoter on a single copy plasmid. This construct was able to rescue the oleate-non-utilizing (onu) phenotype of a pex11Δ strain, indicating that the localization experiments were carried out with a functional form of Pex11p (Erdmann and Blobel, 1995).
Yeast cells expressing Pex11p–HA were processed for indirect immunofluorescence using antibodies specific for the HA tag and antibodies against a peroxisomal matrix protein, thiolase (Figure 1). In wild-type cells, double labelling immunofluorescence microscopy revealed a congruent punctate pattern for thiolase and Pex11p–HA, confirming that Pex11p–HA is targeted to peroxisomes. Pex11p–HA-containing structures were also detected in most pexΔ mutants with the exception of pex3Δ and pex19Δ cells (Figure 1; Table I). These two mutants showed a very weak, diffuse staining when processed for anti-HA immunofluorescence microscopy, suggesting that Pex11p–HA was mislocalized in these cells. To confirm the absence of a particulate staining in pex3Δ and pex19Δ, we also expressed a GFP-tagged version of Pex11p in these cells. The localization of the fusion protein was studied in living cells by confocal fluorescence microscopy. Again, in pex3Δ and pex19Δ cells, a weak, diffuse labelling was observed, whereas in wild-type cells and in all other pexΔ mutants a particulate staining pattern was observed (Table I).
Fig. 1. pex3Δ and pex19Δ cells lack Pex11p–HA-containing membrane structures. Oleic acid-induced wild-type and pexΔ mutant cells expressing HA-tagged Pex11p as a marker for peroxisomal membranes were processed for double immunofluorescence microscopy using rabbit polyclonal antibodies specific for peroxisomal thiolase (Fox3p) and mouse monoclonal antibodies specific for the HA epitope. Secondary antibodies were CY3-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG. Bar, 5 μm.
Table I. PMP localization and stability in pexΔ mutants.
| Strain | IF/confocala | EMb | PMP stabilityc |
|---|---|---|---|
| wild type | + | +d | NAe |
| pex1Δ | + | + | + |
| pex2Δ | + | + | + |
| pex3Δ | − | − | − |
| pex4Δ | + | + | + |
| pex5Δ | + | + | + |
| pex6Δ | + | + | + |
| pex7Δ | + | +d | + |
| pex8Δ | + | + | + |
| pex10Δ | + | + | + |
| pex11Δ | + | +d | + |
| pex12Δ | + | + | + |
| pex13Δ | + | + | + |
| pex14Δ | + | + | + |
| pex15Δ | + | + | + |
| pex17Δ | + | + | + |
| pex19Δ | − | − | − |
aThe presence (+) or absence (−) of punctated structures in cells expressing PEX11–HA or PEX11–GFP as determined by indirect immunofluorescence microscopy and confocal fluorescence microscopy, respectively.
bThe presence (+) or absence (–) of labelled membrane structures in cells expressing PEX11–HA as determined by immunogold labelling with anti-HA.
cSteady-state levels of PMPs in pex mutants as determined by Western blotting. (+), 80–100% of that of wild-type cells; (–), <20% of that of wild-type cells.
dMorphologically normal peroxisomes.
eNA, not applicable.
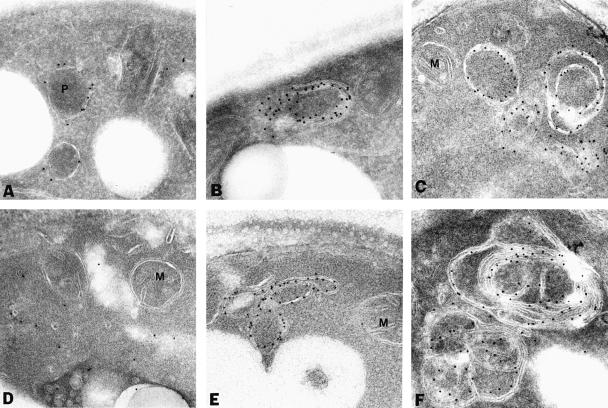
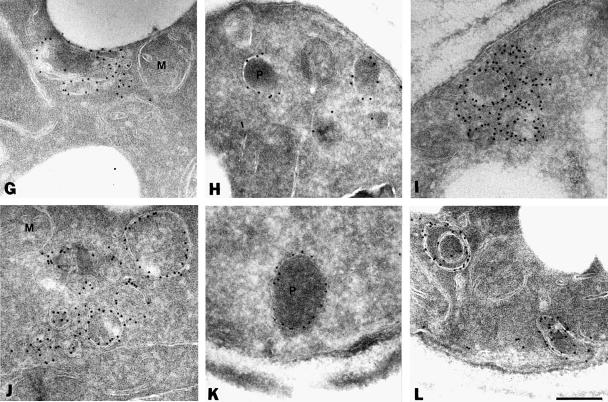
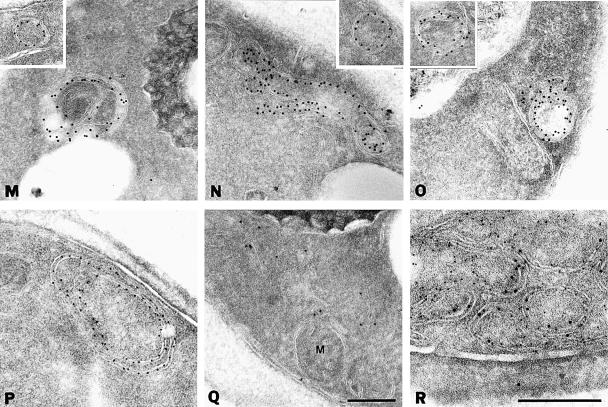
To characterize the PMP-containing structures further, yeast cells expressing Pex11p–HA were processed for immunoelectron microscopy using anti-HA and protein A-conjugated gold. In wild-type cells, gold particles decorated the peroxisomal membranes only, thereby confirming the peroxisomal localization of Pex11p–HA (Figure 2A) (Erdmann and Blobel, 1995). In most pexΔ mutants Pex11p–HA was present in membranous structures (Figure 2B, C and E–P). The appearance of the PMP-containing membrane structures was quite similar in most of the pexΔ mutants, although some variation was observed even within the same pexΔ strain. In addition to large membrane structures that often showed the presence of concentric membranes, smaller vesicular structures were detected (Figure 2, insets). Also, these latter structures were generally bounded by more than one membrane. The appearence of these membrane structures is reminiscent of the ‘ghosts’ described by Motley et al. (1994) in cells of peroxisome biogenesis disorder patients. Double labelling experiments in pex4Δ cells showed that these membranes also contained the peroxisomal integral membrane proteins Pex13p (Figure 2R) and Pex15p (data not shown). Collectively, these results show that the punctate fluorescent structures detected in the majority of pexΔ mutants represented membrane structures that contain several PMPs. Therefore, these mutants were disturbed in matrix protein import but they could still assemble their PMPs into membranes. On the other hand, no immunogold labelling of membranes was observed in pex3Δ and pex19Δ cells but instead the gold particles were randomly distributed over the cytosol (Figure 2D and Q). These data are in agreement with the localization of Pex11p–HA by indirect immunofluorescence miscroscopy (Figure 1).
Fig. 2. Electron microscopic analysis of Pex11p–HA localization. Oleic acid-induced wild-type and pexΔ mutant cells expressing Pex11p–HA were fixed and processed for immunogold electron microscopy using antibodies specific for the HA epitope. (A) Wild-type cells; (B) pex1Δ cells; (C) pex2Δ cells; (D) pex3Δ cells; (E) pex4Δ cells; (F) pex5Δ cells; (G) pex6Δ cells; (H) pex7Δ cells; (I) pex8Δ cells; (J) pex10Δ cells; (K) pex11Δ cells; (L) pex12Δ cells; (M) pex13Δ cells; (N) pex14Δ cells; (O) pex15Δ cells; (P) pex17Δ cells; (Q) pex19Δ cells; (R) colocalization of GFP-tagged Pex13p (anti-GFP, 10 nm gold) and Pex11p–HA (anti-HA, 5 nm gold) in pex4Δ cells. P, peroxisome; M, mitochondrion. Bar, 0.25 μm.
pex3Δ and pex19Δ cells mislocalize their PMPs to the cytosol
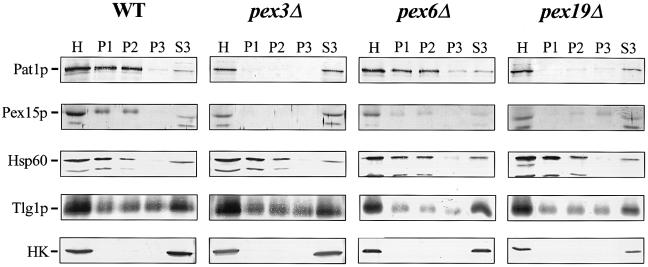
The diffuse staining of Pex11p–HA in pex3Δ and pex19Δ cells suggests that these proteins are mislocalized to the cytosol. To examine the subcellular distribution of PMPs in pex3Δ and pex19Δ cells in more detail, lysates were prepared and subjected to differential centrifugation. Both wild-type cells and pex6Δ cells were included as controls. Sphearoplasts were prepared and lysed gently by osmotic shock in the presence of protease inhibitors. After a centrifugation step at 600 g to remove intact cells and nuclei, the homogenate (H) was fractionated by sequential differential centrifugation generating a 2500 g pellet (P1), a 25000g pellet (P2), a 150000g pellet (P3) and a 150000g supernatant (S3). Equivalent fractions were analysed by immunoblotting with antibodies specific for the PMPs Pat1p and Pex15p and several marker proteins (Figure 3). The mitochondrial protein Hsp60 was recovered primarily from the P1 and P2 fractions. The endosomal syntaxin Tlg1p distributed between the three pellet fractions with a significant fraction residing in S3. The distribution of Tlg1p in our subcellular fractionation procedure is slightly different from that found by others (Holthuis et al., 1998), because of the relatively high sorbitol concentration (0.6M) in our lysis buffer (data not shown). The cytosolic enzyme hexokinase was detected only in the S3 fraction, indicating that the pellet fractions were not contaminated with cytosol or intact cells. In wild-type cells, the PMPs Pat1p and Pex15p distributed between the P1 and P2 fractions, with only a small amount residing in the S3 fraction. Although in pex6Δ cells, the PMPs distributed mainly between the P1 and P2 fractions, a larger amount was recovered in the P3 and S3 fractions when compared with wild-type cells. In pex3Δ cells, PMPs behaved completely as soluble proteins: Pat1p and Pex15p were recovered quantitatively in the S3 fraction and could not be detected in the P1, P2 and P3 fractions. In pex19Δ cells, PMPs were recovered predominantly from the S3 fraction with only minor amounts residing in P2 and P3. These results suggest that pex3Δ and pex19Δ cells mislocalize their PMPs to the cytosol.
Fig. 3. pex3Δ and pex19Δ cells mislocalize their PMPs to the cytosol. Subcellular distribution of PMPs and marker enzymes in oleic acid-induced wild-type and pexΔ mutant cells. After subcellular fractionation equivalent volumes of the 600 g post-nuclear supernatant [homogenate (H)], 2500 g pellet (P1), 25 000 g pellet (P2), 150 000 g pellet (P3) and 150 000 g supernatant (S3) were analysed by immunoblotting. For detection of the PMPs Pat1p and Pex15p in pex3Δ and pex19Δ cells, samples were concentrated 10–fold by trichloroacetic acid precipitation before loading. Antibodies were directed against the proteins as indicated. Cytosolic marker, hexokinase (HK). Mitochondrial and endosomal membrane markers are Hsp60 and Tlg1p, respectively. PMPs are Pat1p and Pex15p.
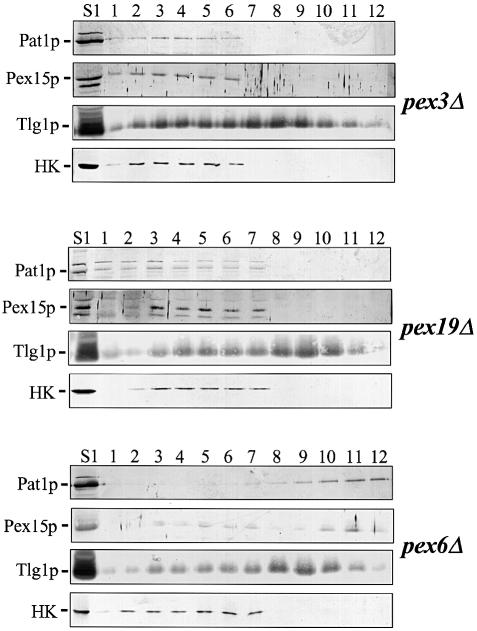
To investigate the possibility that PMPs in pex3Δ and pex19Δ cells are associated with light membrane structures that do not sediment at 150 000 g, the S1 fraction was subjected to floatation analysis. We used the S1 fraction because this fraction does not contain intact cells and nuclei, and lacks most of the mitochondria, but it still contains small membrane structures such as Golgi membranes, transport vesicles, endosomal membranes and peroxisomal membranes (see Figure 3, pex6Δ). Sucrose was added to a final concentration of 55% and the samples were loaded on top of a 60% sucrose cushion and under a 25–46% sucrose gradient (see Materials and methods). The gradients were spun to equilibrium, allowing cellular membranes to migrate from the dense loading fraction into the less dense region of the gradient, due to their intrinsic buoyant densities (Zinser and Daum, 1995). Figure 4 shows that, as expected, Tlg1p floated from the bottom (fractions 1–6) into the less dense region of the gradient (fractions 7–12), whereas the cytosolic enzyme hexokinase remained in the dense fractions. In pex6Δ cells, the PMPs Pat1p and Pex15p floated into the less dense region of the gradient. In contrast, in pex3Δ and pex19Δ cells Pat1p and Pex15p remained in the dense fractions together with the cytosolic marker hexokinase. Although a small fraction of the PMPs was pelleted in pex19Δ cells, these proteins did not float suggesting that they are localized in proteinous aggregates. Floatation gradient analysis of all other pexΔ mutants confirmed that PMPs are associated with membranes in these cells (data not shown). We conclude therefore that these PMPs are mislocalized to the cytosol in pex3Δ and pex19Δ cells, but are associated with membranes in all other pexΔ mutants.
Fig. 4. PMPs are not associated with membranes in pex3Δ and pex19Δ cells. The 2500 g supernatant fraction (S1) prepared from pex3Δ, pex6Δ or pex19Δ cells was adjusted to 55% (w/v) sucrose, layered on top of a 60% sucrose cushion and under a 25–46% sucrose gradient and spun for 18 h at 100 000 g. Fractions obtained from these gradients were analysed by immunoblotting using the antibodies directed against the proteins indicated. Endosomal membrane marker, Tlg1p; cytosolic marker, hexokinase (HK); peroxisomal membrane markers are Pat1p and Pex15p. Fraction 1, bottom; fraction 12, top. Note that the S1 load is located in fractions 2–6 in pex3Δ and in fractions 2–7 in pex6Δ and pex19Δ. Fraction 1 corresponds to the 60% sucrose cushion.
Overproduced Pex15p is associated with ER membranes in pex3Δ cells
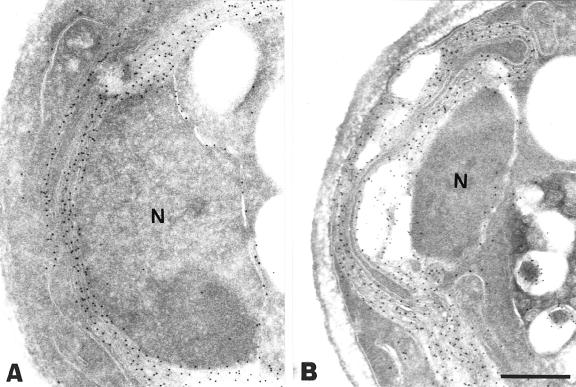
Overexpression of Pex15p in wild-type yeast cells has been reported previously to cause ER membrane proliferation (karmellae formation) with accumulation of the overexpressed protein in these membranes (Elgersma et al., 1997). This observation has led to the hypothesis that Pex15p may be imported into the ER, from where it is further sorted to peroxisomes. However, our data suggest that in both pex3Δ and pex19Δ cells endogenous Pex15p was mislocalized to the cytosol. We therefore determined the localization of overproduced Pex15p in the pex3Δ mutant. As a control, Pex15p was also overexpressed in pex6Δ cells. Immunoelectron microscopy showed that in pex3Δ and pex6Δ cells Pex15p was present in karmellae as well as in the nuclear envelope (Figure 5). These results indicate that the localization of Pex15p in pex3Δ cells depends on its expression level: at physiological levels Pex15p resided in the cytosol whereas overexpressed Pex15p was found in the ER. These results suggest that the ER localization of Pex15p is caused by overexpression of the protein and does not reflect accumulation of a natural Pex15p sorting intermediate.
Fig. 5. Overexpressed Pex15p is associated with ER membranes in both pex3Δ and pex6Δ cells. Immunogold electron microscopy of oleic acid-induced pex3Δ and pex6Δ cells expressing NH-tagged Pex15p under the control of the catalase promoter on a multicopy plasmid. Antibodies directed against the NH tag were used for immunolabelling. (A) pex3Δ cells; (B) pex6Δ cells. Note the labelling of the nuclear envelope in both cells. N, nucleus. Bar, 0.5 μm.
Newly synthesized PMPs are rapidly degraded in pex3Δ and pex19Δ cells
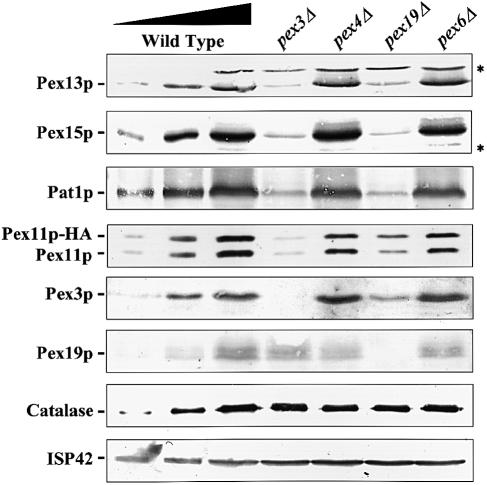
Figure 3 shows that the abundance of PMPs in pex3Δ and pex19Δ cells was low compared with wild-type cells or other pexΔ mutants. To examine the fates of PMPs in pex3Δ and pex19Δ cells in more detail, cell lysates prepared from oleate-grown cells were analysed by immunoblotting with antibodies specific for each of the following proteins: Pex3p, Pex11p, Pex13p, Pex15p and Pat1p (all of these are PMPs). In addition, blots were probed with antibodies directed against Pex19p (a predominantly cytosolic protein), catalase (a peroxisomal matrix protein) and ISP42 (a mitochondrial membrane protein). As a control, total cellular protein was also extracted from wild-type cells and all other pexΔ mutants (Figure 6; Table I). Figure 6 shows that the steady-state levels of the five tested PMPs had decreased drastically in pex3Δ and pex19Δ cells when compared with wild-type cells and the other pexΔ mutants (shown for pex4Δ and pex6Δ). The estimated steady-state levels of PMPs in pex3Δ and pex19Δ cells range between 5 and 20% of the levels in wild-type cells. The expression level of Pex11p–HA was comparable to the level found for the endogenous Pex11p: strongly decreased in pex3Δ and pex19Δ cells, but not affected in wild-type cells and the other pexΔ mutants. Interestingly, the Pex3p level was strongly decreased in pex19Δ cells, whereas the level of Pex19p in pex3Δ cells was not lowered. The steady-state levels of catalase and ISP42 (Figure 6) as well as Pex5p (a predominantly cytosolic protein) and Pex14p (a membrane-associated protein) (data not shown) were not affected in pex3Δ and pex19Δ cells. Our data are consistent with the idea that the deficiency in Pex3p and Pex19p specifically affects the steady-state concentration of PMPs but not that of peroxisomal matrix proteins. The low steady-state concentration of PMPs could be caused by either a decrease in synthesis or an increase in degradation of PMPs. Since these experiments were carried out in a pep4–3 strain that is deficient in multiple vacuolar hydrolases, increased degradation by the vacuole could be excluded. Indeed, comparable results were obtained when the steady-state levels of PMPs were determined in a PEP4 strain (data not shown). Reduced steady-state levels of PMPs were also found in pex3Δ and pex19Δ cells grown either on glucose or on glycerol (data not shown). The reduction of PMP expression therefore seems to be independent of the proliferation state of the organelles, suggesting that in pex3Δ and pex19Δ cells not the synthesis but the stability of PMPs is affected.
Fig. 6. The steady-state levels of PMPs are reduced in pex3Δ and pex19Δ cells. Total cellular protein was extracted from oleic acid-induced wild-type, pex3Δ, pex4Δ, pex6Δ and pex19Δ cells. Equal amounts of protein were separated by SDS–PAGE and blotted with antibodies specific for the PMPs Pat1p, Pex3p, Pex11p, Pex13p and Pex15p. In addition, blots were probed with antibodies specific for catalase (peroxisomal matrix protein), Pex19p (predominantly cytosolic protein) and ISP42 (mitochondrial membrane protein). Lanes 1 and 2 of each blot contain 10 and 30% of protein extract from wild-type cells, respectively. The lysates used for the Pex11p blot were prepared from Pex11p–HA-expressing cells. Asterisks mark cross-reacting bands.
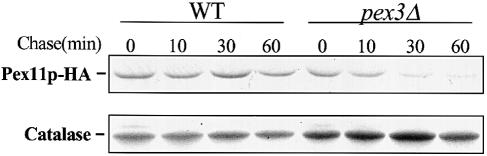
To distinguish further between these two possibilities, we carried out pulse–chase experiments with metabolically labelled cells carrying a pep4–3 mutation. Wild-type and pex3Δ cells expressing Pex11p–HA were pulse-labelled for 10 min with [35S]methionine and [35S]cysteine, and chased up to 60 min with an excess of unlabelled methionine and cysteine. Pex11p–HA and catalase (as a control) were recovered by immunoprecipitation using HA- and catalase-specific antibodies, respectively, and precipitated proteins were visualized by SDS–PAGE and fluorography (Figure 7). Similar amounts of Pex11p–HA were immunoprecipitated from wild-type and pex3Δ cells at 0 min chase, indicating that Pex11p–HA is synthesized at similar rates in both strains. However, during the chase, 35S–labelled Pex11p–HA was severely reduced in pex3Δ cells but not in wild-type cells. Thus, in pex3Δ cells Pex11p–HA was synthesized normally but was then rapidly degraded, whereas in wild-type cells Pex11p–HA was relatively stable during the entire chase period. The amount of radiolabelled catalase, which was immunoprecipitated from the same samples, remained unchanged during the chase in both pex3Δ and wild-type cells. These results imply that the low steady-state levels of PMPs in pex3Δ (and pex19Δ) cells are caused by an increased rate of PMP degradation.
Fig. 7. Newly synthesized Pex11p–HA is rapidly degraded in pex3Δ cells. Wild-type and pex3Δ cells expressing Pex11p–HA were pulse-labelled for 10 min with [35S]methionine and [35S]cysteine, and chased for different times with an excess of unlabelled methionine and cysteine. Pex11p–HA and catalase A were recovered by immunoprecipitation using anti-HA and anti-catalase A antibodies, respectively, and visualized by SDS–PAGE and fluorography.
Discussion
A large collection of pex mutants have previously been selected on the basis of mislocalization of peroxisomal matrix proteins to the cytosol. We constructed 16 different yeast strains, each containing a deletion of a particular PEX gene, and identified among this collection two mutants, pex3Δ and pex19Δ, which also failed to assemble PMPs into membranes. These observations imply that most peroxins found thus far are specifically required for matrix protein localization and are not essential for PMP localization. Furthermore, our results show that all pexΔ mutants (except pex3Δ and pex19Δ) contain peroxisomal membrane structures that are similar in appearance. These observations also suggest that all these mutants have a defect in the same pathway: the import/targeting of matrix proteins into peroxisomes. Indeed, some of these peroxins have been shown to be directly involved in peroxisomal matrix protein targeting. They include the mobile PTS receptors (Pex5p and Pex7p) and components of the peroxisomal membrane complex required for receptor docking (Pex13p, Pex14p and Pex17p) (Erdmann et al., 1997; Subramani, 1998; Hettema et al., 1999; Tabak et al., 1999). Additional peroxins that might be involved in peroxisomal matrix protein import are Pex1p, Pex2p, Pex4p, Pex6p, Pex8p, Pex10p and Pex12p (Dodt and Gould, 1996; Yahraus et al., 1996; Van der Klei et al., 1998; R.Erdmann, unpublished observations). There is experimental support for their function in one of the steps of the PTS–receptor cycle, which involves: (i) binding of newly synthesized PTS-containing proteins in the cytosol; (ii) transport to and docking of receptor–ligand complexes on the peroxisomal membrane; (iii) delivery of the ligand to the translocation machinery and its translocation across the membrane; and (iv) recycling of the receptors back to the cytosol (Tabak et al., 1999). Finally, Pex11p has not been directly linked to the matrix protein import process but has been reported to play a role in peroxisome proliferation in S.cerevisiae (Erdmann and Blobel, 1995; Marshall et al., 1995).
Also, pex3Δ and pex19Δ cells mislocalize matrix proteins to the cytosol. However, it is very likely that the matrix protein import deficiency in these two mutants is a consequence of the failure to assemble PMPs into their membranes. Our data show that the PMPs are mislocalized to the cytosol in pex3Δ and pex19Δ cells where they are rapidly degraded, suggesting that Pex3p and Pex19p are required for proper localization of PMPs. Recently, however, it has been reported that the PMP Pex3p in Pichia pastoris pex19Δ mutant cells might still be associated with membrane structures (Snyder et al., 1999). Since no other membrane proteins besides Pex3p were analysed in these membranes, the subcellular origin of these membrane structures remains unknown. It is even possible that some PMPs might associate with abundant cellular membranes as a consequence of either a lack of a proper target membrane or a targeting machinery (see also below, localization of Pex15p). Unfortunately, the extremely low abundance of Pex3p in our S.cerevisiae pex19Δ cells did not allow analysis of the subcellular localization of this protein. Further studies of the pex19Δ mutant will be required to resolve this issue.
Pex3p is a peroxisomal integral membrane protein. Höhfeld et al. (1991) suggested that Pex3p is anchored in the peroxisomal membrane through an N–terminal hydrophobic region, while the C-terminal portion of the protein is exposed to the cytosol. Pex19p is a farnesylated protein that is predominantly found in the cytosol with only a small fraction found associated with the peroxisomal membrane (Götte et al., 1998). Farnesylation of Pex19p is required both for the biological function of the protein and for its membrane association. Remarkably, Pex3p and Pex19p have been shown to interact in vivo and, thus, they might act in tandem in an as yet undefined step in the assembly of the peroxisomal membrane (Götte et al., 1998). What could be the detailed function of these proteins in this cellular process? Three possible models can be distinguished.
First, Pex3p and Pex19p might function directly in the targeting and insertion of PMPs. Analogous to the mobile PTS1 and PTS2 receptors (Pex5p and Pex7p, respectively), Pex19p might function as a soluble receptor that binds newly synthesized PMPs in the cytosol and subsequently directs the PMPs to the peroxisomal membrane.
Secondly, Pex19p might be a PMP-specific chaperone that keeps PMPs in an import-competent conformation. Both models are consistent with the predominantly cytosolic location of Pex19p. These models also predict that Pex19p interacts with other PMPs, either with other peroxins or with transporters such as Pat1p. Indeed, Pex19p has been reported to interact with the PMP Pex10p (Snyder et al., 1999). This could mean that Pex10p might also be part of the Pex3p- and Pex19p-containing machinery required for the localization of PMPs. However, we show that in contrast to pex3Δ and pex19Δ mutants, cells lacking Pex10p contain typical peroxisomal membrane ghosts (Figures 1 and 2). Therefore, the most simple explanation would be that the observed Pex19p–Pex10p interaction reflects the above mentioned scenarios in which Pex19p acts as either a specific chaperone or a targeting factor for PMPs. In line with this assumption, interactions with several other PMPs have indeed been found for human Pex19p (S.J.Gould, personal communication). In these models the membrane localization of Pex3p therefore suggests a role for this protein at a later stage of the PMP localization process.
Thirdly, Pex3p and Pex19p might play a role in peroxisomal membrane maturation. Recently, peroxisomes were postulated to derive from a pre-peroxisomal structure (South and Gould, 1999). The subcellular origin of these putative pre-peroxisomal structures is not known, but the ER has been suggested (Titorenko and Rachubinski, 1998). In this model, pre-peroxisomal vesicles provide the membranes that first acquire the PMPs and subsequently the matrix proteins, eventually converting the vesicles into mature peroxisomes. The observed mislocalization of PMPs in pex3Δ and pex19Δ cells could, therefore, also be explained by an involvement of Pex3p and Pex19p in the maturation of pre-peroxisomal vesicle into PMP import-competent organelle.
Our differential centrifugation and floatation gradient data revealed that in both pex3Δ and pex19Δ cells PMPs behaved as cytosolic proteins and were not associated with membranes. These observations lend credit to the model postulated by Lazarow and Fujiki (1985), and substantiated by others, that PMPs are synthesized on free polysomes and post-translationally inserted in the peroxisomal membrane. However, our data do not rule out the possibility that some PMPs might travel via the ER, since post-translational membrane protein insertion does occur in the ER of yeast (Kutay et al., 1995). Pex15p is one of the proteins that has been suggested to travel via the ER to the peroxisome (Elgersma et al., 1997). This suggestion was based largely on the observation that overexpression of Pex15p leads to profound ER membrane proliferation (karmellae) and to accumulation of the overexpressed protein in these membranes (Elgersma et al., 1997). Since in the pex3Δ and pex19Δ mutants the endogenous Pex15p is cytosolic and not membrane-bound we determined the localization of overexpressed Pex15p in a pex3Δ mutant. In contrast to the endogenous Pex15p, overexpressed Pex15p was found in karmellae in the pex3Δ mutant. These results suggest that targeting of Pex15p to the ER is caused by overexpression of the protein and does not reflect the import pathway taken by Pex15p at physiological levels. Overexpression of other PMPs in pex3Δ and pex19Δ cells also resulted in membrane proliferation, albeit to a much lesser extent. In pex3Δ and pex19Δ cells, overexpressed Pex13p was found in small membrane vesicles of unknown origin (data not shown). Our experiments show that results obtained upon overexpression of PMPs in pex mutants should be treated with caution.
It is striking that even a very hydrophobic integral membrane protein such as Pat1p, which contains six putative transmembrane regions, does not aggregate in the absence of a membrane. These observations suggest that hydrophobic regions of PMPs are shielded from the aqueous environment during their transport through the cytosol en route to the peroxisome. In addition to a possible function of Pex19p in this step of PMP transport other, more general, chaperones might be involved. Indeed, Pause et al. (1997) showed that in vitro synthesized PMP22 associates with the TCP1 ring complex (TriC), a cytosolic chaperone. pex3Δ and pex19Δ cells will be useful tools for addressing the role of chaperones in PMP targeting and insertion.
While this work was in progress, analysis of cell lines derived from patients suffering from peroxisomal biogenesis disorders indicated that Pex16p and Pex19p are required for PMP import in man (Kinoshita et al., 1998; Honsho et al., 1999; Matsuzono et al., 1999; South and Gould, 1999). Combined with these data, we now have evidence for the essential role of three peroxins in PMP localization: Pex3p, Pex19p and Pex16p. Pex19p has been identified in both yeast and mammals and its function is probably evolutionarily conserved between the two species. A putative mammalian orthologue of Pex3p has been identified as well (Kammerer et al., 1998), but a mammalian cell line deficient for PEX3 is not yet available. Remarkably, the yeast genome seems to lack a PEX16 homologue and the function of Pex16p does not appear to be conserved between different species (Eitzen et al., 1997; Lin et al., 1999; South and Gould, 1999). Further analysis of these three peroxins should provide insight into the molecular mechanism of PMP targeting and insertion.
Materials and methods
Yeast strains and culture conditions
Yeast strains used in this study were S.cerevisiae BJ1991 (Matα, leu2, trp1, ura3–251, prb1–1122, pep4–3, gal2) and UTL–7A (Mata, ura3–52, trp1, leu2–3/112). Mutants were generated by one-step PCR-mediated gene disruption using either the kanMX4 (Wach et al., 1994) or the LEU2 gene as a selectable marker. Deletions were confirmed by Southern blot analysis and by rescue of the onu phenotype of the deletion strain after transformation with the corresponding wild-type PEX gene.
Yeast transformants were selected and grown on minimal medium containing 0.67% yeast nitrogen base without amino acids (YNB–WO) (Difco), 2% glucose and amino acids (20–30 μg/ml) as needed. The liquid media used for culturing of the cells for total protein isolation, subcellular fractionation, immunofluorescence microscopy and immunoelectron microscopy contained 0.5% potassium phosphate buffer pH 6.0, 0.3% yeast extract, 0.5% peptone, 0.1% (v/v) oleate and 0.2% (v/v) Tween–40. Before shifting to this medium, cells were grown on 0.67% YNB–WO containing 0.3% glucose for at least 24 h. Minimal glycerol medium used for pulse–chase experiments contained 0.67% YNB–WO and 3% (w/v) glycerol and amino acids as needed. Oleate plates contained 0.67% YNB–WO, 0.1% yeast extract, 0.1% oleate (v/v), 0.25% (v/v) Tween–40, 2% agar and amino acids as needed.
Immunofluorescence and immunoelectron microscopy
Immunofluorescence microscopy was performed essentially according to the procedure of Rout and Kilmartin (1990) with modifications as previously described (Erdmann and Kunau, 1994). Rabbit antiserum against thiolase was used at dilutions of 1:3000. HA-tagged Pex11p was detected with monoclonal 12CA5 antiserum (BAbCO, Richmond, CA; dilution of 1:1000). For detection, a 6 μg/ml solution of fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit and Cy3-conjugated donkey anti-mouse (Jackson Immuno Research Laboratories, West Grove, PA) was used. For immunogold labelling oleate-induced cells were fixed with 2% paraformaldehyde and 0.5% glutaraldehyde. Ultrathin sections were prepared and incubated as described previously (Distel et al., 1992).
Antibodies
Anti-thiolase (Fox3p) (Erdmann and Kunau, 1994), anti-Pex3p (Höhfeld et al., 1991), anti-Pex13p (Elgersma et al., 1996), anti-Pex15p (Elgersma et al., 1997), anti-catalase (Hettema et al., 1998), anti-Pex19p (Götte et al., 1998) and anti-Pat1p (Hettema et al., 1996) have been described previously. Anti-hexokinase and anti-ISP42 were kindly provided by M.Meijer (Amsterdam, The Netherlands). Anti-Tlg1p was a generous gift of H.Pelham (Cambridge, UK) and J.Holthuis (Amsterdam, The Netherlands). Anti-Pex11p was kindly provided by J.Goodman (Dallas, TX). Anti-NH was a generous gift of P.van der Sluijs (Utrecht, The Netherlands). Anti-GFP was provided by J.Fransen (Nijmegen, The Netherlands).
Pulse–chase experiments
Cells growing exponentially on rich glycerol medium were harvested and resuspended in fresh minimal glycerol medium at OD600 = 0.6 and allowed to grow for 1.5 h at 28°C. Cells were harvested and resuspended in minimal glycerol medium at OD600 = 10. Subsequently, 250 μCi of [35S]methionine/[35S]cysteine were added per eight OD600 units and cells were incubated at 28°C for 10 min. The chase was started by the addition of 6 mM unlabelled methionine and cysteine to the reaction mixtures followed by further incubation at 28°C for 0, 10, 30 or 60 min. Chase reactions were stopped by addition of 0.02% sodium azide and stored on ice. To prepare glass bead lysates, cells were harvested and resuspended in 250 μl of phosphate-buffered saline/1% Triton X–100 containing 1 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail. After addition of glass beads, cells were vortexed for 15 min at maximal speed at 4°C. Glass beads and cell debris were removed by centrifugation at 17 000 g for 2 min at 4°C. Half of the supernatant (100 μl) was used for immunoprecipitation with a monoclonal antibody against HA (12CA5), the second half was used for immunoprecipitation with a polyclonal antibody against catalase A. Antigen–antibody complexes were isolated by the addition of protein A–Sepharose. Precipitates were washed twice with 10 mM Tris–HCl pH 8.6, 300 mM NaCl, 0.05% Triton X–100, 0.05% SDS, and analysed by SDS–PAGE and autoradiography.
Subcellular fractionation experiments
Cells grown overnight on oleate medium were converted to sphaeroplasts with Zymolyase 100T (1 mg/g cells). The sphaeroplasts were washed twice in 1.2 M sorbitol, 5 mM 2(N–morpholino)ethane sulfonic acid (MES) pH 6, 1 mM EDTA and 1 mM KCl. The washed sphaeroplasts were lysed by osmotic shock in 0.65 M sorbitol, 5 mM MES pH 6, 1 mM EDTA and 1 mM KCl (fractionation buffer). Intact cells and nuclei were removed from the homogenate by two centrifugation steps at 600 g for 10 min. The homogenate (H) was further fractionated by sequential differential centrifugation from which we obtained a 2500 g pellet (P1), 25 000 g pellet (P2), 150 000 g pellet (P3) and 150 000 g supernatant (S). Pellet fractions were resuspended in fractionation buffer. Equivalent volumes of these fractions were analysed by SDS–PAGE and immunoblotting.
Floatation gradients
A homogenate (H) was centrifuged at 2500 g for 15 min and 0.5 ml of the supernatant obtained was mixed with 9 vols of 60% sucrose. The sample was loaded on top of a 1 ml 60% sucrose cushion and under a sucrose step gradient consisting of 1 ml fractions of 46, 42, 38, 30 and 25% sucrose (w/v). These gradients were centrifuged for 18 h at 100 000 g in an SW41 rotor at 4°C. Fractions were collected and analysed by SDS–PAGE and immunoblotting. All sucrose solutions were made in fractionation buffer.
Miscellaneous
Protein extracts were prepared by breaking the cells with glass beads and acid precipitation as described by Elgersma et al. (1996). Protein concentrations were determined by the method of Bradford (1976), and verified by SDS–PAGE and protein staining. Construction of HA-tagged PEX11 (Erdmann and Blobel, 1995), NH-tagged PEX13 (Elgersma et al., 1996) and NH-tagged PEX15 (Elgersma et al., 1997) has been described previously. SDS–PAGE and immunoblotting were performed as described by Hettema et al. (1996).
Acknowledgments
Acknowledgements
We are grateful to Jan van Marle and Bart Heuvel for confocal microscopy analysis of Pex11p–GFP-expressing cells, to An Stroobants for her contribution to Pex15p overexpression experiments, to Duco Zonneveld for his help with pulse–chase experiments, and to Henk Tabak, Ineke Braakman, Piet Borst and the members of our laboratories for stimulating discussions. We thank Michiel Meijer, Hugh Pelham, Joost Holthuis, Peter van der Sluijs, Jack Fransen and Joel Goodman for providing us with antibodies. This work was supported by a grant from the Netherlands Organisation of Scientific Research (NWO) and by grants from the Deutsche Forschungsgemeinschaft (ER178/2–2 and SFB 449) and the Fonds der Chemischen Industrie.
References
- Albertini M., Rehling, P., Erdmann, R., Girzalsky, W., Kiel, J.A.K.W., Veenhuis, M. and Kunau, W.-H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell, 89, 1–20. [DOI] [PubMed] [Google Scholar]
- Baerends R.J.S., et al. (1996) The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J. Biol. Chem., 271, 8887–8894. [DOI] [PubMed] [Google Scholar]
- Bodnar A.G. and Rachubinski, R.A. (1991) Characterization of the integral membrane polypeptides of rat liver peroxisomes isolated from untreated and clofibrate-treated rats. Biochem. Cell Biol., 69, 499–508. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilising the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brocard C., Lametschwandtner, G., Koudelka, R. and Hartig, A. (1997) Pex14p is a member of the protein linkage map of Pex5p. EMBO J., 16, 5491–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diestelkötter P. and Just, W.W. (1993) In vitro insertion of the 22 kD peroxisomal membrane protein into isolated rat liver peroxisomes. J. Cell Biol., 123, 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel B., Gould, S., Voorn–Brouwer, T., Van den Berg, M., Tabak, H.F. and Subramani, S. (1992) The carboxyl-terminal tripeptide serine-lysine-leucine of firefly luciferase is necessary but not sufficient for peroxisomal import in yeast. New Biol., 4, 157–165. [PubMed] [Google Scholar]
- Distel B., et al. (1996) A unified nomenclature for peroxisome biogenesis. J. Cell Biol., 135, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G. and Gould, S.J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol., 135, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer J.M., McNew, J.A. and Goodman, J.M. (1996) The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J. Cell Biol., 133, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen G.A., Szilard, R.K. and Rachubinski, R.A. (1997) Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane protein. J. Cell Biol., 137, 1265–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., Kwast, L., Klein, A., Voorn–Brouwer, T., Van den Berg, M., Metzig, B., America, T., Tabak, H.F. and Distel, B. (1996) The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1 containing proteins. J. Cell Biol., 135, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., Kwast, L., Van den Berg, M., Snyder, W.B., Distel, B., Subramani, S. and Tabak, H.F. (1997) Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in Saccharomyces cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J., 16, 7326–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., Elgersma, H.M., Wenzel, T., McCaffery, J.M., Farquhar, M.G. and Subramani, S. (1998) A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris.J. Cell Biol., 140, 807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. and Blobel, G. (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol., 128, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. and Blobel, G. (1996) Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol., 135, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. and Kunau, W.-H. (1994) Purification and immunolocalization of the peroxisomal 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae.Yeast, 10, 1173–1182. [DOI] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis, M. and Kunau, W.H. (1997) Peroxisomes: organelles at the crossroads. Trends Cell Biol., 7, 400–407. [DOI] [PubMed] [Google Scholar]
- Fransen M., Terlecky, S.R. and Subramani, S. (1998) Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl Acad. Sci. USA, 95, 8087–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Rachubinski, R.A. and Lazarow, P.B. (1984) Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc. Natl Acad. Sci. USA, 81, 7127–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky W., Rehling, P., Stein, K., Kipper, J., Blank, L., Kunau, W.-H. and Erdmann, R. (1999) Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J. Cell Biol., 144, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Andrews, D.W. and Rachubinski, R.A. (1994) Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl Acad. Sci. USA, 91, 10541–10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K., Girzalsky, W., Linkert, M., Baumgart, E., Kammerer, S., Kunau, W.-H. and Erdmann, R. (1998) Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol., 18, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Kalish, J.E., Morrell, J.C., Bjorkman, J., Urquhart, A.J. and Crane, D.I. (1996) An SH3 protein in the peroxisome membrane is a docking factor for the PTS1 receptor. J. Cell Biol., 135, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler T., Stierhof, Y.-D., Wirtz, E. and Clayton, E. (1996) Import of a DHFR hybrid protein into glycosomes in vivo is not inhibited by the folate-analogue aminopterin. J. Cell Biol., 132, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Van Roermund, C.W.T., Distel, B., Van den Berg, M., Vilda, C., Rodrigues–Pousada, C., Wanders, R.J.A. and Tabak, H.F. (1996) The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae.EMBO J., 15, 3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Ruigrok, C.C.M., Groot Koerkamp, M., Van den Berg, M., Tabak, H.F., Distel, B. and Braakman, I. (1998) The cytosolic DnaJ-like protein Djp1p is involved specifically in peroxisomal protein import. J. Cell Biol., 142, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Distel, B. and Tabak, H.F. (1999) Import of proteins into peroxisomes. Biochim. Biophys. Acta, 1451, 1–18. [DOI] [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis, M. and Kunau, W.H. (1991) PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol., 114, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C.M., Nichols, B. and Pelham, H.R.B. (1998) The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell, 9, 3383–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho M., Tamura, S., Shimozawa, N., Suzuki, Y., Kondo, N. and Fujiki, Y. (1999) Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome complementation group D. Am. J. Hum. Genet., 63, 1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B., Rehling, P., Albertini, M., Meller, K. and Kunau, W.-H. (1998) Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal translocation machinery. J. Cell Biol., 140, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Shiina, Y., Takano, T., Hashimoto, T. and Osumi, T. (1996) Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro.J. Biol. Chem., 271, 3706–3713. [DOI] [PubMed] [Google Scholar]
- Kammerer S., Holzinger, S., Welsch, U. and Roscher, A.A. (1998) Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. FEBS Lett., 429, 53–60. [DOI] [PubMed] [Google Scholar]
- Kinoshita N., et al. (1998) Newly identified Chinese hamster ovary cell mutants are defective in biogenesis of peroxisomal membrane vesicles (peroxisomal ghosts), representing a novel complementation group in mammals. J. Biol. Chem., 273, 24122–24130. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert, H.G., Hartmann, E., Wiedenmann, B. and Rapoport, T.A. (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J., 14, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P.B. and Fujiki, Y. (1985) Biogenesis of peroxisomes. Annu. Rev. Cell Biol., 1, 489–530. [DOI] [PubMed] [Google Scholar]
- Lin Y., Sun, L., Nguyen, L.V., Rachubinski, R.A. and Goodman, H.M. (1999) The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science, 284, 328–330. [DOI] [PubMed] [Google Scholar]
- Marshall P.A., Krimkevich, Y.I., Lark, R.H., Dyer, J.M., Veenhuis, M. and Goodman, J.M. (1995) PMP27 promotes peroxisome proliferation. J. Cell Biol., 129, 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M., Erdmann, R., Veenhuis, M. and Kunau, W.-H. (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J., 13, 4908–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono Y., et al. (1999) Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome and potential role in peroxisomal membrane assembly. Proc. Natl Acad. Sci. USA, 96, 2116–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A. and Goodman, J.M. (1994) An oligomeric protein is imported into peroxisomes in vivo.J. Cell Biol., 127, 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Hettema, E., Distel, B. and Tabak, H.F. (1994) Differential protein import deficiencies in human peroxisome assembly disorders. J. Cell Biol., 125, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause B., Diestelkötter, P., Heid, H. and Just, W.W. (1997) Cytosolic factors mediate protein insertion into the peroxisomal membrane. FEBS Lett., 414, 95–98. [DOI] [PubMed] [Google Scholar]
- Rehling P., Marzioch, M., Niesen, F., Wittke, E., Veenhuis, M. and Kunau, W.-H. (1996) The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J., 15, 2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Rout M.P. and Kilmartin, J.V. (1990) Components of the yeast spindle pole body. J. Cell Biol., 111, 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.J., Imanaka, T., Shio, H., Small, G.M. and Lazarow, P.B. (1988) Peroxisomal membrane ghosts in Zellweger syndrome: aberrant organelle assembly. Science, 239, 1536–1538. [DOI] [PubMed] [Google Scholar]
- Schliebs W., Saidowsky, J., Agianian, B., Dodt, G., Herberg, F.W. and Kunau, W.-H. (1999) Recombinant human peroxisomal targeting signal receptor PEX5.J. Biol. Chem., 274, 5666–5673. [DOI] [PubMed] [Google Scholar]
- Snyder W.B., Faber, K.N., Wenzel, T., Koller, A., Lüers, G.H., Rangell, L., Keller, G.A. and Subramani, S. (1999) Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris.Mol. Biol. Cell, 10, 1745–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South S.T. and Gould, S.J. (1999) Peroxisome synthesis in the absence of preexisting peroxisomes. J. Cell Biol., 144, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. (1998) Components involved in peroxisome import, biogenesis, proliferation, turnover and movement. Physiol. Rev., 78, 171–188. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Orii, T., Yakiguchi, M., Mori, M., Hijikata, M. and Hashimoto, T. (1987) Biosynthesis of membrane polypeptides of rat liver peroxisomes. J. Biochem., 101, 491–496. [DOI] [PubMed] [Google Scholar]
- Tabak H.F., Braakman, I. and Distel, B. (1999) Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol., 9, 447–453. [DOI] [PubMed] [Google Scholar]
- Titorenko V.I. and Rachubinski, R.A. (1998) The endoplasmic reticulum plays an essential role in peroxisome biogenesis. Trends Biochem. Sci., 23, 231–233. [DOI] [PubMed] [Google Scholar]
- Van der Klei I.J., Hilbrands, R.E., Kiel, J.A., Rasmussen, S.W., Cregg, J.M. and Veenhuis, M. (1998) The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J., 17, 3608–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Pöhlmann, R. and Philippsen, P. (1994) New heterologous modules for classical or PCR-based disruptions in Saccharomyces cerevisiae.Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Walton P.A., Hill, P.E. and Subramani, S. (1995) Import of stably folded proteins into peroxisomes. Mol. Biol. Cell, 6, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer E.A.C., Lüers, G.H., Faber, K.N., Wenzel, T., Veenhuis, M. and Subramani, S. (1996) Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris.J. Biol. Chem., 271, 18973–18980. [DOI] [PubMed] [Google Scholar]
- Yahraus T., Braverman, N., Dodt, G., Kalish, J.E., Morrell, J.C., Moser, H.W., Valle, D. and Gould, S.J. (1996) The peroxisomal biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for the stability of the PTS1 receptor. EMBO J., 15, 2914–2923. [PMC free article] [PubMed] [Google Scholar]
- Zinser E. and Daum, G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae.Yeast, 11, 493–536. [DOI] [PubMed] [Google Scholar]