Abstract
Numerous genetic studies have shown that the CREB-binding protein (CBP) is an essential component of long-term memory formation, through its histone acetyltransferase (HAT) function. E1A-binding protein p300 and p300/CBP-associated factor (PCAF) have also recently been involved in memory formation. By contrast, only a few studies have reported on acetylation modifications during memory formation, and it remains unclear as to how the system is regulated during this dynamic phase. We investigated acetylation-dependent events and the expression profiles of these HATs during a hippocampus-dependent task taxing spatial reference memory in the Morris water maze. We found a specific increase in H2B and H4 acetylation in the rat dorsal hippocampus, while spatial memory was being consolidated. This increase correlated with the degree of specific acetylated histones enrichment on some memory/plasticity-related gene promoters. Overall, a global increase in HAT activity was measured during this memory consolidation phase, together with a global increase of CBP, p300, and PCAF expression. Interestingly, these regulations were altered in a model of hippocampal denervation disrupting spatial memory consolidation, making it impossible for the hippocampus to recruit the CBP pathway (CBP regulation and acetylated-H2B-dependent transcription). CBP has long been thought to be present in limited concentrations in the cells. These results show, for the first time, that CBP, p300, and PCAF are dynamically modulated during the establishment of a spatial memory and are likely to contribute to the induction of a specific epigenetic tagging of the genome for hippocampus-dependent (spatial) memory consolidation. These findings suggest the use of HAT-activating molecules in new therapeutic strategies of pathological aging, Alzheimer's disease, and other neurodegenerative disorders.
Keywords: hippocampus-dependent memory, CREB-binding protein CBP, histone acetyltransferase, H2B histone, Morris water maze, memory consolidation
INTRODUCTION
What we are is defined by the memories we keep from our past experiences. Since Müller and Pilzecker (1900), it has been clear that such memories are not formed instantaneously. Initially labile and sensitive to interference, they consolidate gradually over time to become stable and resistant. This consolidation process is orchestrated by molecular events, which have long-term consequences—structurally and functionally—at both system and cellular levels. At the system level, the hippocampus is a crucial conductor of the process, receiving and, within a protracted dialog involving other cortical regions, reorganizing the recently learned material in order to form a lasting memory trace (Dudai, 2004; Frankland and Bontempi, 2005; Fanselow and Dong, 2010). A prerequisite to systemic consolidation is that particular brain regions engaged in acquisition, such as the hippocampus, firstly integrates the signal by engaging synaptic or cellular consolidation. It occurs early after training and involves subcellular events that follow different timescales (Medina et al, 2008). It is long known that this step depends on transcriptional activation and de novo protein synthesis (Kandel, 2001; McGaugh, 2000), but the mechanisms underlying synaptic consolidation remain to be established.
Histone acetyltransferases (HATs) are key enzymes regulating the chromatin structure to promote gene transcriptional activation through histone acetylation. Many genetic studies point to a particular HAT, the co-activator CREB-binding protein (CBP) as a major contributor to memory formation (reviewed by Barrett and Wood, 2008). Haploinsufficient mice (cbp+/−) were shown to present chromatin acetylation reductions, late phase of hippocampal long-term potentiation (L-LTP) defects, and some forms of long-term memory (LTM) deficits (Alarcon et al, 2004). Spatial- and temporal-restricted expression of a HAT activity-deficient mutant of CBP showed the importance of HAT activity in these processes (L-LTP and LTM formation) (Korzus et al, 2004). Accordingly, several studies evidenced histone acetylation modifications in response to memory formation, in particular on H3 and H4 histones (reviewed by Gräff and Mansuy, 2008 and Roth and Sweatt, 2009). Several domains of CBP, such as the amino terminus (Bourtchouladze et al, 2003; Wood et al, 2005) or the KIX transcription factor-binding site (Wood et al, 2006a; Vecsey et al, 2007), have been defined important for synaptic plasticity and LTM. In addition, other related proteins displaying HAT function, such as E1A-binding protein p300 (p300) and p300/CBP-associated factor (PCAF), were recently implicated in memory processes as well (Oliveira et al, 2007; Maurice et al, 2008).
To further illuminate the role of HATs in memory, attempts to pharmacologically enhance histone acetylation levels have been made. This was made possible through the development of histone deacetylase (HDAC) inhibitors (HDACi) (Dokmanovic et al, 2007). HDACi molecules have been successful in enhancing memory-related functions such as LTP (Levenson et al, 2004; Alarcon et al, 2004), or improving memory processing involved in cued or contextual fear conditioning (Bredy et al, 2007; Bredy and Barad, 2008; Lattal et al, 2007; Vecsey et al, 2007; Fischer et al, 2007), spatial learning in the Morris water maze (MWM) (Fischer et al, 2007; reviewed by Barrett and Wood, 2008 and Grissom and Lubin, 2009), or novel object recognition (Stefanko et al, 2009). Importantly, this strategy also worked in mice subjected to experimental brain damage (Fischer et al, 2007; Dash et al, 2009). The molecular mechanisms underlying these effects is not clear, but is based on the hypothesis that, if ‘global' acetylation levels are increased and/or the activated-acetylation status of histones is maintained over time, this could potentially induce genetic programs at least partly related to memory and then sustain or enhance memory functions. Recently, HDAC2 was found associated with promoters of genes implicated in synaptic plasticity and memory formation, and Hdac2 knockout mice show memory improvement (Guan et al, 2009). Such findings are important to guide the development of new specific HDACi molecules. It is worth noting that different class I HDACi were recently found to be efficient in restoring some memory functions in animal models of Alzheimer's disease (AD) (Ricobaraza et al, 2009; Francis et al, 2009; Kilgore et al, 2010). Importantly, Vecsey et al (2007) showed that the memory-enhancing effects of HDACi were dependent on the CBP pathway. Such data further emphasize that KIX-interacting transcription factors like CREB recruit CBP HAT activity in the hippocampus during memory formation.
Despite the evidence suggesting that HATs contribute to memory formation and that HDACs enhance some memory functions, a clear picture of HAT enzyme modulation during the ongoing process of memory formation—and more particularly during a consolidation phase—is still lacking. Indeed, their expression profile has never been investigated during the formation of a memory. This is likely because a particular modification occurring in perhaps only a subregion of the hippocampus, which represents a mixed cellular population, will probably come up very discretely at the global level. In this report, we conducted a thorough examination of the four core histones' acetylation pattern, as well as the expression profile of different HATs, in the rat dorsal hippocampus during the ongoing phase of spatial memory formation in an MWM task. We focused on the dorsal hippocampus because this hippocampal subregion, unlike the ventral one, has a crucial role in spatial memory functions (Fanselow and Dong, 2010). Taken together, our data reveal a dynamic upregulation of the HAT enzymes during the formation and consolidation of a memory trace. This newly generated HAT activity is likely to further ensure specific acetylated-histone-dependent events, in particular on H4 and H2B histones, which we found to be regulated on memory/plasticity-related genes during the ongoing process of memory formation. In addition, regulations of CBP and histone acetylation were lost in a rat model of hippocampal denervation presenting two major degenerative features found in AD, namely neuronal loss in the basal forebrain (BF) and the entorhinal cortex (EC), and in which spatial memory consolidation does not occur (Cassel et al, 2008; Traissard et al, 2007). Our findings emphasize that spatial memory dysfunctions, as seen in AD, could be the result of altered genetic (CBP expression) and epigenetic (histone acetylation) regulations in the hippocampus, otherwise controlled by these neuronal inputs (ie, glutamatergic and cholinergic). This work opens the interesting possibility of reinstating memory consolidation processes by directly modulating the expression of specific HATs during neurodegenerative conditions.
MATERIALS AND METHODS
Animals
The study adhered to the regulations specified by the European Directive of 24 November 1986 (86/609/EEC) and the French Department of Agriculture (authorization no. 67-215 for J-CC and no. 67-7 for APV). Male Long–Evans rats (n=126), about 3–4 months old at the time of behavioral testing, were used. They were housed individually in a quiet room, under a 12 h light/dark cycle (light on at 0700 hours) with ad libitum access to food and water. For animal surgery and histological verifications in double lesion model, see Supplementary Materials and methods.
Spatial Testing in the MWM
The specifications of the water maze and the testing procedures have been described previously (Traissard et al, 2007). Briefly, for all rats, the first part of training was a four-trial session per day using a visible platform (VPf) located in the south-east quadrant of the pool, starting randomly from each of the four cardinal points at the edge of the pool. In the rats that were subjected to surgery (lesions and sham), training with the VPf was performed over 2 days, with four trials each day. The learning rats (hidden platform (HPf) group) were then given four successive acquisition trials per day (inter-trial interval, 10–15 s), starting randomly from each of the four cardinal points from the edge of the pool, for 1–5 consecutive days to learn the location of a platform hidden 1 cm below the water surface in the north-west quadrant. The distance traveled before reaching the platform was recorded by a video tracking system (Ethovision, Noldus, the Netherlands). Control rats (VPf group) were swimming toward a VPf, of which the location was changed from trial to trial on each day to make sure rats did not associate a spatial cue with the platform. In the lesion experiments, control rats were sham-operated rats that had to learn the location of an HPf. At the end of acquisition, all rats trained with the HPf were tested for retention in a probe trial administered 1 h after the last acquisition trial. The platform was removed and all rats from the HPf group were released from the north-east start point and given 60 s to swim. The variable taken into account for the probe trial was the time spent in each of the four quadrants. Control groups (VPf) had to swim to a VPf during the probe trial, as previous experiments showed that removing the platform after training with a VPf was an extremely stressful experiment.
Statistical Analyses (Behavioral Studies)
Performance recorded during acquisition was evaluated using a two-way ANOVA with repeated measures considering days (1–3 or 1–5) and platform visibility HPf (learning) vs VPf (control), or treatment (sham vs lesion) as factors. Probe trial performance was analyzed using a one-way ANOVA or/and t-test to compare the time spent in the target quadrant to chance (ie, 15 s). When appropriate, post hoc comparisons used the Newman–Keuls multiple range statistic. Values of p<0.05 were considered significant.
Preparation of Tissues for Biochemical Studies
Animals were decapitated and their brains rapidly removed from the skull and transferred on ice. For rats of the MWM experiments, euthanasia occurred right after the end of the probe trial (HPf) or the one-trial session to the VPf. Different rat groups have been made for each different preparation of tissues. Freshly dissected tissues were then immediately frozen at −80°C, until being processed for biochemical studies (RNA, total protein, and histone extractions) or were immediately processed (chromatin and nuclear protein extractions). In each experiment, the cerebellum was used as control tissue.
Real-Time Reverse Transcription-PCR
Total RNA was extracted from hippocampal tissues using Trizol reagent (Invitrogen, Cergy-Pontoise, France). Samples were homogenized in a Tissue Lyser (Qiagen, Courtaboeuf, France). RNAs were ethanol-precipitated twice. Only RNA samples with optical density (OD)260/280 and OD260/230 ratio close to 2.0 in TE (pH 8.0) were selected for reverse transcription (n=5 in each group). RNA samples were denaturated 10 min at 70°C and cDNA synthesis was performed on 1 μg of total RNA (iScript cDNA Synthesis kit; Bio-Rad, Marne La Coquette, France). Quantitative PCR (qPCR) analysis was carried out on a Bio-Rad iCycler System using iQSYBR Green Supermix. A specific standard curve was performed in parallel for each gene, and each sample was quantified in duplicate (n=5). PCR conditions were 3 min at 94°C, followed by 40 cycles of 45 s at 94°C and 10 s at 60°C. Data were analyzed using the iCycler software, and normalized to the 18S ribosomal subunit RNA. Informations with regard to primers and PCR products are presented in Supplementary Materials and methods.
Protein Preparation and Western Blot Analyses
Total protein preparation
Tissues were lysed and homogenized in Laemmli buffer. After 10 min at 70°C, samples were sonicated for 10 s (Ultrasonic Processor, power 60%), boiled 5 min, centrifuged (20 000 g, 5 min), and supernatant frozen at −20°C.
Histone extraction
All procedures were performed on ice. Frozen tissues (15–20 mg) were incubated in 500 μl of lysis buffer (10 mM Tris-HCl (pH 6.5), 50 mM sodium bisulfite, 1% Triton, 10 mM MgCl2, 8.6% sucrose). Protease inhibitor cocktail and sodium butyrate (1 mM) were added before use. Samples were homogenized with Dounce glass homogenizer for 10 strokes. Tissue homogenates were centrifuged (7700 g, 1 min, 4°C). Histones were acid extracted from the nuclear fraction in 150 μl of 0.4 N H2SO4, sonicated for 10 s (Ultrasonic Processor, power 40%), and then incubated 1 h at 4°C. Acid extracts were centrifuged at 14 000 g for 10 min. Histones (supernatant) were transferred to a fresh tube and precipitated with 1.5 ml acetone over night at −20°C. The resulting purified proteins were resuspended in Laemmli buffer.
Nuclear protein extractions were prepared according to Dignam et al (1983). Briefly, the regular protocol is followed until the nuclear pellets are collected in buffer C, supplemented with protease inhibitor cocktail and sodium butyrate (1 mM). They are then softly dounce homogenized, and after centrifugation, the soluble nuclear fraction is quickly frozen and kept at −80°C. DTT was avoided to preserve HAT activities in the samples.
Western blot analyses
Protein concentration was measured using the RC-DC Protein Assay (Bio-Rad). Western blots were performed as described previously (Rouaux et al, 2007), with polyclonal antibodies against CBP (06-297), p300 (05-257), acetyl-histone H2B (07-373), acetyl-histone H3 (06-599) (Upstate Biotechnology, New York, NY, USA), histone H3 (39163) (Active Motif, Carlsbad, CA, USA), histone H2B (ab18977) (Abcam, Cambridge, UK), and PCAF (3378) (Cell Signaling). The secondary antibody used was a horseradish peroxidase-conjugated whole-goat anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Blots were revealed with ECL and exposed with Amersham hyperfilm ECL (GE Healthcare, Chalfont St Giles, UK). Results were quantified using the ImageJ software.
HAT Activity
HAT activity was measured an nuclear protein extracts with a colorimetric assay kit from Biovision (Clinisciences). A standard curve was performed with HeLa nuclear extracts. All samples were tested in duplicates. The activity was analyzed in a microplate reader at absorbance A=440 nm. HAT activity is expressed in nmol/min/μg sample according to A=ɛLC, with ɛ=37 000/M/cm and L=0.5 cm.
CBP Immunohistochemistry
A new group of rats was trained as described above (3 days in the MWM, VPf, and HPf conditions). Animals were deeply anesthetized after the probe trial with an intraperitoneal injection of pentobarbital (200 mg/kg) and perfused transcardially with 150 ml of ice-cold paraformaldehyde (4% in 0.1 M PB, 4°C). Brains were rapidly removed from the skull and post-fixed for 2 h in the same fixative at +4°C. Brains were immersed in a PBS solution and kept at +4°C until vibratome sectioning. Coronal hemisections, 40 μm in thickness, were made through the dorsal hippocampus (from −2.30 to −4.16 mm from Bregma, according to Paxinos and Watson, 1998) using the vibratome (Leica VT1000M), and the floating sections were kept at −20°C in a cryoprotectant before use. After permeabilization (PBS1X/Triton 2%, 15 min), unspecific labeling was blocked (PBS1X/Triton 0.1%/horse serum 5%, 30 min, 37°C) and slices were incubated overnight with polyclonal anti-CBP antibody (sc-369; Santa Cruz Biotechnology), washed, and further incubated with anti-rabbit horseradish peroxidase-conjugated (1/500) antibody (sc2004; Santa Cruz Biotechnology) for 1 h. After three washes with PBS1X/Triton 0.1%, the revelation was carried out with diaminobenzidine (DAB 0.05%, Tris 0.04 M, pH 7.5, H2O2 0.03%) and mounted with Roti Histokit II (Roth, Karlsruhe, Germany). OD and area measurements were performed as described in Supplementary Materials and methods.
Chromatin Immunoprecipitation
Chromatin Preparation
After decapitation, freshly dissected tissues were chopped with a razor blade and rapidly put in 5 ml PBS containing 1% formaldehyde for 15 min at room temperature. The fixation step was stopped by the addition of glycine (0.125 M final concentration). Samples were prepared as described in Panteleeva et al (2007), snap frozen in liquid nitrogen, and stored at −80°C until needed.
Immunoprecipitation
An equal amount (100 μg) of DNA/protein complex was assayed and brought to a volume of 500 μl with chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl, pH 8.1, 167 mM NaCl). A fraction of supernatants (20 μl) from each sample were saved before IP and served as ‘total input chromatin'. The remaining supernatant was incubated overnight at 4°C with 3–5 μg of primary antibodies (acetylated-H3 (06-599; Upstate Biotechnology); acetylated-H2B, (ab1759; Abcam); CBP (A-22) sc-369/CBP (C-1) sc-7300 (Santa Cruz Biotechnology), negative control IgG antibody (Active Motif), or no antibody. This was followed by incubation with salmon sperm DNA/protein A agarose (Upstate Biotechnology) for 2 h. One microgram of rabbit anti-mouse IgG (Euromedex) was added for 30 min before the salmon sperm DNA/protein A agarose beads when a monoclonal antibody was used. After several wash steps (low salt, high salt, LiCl, and TE buffers), complexes were eluted in 250 μl of buffer (1% SDS, 0.1 M NaHCO3). The crosslinking was reversed (overnight at 65°C) and the DNA was subsequently purified with RNAse (45 min, 37°C), proteinase K (2 h, 45°C), phenol/chloroform extraction, and ethanol precipitation. After a last wash with 70% ethanol, pellets were resuspended in 50 μl of nuclease-free milliQ water.
PCR amplification
Semiquantitative PCR amplifications were performed with a PCR/Express from Hybaid (94°C for 2 min, and 94°C for 30 s; 60°C for 30 s; and 72°C for 30 s for 30–32 cycles). The different primers for promoter PCR amplification are described in Supplementary Materials and methods.
Statistical Analysis (Biochemical Studies)
Statistical analyses were performed using Student's t-test (PRISM version 4.0b; GraphPad, San Diego, CA, USA). Data are expressed as the mean±SEM. Differences at p<0.05 were considered significant.
RESULTS
Determining the Time Frame in which an Efficient Spatial Memory is being Formed During Acquisition
The aim of this study was to characterize acetylation-related events (histone acetylation and HAT expression profiles) associated with spatial memory consolidation. To this end, we used a standard reference memory task in the MWM, in which rats are trained during a 5-day acquisition time course. The rats from the learning group had to search for an escape platform that was hidden at a fixed location underneath the water surface (HPf). As memory forms over several training periods, we first needed to identify a time point at which most of the signaling corresponds to consolidation-related events. This can be defined by the time at which the memory is efficient enough so that there is a marked improvement in performance over trials, but before the memory trace is formed and reliably efficient. Behavioral data are presented in Supplementary Figure S1 (A, training; B, probe trial; and C, typical swim tracks). In this spatial learning group (HPf), there was a marked improvement between days 1 and 2 (3-day group: p<0.01; 5-day group: p<0.001) and an almost maximal performance level was reached on day 3 (Supplementary Figure S1A). The two-way ANOVA (platform visibility × day) showed a significant interaction between both factors in the 3-day group (F(2,16)=7.02, p<0.01) and a tendency toward an interaction in the 5-day group (F(4,32)=2.14, p=0.098). That there was no further improvement in performance between days 3 and 5 confirmed that a reliable memory trace was established already during/after the third training day. A probe trial was given 1 h after the last training trial (either VPf or HPf) of day 3 to measure rat performances in the MWM. Analysis of the probe trial performance after 1 day of training indicated that the memory trace was not established yet, as rats performed by chance (ie, time in target quadrant vs 15 s: t(4)=0.7, NS). Interestingly, in the 3-day training group, probe trial performance was significantly above chance (t(4)=3.0, p<0.05), as was also the case in the 5-day group (t(4)=8.0, p<0.01), in which the time spent in the target quadrant was also longer than in the 3-day group. Thus, we determined that in our specific experimental conditions (room, pool, spatial cues, training schedule, etc), the critical time frame for efficient memory consolidation is likely to occur during day 3. Therefore, we conducted our investigations at the end of a training period of 3 days. The rats were euthanized 1 h after the last training trial in order to allow signal transduction and transcriptional regulations to take place, but less than 45 s after a probe trial, thereby minimizing the molecular impact of the latter. Thus, at this time point, we can conceive that the corresponding transcriptional events are related to molecular events occurring on day 3 both in terms of memory function and cumulative and ongoing changes in consolidation that extended over the 3 days of training. The probe trial was given to check for the presence of an efficient memory trace.
To compare the net expression levels measured during the consolidation of the memory trace, we formed a control group of rats trained under similar experimental conditions, except that they did not have to learn a particular location as they swam to a VPf. This controls for any molecular modification occurring during the water maze task, which is relevant to the context processing (navigation, stress, swimming, etc), but not to spatial learning. Expression levels measured in the HPf group relative to the VPf group should thus give a net change relevant to spatial learning.
For all subsequent experiments focusing on acetylation-dependent regulations, we trained three different groups of rats using the same 3-day protocol. Results of acquisition and retention measured for the first group (n=15 per group for histone extractions and RNA/protein experiments) are representative of subsequent trainings and are shown in Figure 1a. Specific results obtained for the two other groups are shown in Supplementary Figure S1D (n=8 per group for nuclear extractions and ChIP experiments) and Supplementary Figure S1E (n=5 per group for immunohistochemistry experiments). With the VPf (open circles), distance to reach the platform was significantly shorter than that with the hidden one (HPf, black circles) on day 1 (p<0.001). In the HPf condition, there was a significant improvement of performance over days (p<0.001). Probe trials showed that the HPf rat group performed significantly above chance in the target quadrant, but not differently from or even below chance in the other quadrants (time in target vs 15 s: t(15)=7.3, p<0.001).
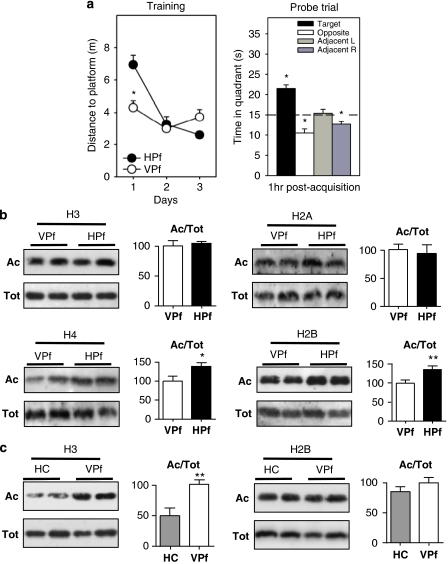
Figure 1.
Specific increase of H2B/H4 acetylation levels in rat hippocampus during spatial memory formation. (a) Water maze performance in rats trained over 3 consecutive days (four trials per day) for spatial reference memory. Control rats had to swim toward a visible platform (VPf) whose location was changed from trial to trial. Learning rats had to swim to a hidden platform (HPf) at a fixed location. Left: Acquisition data are expressed as the mean (±SEM) distances to reach the platform. HPf rats showed a marked improvement between days 1 and 2; *p<0.001. Right: Probe trial performance in the HPf rats given as the mean time in the far quadrants (±SEM); performance was significantly above chance level (ie, 15 s) only in the target quadrant, whereas time in the three other quadrants was either at chance level or significantly under chance. *p<0.05, significantly different from chance. (b) Changes in histone acetylation levels during the formation of a spatial memory. Acetylated (Ac) and total (Tot) histone levels were measured by western blot analyses for each histone core in histone extracts obtained from the dorsal hippocampus of 3-day-trained rats (HPf vs VPf; n=7 per group). Lysine acetylations measured here are H3K9K14, H4K12, H2AK9, and H2BK5K12K15K20. Typical western blots are represented in duplicates. Quantified results are represented as fold induction of the Ac/Tot ratio for each histone, the ratio obtained in the control condition being arbitrarily set at 100%. Student's t-test: **p<0.01,*p<0.05, as compared with VPf group. Significant increases in acetylation of H2B (1.35-fold) and H4 (1.38-fold) levels, but not of H3 and H2A, were measured in the rat hippocampus after 3 days of spatial learning. (c) Histone acetylation level comparison between home cage (HC) vs VPf control rats. Rats were either trained in the visible version of the water maze (VPf, n=6) or left in their HC (n=5)—but manipulated daily for a few minutes—over 3 consecutive days. Histone extracts were prepared and the acetylated and total histone levels are shown in duplicate for H3K9K14 and H2BK5K12K15K20. A significant increase in H3 acetylation is measured between both groups (twofold), whereas H2B acetylation ratio remains unchanged. Student's t-test: **p<0.005, as compared with the HC group.
Spatial Learning Induces Differential Histone Acetylation in the Hippocampus
Acetylation levels on different lysine residues of the four core histones were measured in our experimental conditions, using histone protein extracts obtained from the dorsal hippocampus (Figure 1b) and the cerebellum (control structure unrelated to spatial memorization; Supplementary Figure S2). At this global level, we detected a significant increase of acetylation levels for H2B histone (tetra-acetylated-H2BK5K12K15K20; 1.35-fold, p<0.01) and H4 (H4K12; 1.38-fold, p<0.05), but not for H3 (H3K9K14), or for H2A (H2AK9) histones, in HPf rats compared with VPf rats (Figure 1b). The same results were obtained when testing acetylated-H2BK12, which was activated upon learning at about the same range, whereas H3K14 and H4K5 acetylation levels remained equivalent to those found in the control group (data not shown). No significant changes between the two groups were noted in the cerebellum (Supplementary Figure S2). It is worth noting that, at this global level, acetylation induction remained very mild in the dorsal hippocampus of the learning group. Observation of acetylated-H2B histone with the neuronal marker MAP2 co-immunolabelings revealed that acetylated-H2B was present in a larger cell population than only hippocampal neurons (Supplementary Figure S3A), suggesting that the specific signal from neurons might have been underestimated when tested by western blot analyses.
Hence, from this experiment, we can clearly confirm that H2B and H4 acetylation levels are increased during memory formation. Acetylation of other histones, particularly acetylation of H3, is not changed upon learning. As the test conditions, especially when involving fear, anxiety, navigation, and/or physical exercise (swimming), could have dramatic repercussion on transcriptional events as well, we then undertook to compare H3- and H2B-acetylated levels in the dorsal hippocampus of two additional control groups that were not subjected to any spatial memorization task: a home cage (HC) control group and a group of rat tested with a VPf. The rats from the HC control group had been manipulated daily for a few minutes over 3 consecutive days before euthanasia, whereas the rats from the VPf group received 3-day training in the VPf version of the water maze (same as the control rat group used throughout this study, same experimental room). Strikingly, our results show that H3 acetylation levels were greatly enhanced in the VPf group compared with HC (Figure 1c; about twofold, p=0.0056), whereas no significant changes were detected for H2B acetylation. It is therefore likely that H3 acetylation levels are already optimal for transcriptional processes to occur after 3 days in the water maze, whatever the task, ie, swimming to a VPf or learning the localization of an HPf, and this situation appeared very different from that of HC rats (see Discussion).
Histone Acetylation is Differentially Enriched on Promoters of Memory/Plasticity-Related Genes upon Formation of Spatial Memory
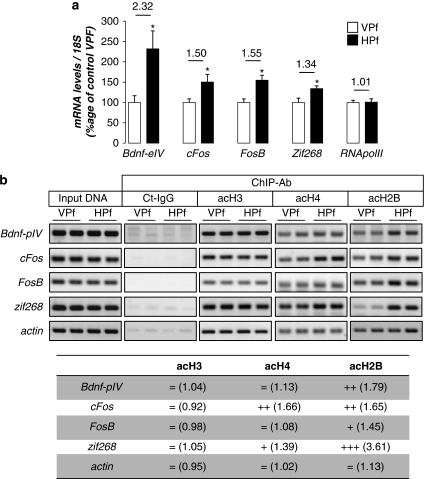
It was important next to establish a functional link between acetylated-histone modulations and specific gene transcription. The expression of four genes related to synaptic plasticity and/or memory was measured by RT-qPCR in our first groups of trained rats (see behavioral data in Figure 1a). We measured the expression of bdnf-eIV (brain-derived neurotrophic factor, exon IV; Aid et al, 2007), cFos, and FosB, as well as the well-documented immediate-early gene activated during spatial memorization: zif268 (early growth response 1; Guzowski et al, 2001). They have all been shown to bear canonical CREB-binding sites (Shieh et al, 1998; Davis et al, 2000; Impey et al, 2004). Figure 2a shows that transcription of all of these genes was significantly increased in the HPf group compared with the control VPf group: 2.32-fold (p<0.05) for bdnf-eIV; 1.50-fold (p<0.05) for cFos, 1.55-fold (p<0.05) for FosB, and 1.34-fold (p<0.05) for zif268. We then sought to determine the enrichment in acetylated-H3, -H4, and -H2B histones on their proximal promoter (Figure 2b). To this end, ChIP experiments were performed on dorsal hippocampi freshly dissociated from newly trained rat groups (see behavior in Supplementary Figure S1D) with antibodies directed against the modified histones or a non-related IgG control antibody. Semiquantitative PCR results are shown in duplicates and their quantification is presented in the table at the bottom of Figure 2b. An enrichment of acetylated-H2B histone was clearly detectable on the promoter of the four genes (bdnf-pIV: 1.79-fold; cFos: 1.65-fold; FosB: 1.45-fold; and zif268: 3.61-fold) in the HPf-trained rats compared with the control group (VPf). Acetylated-H2B histones are present on the actin promoter, but levels do not change upon learning. Acetylated-H4 was found enriched on cFos and zif268 only (1.66- and 1.39-fold, respectively). Acetylated-H3 histone is present on all these genes, but no difference was seen between the two groups. This is reminiscent of the fact that global levels of acetylated-H3 did not change in the learning HPf group when compared with the control VPf group. Altogether, these data show that spatial learning induces the transcription of specific memory/plasticity-related genes, in part through a differential increase of acetylated histones onto their proximal promoter. It is worth noting for the first time that acetylated-H2B is increased by learning and actually participates in this process by binding the promoter of such genes. The further characterization of other acetylated-H2B target genes is currently under investigation.
Figure 2.
Several memory/plasticity-related genes present an acetylated-H2B/H4 enrichment on their promoters during the consolidation of a spatial memory. (a) mRNA expression levels of several CBP/memory-related genes: bdnf-eIV, cFos, FosB, and zif268 were evaluated by RT-qPCR in the dorsal hippocampus of control (visible platform, VPf) and learning (hidden platform, HPf) groups trained for 3 days. RNA polymerase II was tested as an internal negative control. Values were normalized to the 18S ribosomal subunit. Data are expressed as mean (±SEM) (n=6 per group), with percentage relative to the control group (VPf) arbitrarily set at 100%. Fold inductions are indicated above histograms. Student's t-test: *p<0.05, as compared with VPf. (b) Chromatin immunoprecipitation were performed on 3-day-trained rats from the VPf and HPf groups (n=3). Semiquantitative PCR were performed to analyze the presence of bdnf-pIV, cFos, FosB, and zif268 proximal promoters in the immunoprecipitates. Controls for immunoprecipitation were performed with an unrelated control antibody (Ct-IgG) or no antibody (noAb). Input DNA corresponding to the total of chromatin before immunoprecipitation was run in parallel as positive control. Representative results are shown in duplicate for each gene tested. β-Actin was used as internal control. Bands were quantified (ImageJ) and fold inductions (f.i.) are indicated in the table below. An activation score was arbitrarily affected by the HPf vs VPf condition: (+++) when f.i. >2.00; (++) when f.i. >1.50; (+) when f.i. >1.25; and (=) when −0.25<f.i.>+0.25. acH3, acetylated K9, K14 H3 histone; acH4, acetylated K12 H4 histone; acH2B, acetylated K12, K15 H2B histone; Bdnf-eIV, bdnf exon IV; bdnf-pIV, bdnf promoter IV.
Global HAT Expression and Activity are Upregulated in the Dorsal Hippocampus during the Formation of a Spatial Memory
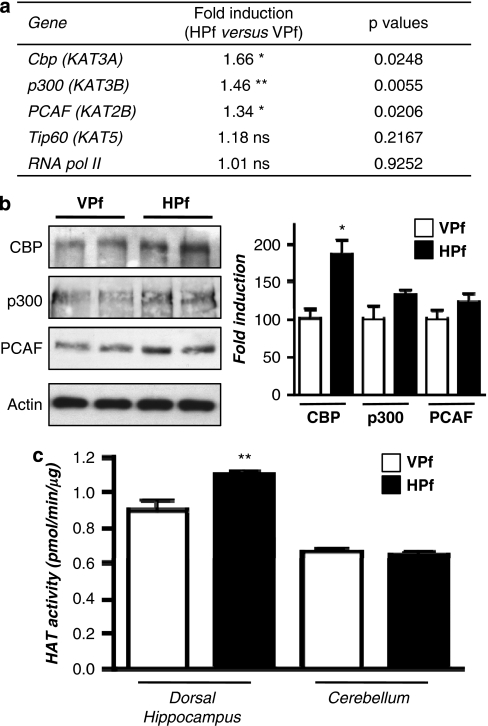
Gene expression was then evaluated for several HATs: p300 (KAT3B), a CBP-related protein that shares many of its functions, the GNAT family member PCAF (p300/CBP-associated factor), and the MYST family member Tip60 (KAT5). Results are shown in Figure 3a. The results of this experiment showed a 1.66-fold (p<0.05) increase of cbp mRNA expression levels in the learning group relative to the control group. A moderate but significant increase was also observed for p300: 1.46-fold (p<0.01) and PCAF: 1.34-fold (p<0.05), whereas no change in Tip60 was measured. Levels of the RNA polymerase II subunit, used here as a second internal control, remained unchanged. In addition, when measured in a group of rats trained for only one day (instead of 3) in the MWM, the level of these HATs were not significantly different in the HPf group from that in the VPf group (Supplementary Figure S4A), suggesting that increased HAT expression is induced and accumulates over repeated training (ie, reference memory and not working memory).
Figure 3.
Increased expression level of several HATs and their activity during consolidation of a spatial memory. (a) HAT gene expression. mRNA expression levels of different genes were evaluated by RT-qPCR in the dorsal hippocampus of control (visible platform, VPf) and learning (hidden platform, HPf) groups (n=5 per group). Values were normalized to the 18S ribosomal subunit and the fold inductions and p-values are presented. Student's t-test: *p<0.05, **p<0.01, and NS, nonsignificant. Cbp, p300, and PCAF mRNA levels are significantly increased in the hippocampi of learners (HPf). CBP, CREB-binding protein; p300, E1A-binding protein p300; PCAF, p300/CBP-associated factor; Tip60, HIV-1 tat interactive protein; and RNA pol II, polymerase (RNA) II. The new nomenclature of HATs as lysine(K)-acetyltransferases (KATs) is mentioned within parenthesis (Allis et al, 2007). (b) HAT protein expression. HAT levels were assessed by western blot in nuclear protein extracts obtained from the dorsal hippocampus of rats trained as in (a). Typical blots are shown in duplicates. Blots were quantified (n=7 per group) and results normalized against actin are shown (right). *p<0.05, as compared with VPf. CBP was increased by 1.86-fold in the dorsal hippocampus during consolidation of spatial learning. P300 and PCAF show a nonsignificant trend to increase in the same conditions. (c) HAT activity. The same nuclear protein extracts as in (b) were used. HAT activity was measured on 30 μg of dorsal hippocampus or cerebellum nuclear protein extracts from the two rat groups (VPf and HPf). HAT activity is expressed in nmol/min/μg protein according to A=ɛLC. No significant change is found in the cerebellum, whereas global HAT activity was increased in the dorsal hippocampus. **p<0.01, HPf vs VPf.
The different HAT protein levels were measured from total protein extracts and nuclear protein extracts prepared from different animals from the secondly trained group (see behavioral data in Supplementary Figure S1D). We found that CBP protein levels significantly increased in total extracts (1.68-fold, p<0.05) and nuclear protein extracts (1.86-fold, p<0.05) from the dorsal hippocampus of learning compared with control rats (Figure 3b). P300 and PCAF proteins also showed a trend to, but not significant, increase in these latter extracts (1.31- and 1.21-fold respectively, Figure 3B). No change was observed between the HPf and VPf groups in the cerebellum (Supplementary Figure S4B). Interestingly, when tested in an in vitro assay, these nuclear extracts obtained from the dorsal hippocampus of learning rats displayed a significant increase in global HAT activity when compared with control rats (18.5%, p<0.01). No change was seen between the two groups in the cerebellum (Figure 3c).
The CBP-Dependent Pathway is Specifically Upregulated in the Dorsal Hippocampus during the Formation of a Spatial Memory
As the dorsal hippocampus is a mixture of different cell populations, we also investigated CBP localization by immunohistochemistry in normal rats. As revealed by co-labeling of CBP (−FITC) with the neuronal marker MAP2 (−Cy3), we observed that CBP is expressed at higher levels in pyramidal neurons of the CA1 region (Supplementary Figure S3B), and in granular neurons of the dentate gyrus (DG) (data not shown), compared with the surrounding non-neuronal cells. This suggests that our studies are not particularly biased by the expression of CBP in non-neuronal cells.
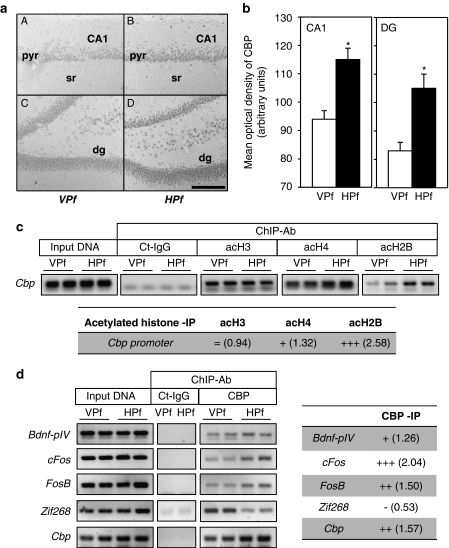
In order to quantify CBP levels between both groups (VPf and HPf) at the subregion level of the dorsal hippocampus (CA1 vs DG), CBP immunohistochemical staining followed by DAB labeling was performed in a last rat group (see behavioral data in Supplementary Figure S1E). A typical labeling is presented in Figure 4a. Densitometry counting is shown in Figure 4b. A two-way ANOVA of the ODs of CBP immunoreactivity in CA1 (left panel) and in the DG (right panel) revealed a significant effect of platform visibility (F(1,9)=17.6, p<0.01) and an overall effect of the region (F(1,9)=91.1, p<0.001) that was due to stronger ODs in CA1 as compared with the DG; the interaction between both factors was not significant (F(1,9)<1.0, NS). Multiple comparisons showed that the ODs were significantly larger in learners (HPf) vs controls (VPf) in each region (p<0.01). In a control structure, the auditory cortex, we found similar CBP levels in both groups (HPf=100.4±6.6 vs VPf=100.5±5.2; F(1,9)<1, NS), confirming the region-specific enhancement of CBP immunoreactivity in the dorsal hippocampus during learning.
Figure 4.
CBP-dependent regulations in the rat dorsal hippocampus during the formation of a spatial memory. (a) Representative images of CBP immunoreactive staining revealed with DAB staining in regions CA1 (A, B) and in the dentate gyrus (dg; C, D) of the dorsal hippocampus in representative VPf rats (A, C) and HPf (B, D) rats. Pyr: pyramidal neuron layer; sr: stratum radiatum; dg: dentate gyrus. Scale bar=250 μm. (b) Mean (±SEM) optical density of CBP immunoreactive staining in the CA1 pyramidal cell layer (CA1) and in the dentate gyrus granular layer (DG). In HPf rats (n=5), whatever the region, the optical density of CBP immunoreactivity was significantly larger than in VPf rats (n=5). *p<0.05. (c) Chromatin immunoprecipitation of acetylated histones were performed on chromatin obtained from 3-day-trained rats from the VPf and HPf groups (n=3). Semiquantitative PCR were performed to analyze the presence of cbp proximal promoter in the immunoprecipitates. Controls for immunoprecipitation were performed with an unrelated control antibody (Ct-IgG) or no antibody (noAb, not shown). Representative results are shown in duplicate for each gene tested. Bands were quantified (ImageJ) and fold inductions (f.i.) are indicated in the table below. acH3: acetylated K9, K14 H3 histone; acH4: acetylated K12 H4 histone; and acH2B: acetylated K12, K15 H2B histone. (d) Chromatin immunoprecipitation of CBP were performed on chromatin obtained from 3-day-trained rats from the VPf and HPf groups (n=3). Semiquantitative PCR were performed to analyze the presence of bdnf-pIV, cFos, FosB, zif268, and cbp proximal promoters in the immunoprecipitates. Controls for immunoprecipitation were performed with an unrelated control antibody (Ct-IgG, shown in simplicate) or no antibody (noAb, not shown). Representative results are shown in duplicate for each gene tested. Bands were quantified (ImageJ) and fold inductions (f.i.) are indicated in the table on the right. Bdnf-eIV: bdnf exon IV; bdnf-pIV: bdnf promoter IV. (c, d) Input DNA corresponding to the total of chromatin before immunoprecipitation was run in parallel as positive control. An activation score was arbitrarily affected by the HPf vs VPf condition: (+++) when f.i. >2.00; (++) when f.i.>1.50; (+) when f.i. >1.25; (=) when 1.25<f.i.>0.75; and (−) when f.i.<0.75.
If cbp transcription was significantly enhanced in the hippocampus of learning rats, there should be changes at the chromatin level of its promoter. We tested this using ChIP experiments with the different acetylated histones on the cbp promoter. Primers were designed spanning 110 bp of the proximal region of the rat cbp promoter sequence (see Materials and methods) and we found a strong enrichment of acetylated-H2B in this region in HPf vs VPf groups (Figure 4c). In addition, there was a moderate enrichment of H4 in HPf as compared with VPf rats, as for the other target genes tested, and no enrichment of H3; yet, the binding was already strongly detected in the VPf control situation (Figure 4c). These results are in line with our previous data showing differential acetylation of histones in response to learning (Figure 1b) at specific genes (Figure 2b) and further outline that the cbp promoter is favorable to transcriptional upregulation of the cbp gene at that time of memory formation. Finally, we expected to be able to immunoprecipitate the different HATs, but could get reliable results only for CBP (Figure 4d). We found a differential CBP occupancy on the different genes. Surprisingly, bdnf-pIV, one of the most activated genes in the HPf conditions (see qPCR results in Figure 2a) displayed a very moderate enrichment of CBP occupancy upon learning. A possibility is that CBP could also be present on the bdnf promoter IV of rats swimming to the VPf and that another HAT (such as p300 or PCAF) has increased histone acetylation on the bdnf promoter IV in relation to the memory of the platform location (see Discussion). Again, the result obtained for zif268 is very surprising as CBP occupancy is clearly decreased in the learning group, whereas this same promoter region displayed a strong acetylated-H2B enrichment (see Figure 2b). By contrast, CBP occupancy is increased in both cFos and FosB gene promoters. These interesting results emphasize a model in which several HATs are in play for modulation of specific memory/plasticity-related genes. This interplay between the different HATs that control such gene expression needs further investigation.
Overall, these experiments point to a functional increased expression of several HATs that can regulate specific gene transcription via H2B and H4 acetylation during an ongoing process of spatial memory formation and suggest that these newly formed HAT molecules could contribute to the consolidation of the trace by inducing new genetic programs.
The CBP/Acetylation Pathway is Downregulated after Hippocampal Denervation Inducing Spatial Memory Deficits
If our data obtained on the CBP/histone acetylation-dependent signaling pathway are relevant to memory formation and consolidation, one should expect to find them altered in an animal model in which consolidation is impaired. Traissard et al (2007) recently reported on a rat model that is completely unable to learn a reference memory task in the MWM (Traissard et al, 2007). These rats are double-lesioned on brain regions that typically degenerate during the early course of AD, a neurodegenerative disease characterized, at the cognitive level, by progressive memory disruption ending up in dementia. Of note, spatial memorization processes are also particularly affected in this disease. Lesioned regions in the rat model include the cholinergic component of the BF and the connections between the EC. Verification of the lesions in lesion or sham-operated rats are presented in Figure 5a and behavioral scores (training and probe trial) are shown in Figure 5b.
Figure 5.
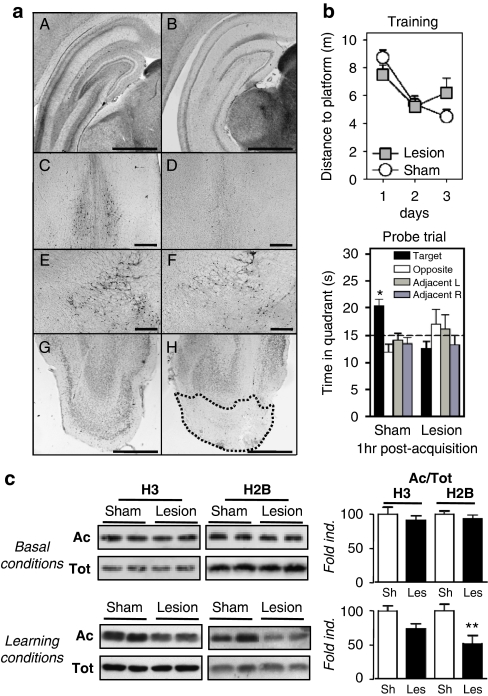
Histone acetylation levels in a cognitively impaired rat model. (a) Histological verification of the location and extent of cholinergic and entorhinal cortex (EC) lesions. A, C, E, and G are representative pictures of one sham-operated rat; B, D, F, and H are representative pictures of one rat subjected to the double lesions. A, B are typical examples of AChE-positive staining in the posterior hippocampus as observed at about −5.5 mm from Bregma (notice the marked decrease of dark staining accounting for cholinergic denervation in B); C, D are typical examples of ChAT-positive immunostaining in the medial septum at about +0.2 mm from Bregma (notice the dramatic reduction in D); E, F are examples of ChAT-positive immunostaining in the nucleus basalis magnocellularis and the substantia innominata (notice that the decrease is much less pronounced than in the septal region in F). G, H are examples of NeuN-positive staining in the posterior region of the cortex (coronal sections) illustrating the lesion extent in the medial and lateral entorhinal cortex at about Bregma 7.6 mm (notice the almost total loss of neuronal staining within the area delimited by the stippled line in H). All coordinates indicated in this caption are according to Paxinos and Watson (1998). Scale bar=1000 μm in A, B, G, and H, and scale bar=500 μm in C, D, E, and F. (b) Effects of double lesions on water maze performance. Upper panel: Acquisition data during the 3-day-training period are expressed as the mean (±SEM) of the distances to reach the platform in rats subjected to a lesion of septal cholinergic neurons and a fiber-sparing lesion of neurons in the EC (lesion, n=10) as compared with performance in the sham-operated controls (sham, n=11). Lower panel: Probe trial performance indicated as the mean time in the far quadrants (±SEM). Sham-operated control rats showed a performance in the target quadrant that was significantly above chance (ie, 15 s). In contrast, the double lesion affected spatial memory functions, as the lesioned rats did not focus their search in the appropriate quadrant. *p<0.05, significantly above chance. (c) Acetylated (Ac) and total (Tot) histone levels were measured by western blot analyses for H3 and H2B histones in total extracts obtained from the dorsal hippocampus of home cage rats (basal conditions; sham, n=6; lesion, n=8) or of trained rats (learning conditions: HPf group during 3 days for all rat groups; sham, n=7; lesion, n=7). Lysine acetylations measured are H3K9K14 and H2BK5K12K15K20. Typical western blots are represented in duplicates. Quantified results are represented as fold induction of the Ac/Tot ratio for each histone. The ratio obtained in the control condition is arbitrarily set at 100%. Sh, sham; Les, lesion. Student's t-test: **p<0.01. A significant decrease in acetylation of H2B is observed in the hippocampus of trained rats, but not in that of home cage rats. Acetylated-H3 levels tend to decrease as well only in learning conditions.
At the anatomical level, intraseptal injections of 192 IgG-saporin induced a massive decrease of AChE activity in the posterior hippocampus (Figure 5a, A and B), as well as of the number of ChAT-positive neurons in the medial septum (Figure 5a, C and D) and, although to a much lesser extent, in nucleus basalis magnocellularis (Figure 5a, E and F). We also performed semiquantitative assessment of the OD of AChE staining in several brain structures and found a specific and significant decrease (−47%) in the ODs in the posterior hippocampus (Supplementary Figure S5A). Likewise, the number of ChAT-positive neurons was significantly decreased in the BF (−85 and −83% in the medial septum and the diagonal band of Broca, respectively, and −31% in the nucleus basalis magnocellularis), whereas Parv-immunopositive neurons (and thus GABAergic) were only significantly decreased (−48%) in the diagonal band of Broca (see Supplementary Figure S5A). The lesion extent within the medial and lateral EC, as well as in surrounding regions, was evidenced on sections from the posterior region of the cortex that were immunostained for NeuN (Figure 5a, G and H). Basically, the EC lesions were very similar in location and extent to those reported by Traissard et al (2007); both the hippocampus and the septal region showed evidence for extensive cholinergic damage.
At the behavioral level, the capacity of sham-operated vs lesioned rats to swim to a VPf was evaluated on the first day. The average performance over four consecutive trials did not differ significantly between the two groups, although a tendency to higher distance to reach the platform was evidenced in the lesion group as compared with sham-operated rats (Supplementary Figure S5B, p=0.06). Both groups, however, showed a trial-dependent decrease in the distance to platform as expected, although in the lesion group the distance was moderately but significantly increased (p<0.005) on the fourth trial as compared with sham (Supplementary Figure S5B). These data showed that lesion rats presented no major deficit in navigation, motivation, and/or sensorimotor aspects in an MWM, which could impact performance in the HPf condition. Rats were then trained for 3 days in reference memory. Analysis of the training scores showed no significant group effect (F(1,19)=0.03) or a significant day effect (F(2,38)=13.6, p<0.001), and there was a strong tendency toward a significant interaction between both factors (F(2,38)=2.9, p=0.06). There was an overall improvement (both groups collapsed) owing to performances that were better on days 2 and 3 as compared with day 1 (p<0.001 for each comparison). However, rats bearing the combination of both lesions did not remember the platform location in the probe trial performed after an 1-h delay, whereas sham rats still remembered it (Figure 5b, below). During this trial, their time in the target quadrant was significantly above chance (ie, 15 s; t(10)=4.7, p<0.001), conversely to the time recorded in the lesion rats; in fact, the latter did not differ significantly from chance level (t(9)=1.7, NS). Furthermore, the scores of sham rats were significantly above those found in lesion rats (F(1,19)=19.6, p<0.001). Typical swim tracks observed in the probe trial after the 3-day training session are presented in Supplementary Figure S5C. These analyses clearly show that while lesion rats seemed to improve performance during the training phase, there was no evidence for memory consolidation.
The functional response of the CBP/histone acetylation pathway was then evaluated in such a pathological rat lesion model. We first verified the levels of histone acetylation in basal conditions (ie, HC) in the sham-operated rats vs the lesioned ones. Figure 5c (upper panel) shows that, in the dorsal hippocampus, there were no significant modifications at the global level on the ratio of acetylated/total histone acetylation for H3 or H2B. Similar results were obtained in the cerebellum extracts from the same rats (Supplementary Figure S6, upper panel). These data suggest that the combination of the two lesions did not impact on basal hippocampal (or cerebellar) functions regulated by these specific histone acetylation modifications. Second, we investigated these regulations in sham and lesion rats that had been subjected to the learning task. We found that acetylated-H2B histone levels remained significantly lower by almost 50% (p<0.01) in the lesion group compared with the sham group (Figure 5c, lower panel). This clearly shows that the dynamic regulation of H2B acetylation cannot occur normally in the denervated hippocampus. It is worth noting that acetylated levels of H3 histone, even if not statistically significant, also tended to decrease in the lesion compared with sham rats (26%, NS; Figure 5c). These data suggest that BF cholinergic and EC glutamatergic projections to the hippocampus interfered with the acetylated histone regulation induced by the MWM tasks (context processing and learning of the task). Importantly, such alterations in H2B and H3 acetylation levels found in the rat dorsal hippocampus were not observed in control cerebellar tissue of the rats that had learnt the task (Supplementary Figure S6, lower panel), evidencing a lack of histone acetylation regulation specifically in the denervated hippocampus. We can thus conclude that histone acetylation regulation necessary for memory functions (at least for H2B and H3) in the hippocampus is under the dynamic control of at least one of both BF cholinergic and EC glutamatergic projections.
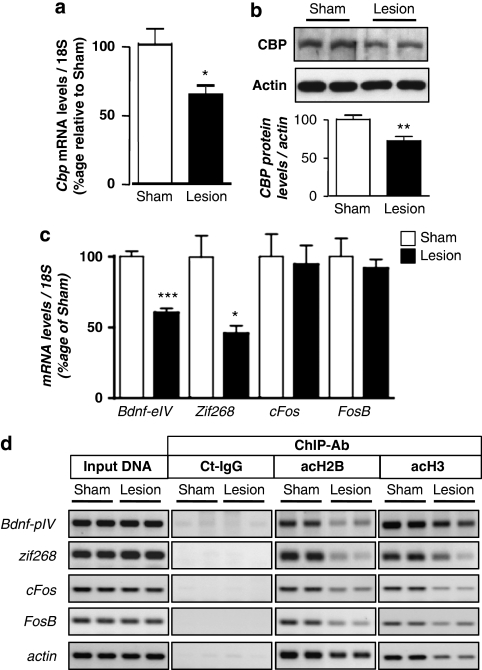
Subsequently, CBP levels, target gene expression, and acetylated-H3/H2B histone enrichment on their promoter were verified after training in the MWM (Figure 6). In the lesion group, CBP was neither upregulated at the mRNA level (CBP levels are 37% lower in lesion compared with sham group, p<0.05; Figure 6a) nor at the protein level (29% lower in lesion compared with sham group, p<0.01; Figure 6b) upon spatial learning, relative to the sham group. Bdnf-eIV and zif268 mRNA levels were not induced in the lesion group in response to spatial training (respectively, 40% lower than in the sham rats, p<0.001 and 54%, p<0.05; Figure 6c). This was further supported by ChIP experiments (Figure 6d) showing that acetylated-H2B histone levels were dramatically reduced on their promoters (bdnf-pIV: 66% and zif268: 64%). These data indicate that, in this lesion model, signaling in the dorsal hippocampus is not sufficient to achieve the gene regulation necessary for memorization. This is consistent with our previous findings and further supports the importance of the CBP/acetylation signaling pathway for memory consolidation. In addition, acetylated-H3 histone enrichment was also decreased on these genes, suggesting that the denervated hippocampus could not integrate context processing-dependent acetylation either. Intriguingly, acetylated histone occupancy was reduced on actin, cFos, and FosB, whereas their expression levels were not significantly altered (Figure 6b and c). A possible explanation could be that in such situation, the transcription of these genes is less dependent on histone acetylation. β-Actin may also be more stable. The molecular mechanisms by which this happens remain unclear, but it is worth noting that if they could compensate for actin, cFos, and FosB regulations, they could not compensate for the specific regulations on bdnf-eIV and zif268 engaged during memory formation.
Figure 6.
CBP- and histone acetylation-dependent regulations during formation of a spatial memory in a cognitively impaired rat model. Cbp transcripts (a) were evaluated by real-time RT-PCR (n=5) and CBP protein (b) by western blot analyses (n=7) in the dorsal hippocampus from sham-operated (sham) or double lesion (lesion) rats group after a 3-day-training period. CBP levels were not increased by the spatial memory training in lesion rats, *p<0.05, **p<0.01, as compared with sham, (c) Real-time RT-PCR analysis of bdnf-eIV, zif268, cFos, and FosB genes in the same conditions as in (a), *p<0.05, ***p<0.001, as compared with sham (n=5). (d) Chromatin immunoprecipitation were performed on 3-day-trained rats from sham and lesion groups in the HPf version of the water maze (n=3 per group). Semiquantitative PCR were performed to analyze the presence of bdnf-pIV, cFos, FosB, and zif268 proximal promoters in the immunoprecipitates. Controls for immunoprecipitation were performed with an unrelated control antibody (Ct-IgG) or no antibody (noAb). Input DNA corresponding to the total of chromatin before immunoprecipitation was run in parallel as positive control. Representative results are shown in duplicate for each gene tested. β-Actin was used as internal control. acH3: acetylated K9, K14 H3 histone; acH2B: acetylated K12, K15 H2B histone; bdnf-eIV: bdnf exon IV; and bdnf-pIV: bdnf promoter IV. After 3 days of spatial training, none of the promoters are enriched in acetylated-H2B or in acetylated-H3 histones in the lesion rat group as compared with sham-operated rats.
Downregulation of the CBP/histone acetylation-dependent pathway is observed in a rat model in which consolidation of a spatial memory is compromised. This probably underlies the lack of proper transcriptional regulation of specific memory genes, as we show for the bdnf and zif268 genes. We hypothesize that a specific genetic program necessary for or underlying memory consolidation is not recruited because a certain HAT/acetylation threshold has not been achieved in the lesion rat (see model in Figure 7). In addition, these data point to the importance of the EC glutamatergic and BF cholinergic projections role in controlling CBP/histone acetylation-dependent regulations in the context of (spatial) memory formation.
Figure 7.
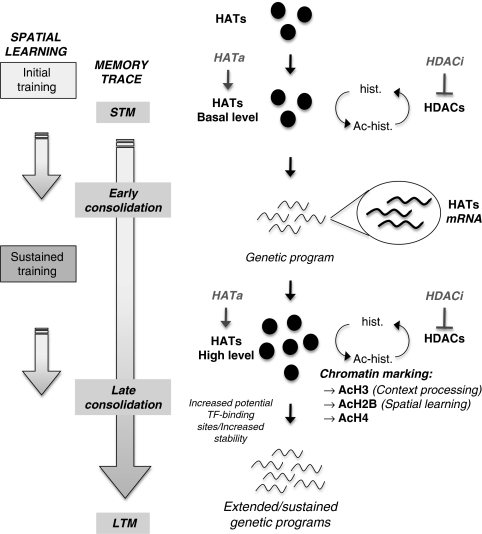
Hypothetical model of molecular interplay underlying consolidation of spatial memory. During the initial steps of a training aimed at acquiring a spatial task, the memory trace is progressively setup following a cascade of intracellular signals involved in short-term memory (STM) processing. These processes are non-transcriptional events (Kandel, 2001; Medina et al, 2008) as existing co-activators (HATs, basal levels) and transcription factors present in neurons are likely in sufficient quantities to promote a genetic program necessary for early consolidation signals. Here we show that, while training is pursued, the expression of several HATs (CBP, p300, and p300/CBP-associated factor (PCAF)) is increased, thus generating new HAT molecules (HATs, high level)—and CBP in particular—during a critical time window of spatial memory formation. We propose that reaching a new HAT threshold could allow a widening of HAT-binding partners to a larger panel of transcription factors and/or increase HAT stability over time, thereby potently triggering extended and/or sustained specific genetic programs for late consolidation. We showed that repetition of training (‘sustained training') is associated with increased HAT activity and new chromatin marking: histone H3 is acetylated in response to context-related processing (visible platform), whereas H2B histone is acetylated during spatial learning (hidden platform). H4 histone acetylation is also increased during the late consolidation phase. Each acetylation step can be reversed under the action of histone deacetylase (HDAC) enzymes, probably underlying the memory enhancer effect of some HDAC inhibitors (HDACi) (for reviews see, Barrett and Wood, 2008). Taken together, our results suggest that an epigenetic tagging of the genome takes place under the influence of HAT expression at specific memory/plasticity genes throughout the formation of spatial long-term memory (LTM). They also emphasize that directly promoting this chromatin marking pattern with HAT activators could be a new therapeutic strategy for memory enhancers, potently more specific than HDAC inhibition. Ac, acetylated; hist., histone; HATa, HAT activator; TF, transcription factor.
DISCUSSION
These experiments reveal three fundamental points in memory formation and consolidation that suggest promising therapeutic strategies. First, there is a differential regulation of the histone acetylation profile in the hippocampus of normal rats while a trace memory is being established. We report for the first time an increase in acetylation of the H2B histone during spatial learning, which can be functionally related to activated memory/plasticity gene transcription. Conversely, H3 acetylation appeared reactive to the environmental context rather than to the spatial learning task itself. This means that H2B acetylation could represent an epigenetic marker of LTM for spatial information. Second, we found that several HATs were upregulated at the transcriptional levels in the dorsal hippocampus during the ongoing process of memory formation. Notably, CBP protein levels were increased after repeated trainings, suggesting a potential role in controlling the length and intensity of specific genetic programs during memory consolidation. Third, we found that the CBP/histone acetylation pathway had lost its responsiveness to training in a rat model that mimics some of the lesions described in AD, thus accounting for impaired memory function and consolidation processes in particular. Currently, there is a great deal of research on therapeutic strategies for memory enhancers based on acetylation modulators, notably with the use of HDACi. These findings could have interesting therapeutic repercussions for treating neurodegenerative disorders such as AD and other memory-related disorders, as they bring compelling evidence predicting that agents able to increase HAT/CBP expression will be more specific than HDACi.
A Potential Link between H2B Acetylation and Memory Formation?
Specific epigenetic tagging of the genome in relation to LTM is an intriguing hypothesis (Wood et al, 2006b; Sweatt, 2009) and different works have focused on histone post-translational modifications, mainly acetylations, during task acquisition (reviewed by Barrett and Wood (2008)). Recent studies have also evidenced changes in histone methylation, at a site marking gene activation, during contextual fear memory (Gupta et al, 2010). We show that, while no change in acetylation levels was detected on H3 and H2A histones, the acetylated forms of H2B and H4 histones were significantly increased in the dorsal hippocampus of rats during spatial memory formation. H3 acetylations were upregulated after 3 days of training in the MWM (HPf and VPf) as compared with untrained controls. This upregulation could be a consequence of adaptations to the testing situation, rats having to cope with stress, fear, swimming, and other basic navigation-related processes in both task versions. Accordingly, H3 acetylation might have epigenetically tagged the genome in order to promote transcription of specific genes recruited by such adaptive processes, or to prepare the chromatin for the subsequent gene transcription required in memorization processes (ie, bring a first degree of chromatin opening). Nevertheless, in the same experimental conditions, H2B acetylation levels remained unchanged (Figure 1c), supporting that the increase observed between learning and control rats (Figure 1b) was highly relevant to the acquisition of a spatial memory.
This is the first work describing H2B acetylation modulation in mammals during a physiological process. Indeed, so far, functional studies on acetylation of H2B have been mostly limited to yeast (reviewed by Wyrick and Parra (2009)). Early biochemical experiments showed that acetylation of the histone H2A–H2B dimer facilitated transcription in vitro (Puerta et al, 1995). H2B bears a conserved repression (HBR) domain for specific gene transcription and acetylation of the N-terminal lysines of H2B can partially overcome this repression (Parra et al, 2006). Our prime interest is now to identify acetylated-H2B target genes and their relevance to spatial memory consolidation in hippocampal neurons. Interestingly, a downregulation of only H2B histone acetylation was found in the hippocampus of cbp+/− mice (Alarcon et al, 2004), and H2B was also identified as a substrate for p300 and PCAF, but not Tip60 (Roth et al, 2001; Allis et al, 2007). At the mechanistic level, a recent paper described the acetylation of H2B in response to depolarization in hippocampal slices in vitro (Maharana et al, 2010). This is in line with our study enlightening the implication of glutamatergic—at least in part with cholinergic—projections in the control of H2B acetylation and further strengthened its potential role in memory formation. We show that acetylated-H2B are enriched at selected gene promoters related to (spatial) memory function (Figure 2B), as well as to the cbp gene itself (Figure 4c). Recently, the HDAC2 isoform was shown to regulate memory formation and synaptic plasticity through deacetylation at some of these gene promoters (Guan et al, 2009). Interestingly, among the four core histones, H2B and H4, but not H3 and H2A, were hyperacetylated in hippocampal histone extracts from HDAC2 knockout mice (Guan et al, 2009). Our findings showing the dynamic H2B/H4 acetylation regulation during spatial memory consolidation add up to the challenging hypothesis that a specific epigenetic code (specific histone modifications marks) could participate in the regulation of specific memory formation (for reviews see, Wood et al, 2006b; Sweatt, 2009).
Increased HATs Expression and Memory Formation
The importance of HAT activity in the formation of LTM is well documented (reviewed by Barrett and Wood (2008)). However, all studies so far reported were performed on mutated mice, and none of them focused on regulations in the normal hippocampus during memory formation. Interestingly, we measured a significant increase in several HATs expression and activity in hippocampal extracts during memory acquisition (Figure 3b and c). It was shown that spatial memorization deficits present in conditional mice expressing a HAT-deficient CBP molecule could be counteracted by extended training (Korzus et al, 2004) and that spatial deficits due to brain damage could be reduced by environmental enrichment that correlated with increased histone-tail acetylation (Fischer et al, 2007). It is thus possible that upon certain signals, co-activators like CBP, p300, and PCAF might be able to compensate for each other in spatial learning. Unexpectedly, we found that CBP occupancy was not maximal on memory-related genes (such as bdnf-pIV), and was even decreased on the zif268 promoter, otherwise clearly enriched in H2B acetylation (Figure 4). These results suggest an interaction between these related HATs (CBP, p300, and PCAF) during memory formation to orchestrate the modulation of specific memory/plasticity genes necessary for long-term memorization processes. Notably, p300 has a critical role in long-term recognition memory, but not in spatial memory formation (Oliveira et al, 2007), whereas PCAF-KO mice present short-term memory and LTM deficits in spatial memorization (Maurice et al, 2008). Thus, the way the different HATs actually interfere with each other during (spatial) memorization processes remains an open question. Further investigation is needed with regard to the development of additional technical tools (eg, ChIP antibodies) and studies in other experimental situations (investigation of other memory tasks and time courses).
The final outcome of HAT activation is the induction of histone acetylation on specific genes, which is possible to perform with the use of deacetylase inhibitors. The potential beneficial role of HDACi has been shown in diverse forms of memory formation (Levenson et al, 2004; Alarcon et al, 2004; Bredy et al, 2007; Bredy and Barad, 2008; Lattal et al, 2007;Vecsey et al, 2007; Fischer et al, 2007; Grissom and Lubin, 2009; Stefanko et al, 2009). Recently, such molecules have also been successfully tested in animal models of AD (Fischer et al, 2007; Ricobaraza et al, 2009; Francis et al, 2009; Kilgore et al, 2010). In spite of the clinical trials of HDACi in cancer therapeutics, there are widespread speculations with regard to the target specificity of these molecules. The HDACs are functional as a class of proteins and not as individual members, and most often they have been shown to compensate for each other's function. In addition, the use of pan-HDACi, by leading to a global increase in acetylation, would produce an imbalance in acetylation levels within the cells. Most importantly, the use of HDACi might also lead to a loss in the tissue- and signal-specific responses. Hence, several strategies targeted toward direct activation of acetyltransferases have to be attempted. Indeed, by directly placing the correct acetylation marks on chromatin at specific target genes, as we show this is the case during memory formation in this study, these molecules should be able to promote or improve memory consolidation processes necessary for long-term memorization with more specificity than HDACi. Interestingly, potent permeable HAT-activating molecules are being developed (Selvi et al, 2008).
CBP Expression and Sensitivity in Neuronal Functions
We show that hippocampal CBP levels increased over the acquisition of a memory task using different techniques (Figures 3 and 4). This appears to be the first report describing a physiological regulation of CBP expression at the system level, which could be relevant to memory formation. So far, CBP upregulations were only described in pathological conditions in some forms of cancer (Karamouzis et al, 2002; Comuzzi et al, 2004). In fact, CBP levels must be finely tuned as a lowered CBP expression level is deleterious: embryos nullizygous for cbp die by embryonic day 10.5 (Oike et al, 1999). In humans, the monoallelic mutation of the CBP locus is the genetic basis for Rubinstein–Taybi syndrome (Petrij et al, 1995). In mature neurons, loss of the CBP protein or inactivation of its enzymatic activity is a mark of neurodegenerative condition (Rouaux et al, 2004; Saha and Pahan, 2006). Thus, competition for recruitment of CBP might provide a potential mechanism for cross-talk between different neuronal functions based on the idea that CBP levels are present in limited concentrations in the cells (Vo and Goodman, 2001).
We propose that increasing the CBP amount during memory formation will increase the CBP-binding possibilities to other transcription factors, thereby widening the expression profile to other target genes, likely to contribute to memory trace consolidation (see model in Figure 7). Moreover, it might increase CBP stability over time, allowing activation of target genes over lengthened delays. The presence of bdnf, peaking 12 h after a one-trial acquisition (inhibitory avoidance), is an example of late molecular activation necessary for consolidation (Bekinschtein et al, 2007). Its specific promoter regulation requires histone acetylation modulation (Bredy et al, 2007; Lubin et al, 2008).
In this report, we show that the CBP/histone acetylation-dependent pathway, at all levels assessed is no longer recruited during a learning task after a combination of cholinergic and glutamatergic hippocampal denervations, suggesting an active role of these projections in regulating neuronal HAT activity and histone acetylation during memory formation. The combination of these lesions impairs declarative-like memory functions in rats and model aspects of anterograde amnesia, as described in AD patients (Cassel et al, 2008). Thus, any method of physiologically modulating CBP levels is of prime interest for memory-related diseases. Indeed, drugs targeted to increase specific CBP expression could reach the threshold of CBP molecules necessary to activate correct genetic programs with more precisions than solely modulating histone acetylation levels. Pharmacological modulation of CBP levels has been reported recently (Saha et al, 2009): preconditioning of neurons with TNF-α protected them from degenerative conditions (and particularly Aβ toxicity) through a neuron-specific increase of CBP expression. The authors suggested that an ‘adequate' amount of CBP is required at a basal level to ensure pro-survival functions. We have previously shown that CBP was a caspase-6 substrate (Rouaux et al, 2003), a caspase that is activated in AD and MCI patients neurons, as well as during aging (Guo et al, 2004; Albrecht et al, 2007, 2009). Herein, we report that the formation of a new memory is accompanied by dynamic regulations of CBP levels—as well as several other related HATs—in the hippocampus. These data may suggest that CBP levels could actually be modulated throughout our lifetime by environmental cues (including LTM formation), thus enabling normal transcriptional processes required for survival and memory functions.
Epigenetic deregulations are increasingly associated with cognitive disorders (Graff and Mansuy, 2009), including AD (Chouliaras et al, 2010). HAT-activating molecules, not only at the level of their activity, but also at that level of their expression, could result in significant advances in therapeutic strategies aimed at tackling the problem of pathological aging. These discoveries may be particularly important in the treatment of AD-associated illnesses, as therapies that delay memory impairment will decrease healthcare costs and increase quality of life.
Acknowledgments
RN is a recipient of a fellowship from the French Ministère de la Recherche et de l′Enseignement Supérieur. IP obtained a 2-year grant from Inserm (‘poste vert'). We wish to thank the Fédération de la Recherche sur le Cerveau (FRC), who brought the research fundings (to ALB) necessary to initiate and support part of this project. Dr K Fisher Ratan (CA, USA) is thanked for critical reading and English proof-reading of the manuscript. This work was supported by the Inserm, ‘Association pour la Recherche sur les Maladies Neurodégénératives' (AREMANE) and ‘France Alzheimer 67' association for UMRS692 (to JPL), and by the University of Strasbourg and the CNRS for FRE 3289 (to JCC). The authors are grateful to O Bildstein, D Egesi, and G Edomwonyi (FRE 3289) for their assistance in animal care; MJ Ruivo (U692) for her technical assistance; and to S Lithfous for help with regard to the counting of ChAT- and Parv-positive cells.
The authors declare that they have no financial or any other type of involvement that might potentially bias their work and be perceived as constituting a potential conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein–Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Albrecht S, Bogdanovic N, Ghetti B, Winblad B, LeBlanc AC. Caspase-6 activation in familial Alzheimer disease brains carrying amyloid precursor protein or presenilin I or presenilin II mutations. J Neuropathol Exp Neurol. 2009;68:1282–1293. doi: 10.1097/NEN.0b013e3181c1da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170:1200–1209. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Lidge R, Catapano R, Stanley J, Gossweiler S, Romashko D, et al. A mouse model of Rubinstein–Taybi syndrome: defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc Natl Acad Sci USA. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel JC, Mathis C, Majchrzak M, Moreau PH, Dalrymple-Alford JC. Coexisting cholinergic and parahippocampal degeneration: a key to memory loss in dementia and a challenge for transgenic models. Neurodegener Dis. 2008;5:304–317. doi: 10.1159/000135615. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, et al. Epigenetic regulation in the pathophysiology of Alzheimer's disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Comuzzi B, Nemes C, Schmidt S, Jasarevic Z, Lodde M, Pycha A, et al. The androgen receptor co-activator CBP is up-regulated following androgen withdrawal and is highly expressed in advanced prostate cancer. J Pathol. 2004;204:159–166. doi: 10.1002/path.1609. [DOI] [PubMed] [Google Scholar]
- Dash PK, Orsi SA, Moore AN. Histone deactylase inhibition combined with behavioral therapy enhances learning and memory following traumatic brain injury. Neuroscience. 2009;163:1–8. doi: 10.1016/j.neuroscience.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J Neurosci. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram. Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures. Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Francis YI, Fa M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, et al. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer's disease. J Alzheimers Dis. 2009;18:131–139. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B. The organization of recent and remote memories. Nat Rev Neurosci. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- Gräff J, Mansuy IM. Epigenetic codes in cognition and behavior. J Neurosci Res. 2008;85:525–535. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci. 2009;30:1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- Grissom NM, Lubin FD. The dynamics of HDAC activity on memory formation. Cell Science. 2009;6:44–48. [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Karamouzis MV, Papadas T, Varakis I, Sotiropoulou-Bonikou G, Papavassiliou AG. Induction of the CBP transcriptional co-activator early during laryngeal carcinogenesis. J Cancer Res Clin Oncol. 2002;128:135–140. doi: 10.1007/s00432-001-0317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana C, Sharma KP, Sharma SK. Depolarization induces acetylation of histone H2B in the hippocampus. Neuroscience. 2010;167:354–360. doi: 10.1016/j.neuroscience.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. Memory—a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- Medina JH, Bekinschtein P, Cammarota M, Izquierdo I. Do memories consolidate to persist or do they persist to consolidate. Behav Brain Res. 2008;192:61–69. doi: 10.1016/j.bbr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Müller GE, Pilzecker A. Experimentalle beitrage zur lehre vom gedächtnis. Z Psychol. 1900;l:1–288. [Google Scholar]
- Oike Y, Takakura N, Hata A, Kaname T, et al. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleeva I, Boutillier S, See V, Spiller DG, Rouaux C, Almouzni G, et al. HP1α guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. The Embo J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MA, Kerr D, Fahy D, Pouchnik DJ, Wyrick JJ. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol Cell Biol. 2006;26:3842–3852. doi: 10.1128/MCB.26.10.3842-3852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Puerta C, Hernandez F, Lopez-Alarcon L, Palacian E. Acetylation of histone H2A.H2B dimers facilitates transcription. Biochem Biophys Res Commun. 1995;210:409–416. doi: 10.1006/bbrc.1995.1676. [DOI] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, Loeffler JP, Boutillier AL. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem Pharmacol. 2004;68:1157–1164. doi: 10.1016/j.bcp.2004.05.035. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Panteleeva I, Rene F, Gonzalez de Aguilar JL, Echaniz-Laguna A, Dupuis L, et al. Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci. 2007;27:5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Ghosh A, Palencia CA, Fung YK, Dudek SM, Pahan K. TNF-alpha preconditioning protects neurons via neuron-specific up-regulation of CREB-binding protein. J Immunol. 2009;183:2068–2078. doi: 10.4049/jimmunol.0801892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi BR, Jagadeesan D, Suma BS, Nagashankar G, Arif M, Balasubramanyam K, et al. Intrinsically fluorescent carbon nanospheres as a nuclear targeting vector: delivery of membrane-impermeable molecule to modulate gene expression in vivo. Nano Lett. 2008;8:3182–3188. doi: 10.1021/nl801503m. [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci USA. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traissard N, Herbeaux K, Cosquer B, Jeltsch H, Ferry B, Galani R, et al. Combined damage to entorhinal cortex and cholinergic basal forebrain neurons, two early neurodegenerative features accompanying Alzheimer's disease: effects on locomotor activity and memory functions in rats. Neuropsychopharmacology. 2007;32:851–871. doi: 10.1038/sj.npp.1301116. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006a;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory. Learn Mem. 2006b;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Parra MA. The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective. Biochim Biophys Acta. 2009;1789:37–44. doi: 10.1016/j.bbagrm.2008.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.