Abstract
EmrE is an Escherichia coli multidrug transporter that confers resistance to a variety of toxins by removing them in exchange for hydrogen ions. The detergent-solubilized protein binds tetraphenylphosphonium (TPP+) with a KD of 10 nM. One mole of ligand is bound per ∼3 mol of EmrE, suggesting that there is one binding site per trimer. The steep pH dependence of binding suggests that one or more residues, with an apparent pK of ∼7.5, release protons prior to ligand binding. A conservative Asp replacement (E14D) at position 14 of the only membrane-embedded charged residue shows little transport activity, but binds TPP+ at levels similar to those of the wild-type protein. The apparent pK of the Asp shifts to <5.0. The data are consistent with a mechanism requiring Glu14 for both substrate and proton recognition. We propose a model in which two of the three Glu14s in the postulated trimeric EmrE homooligomer deprotonate upon ligand binding. The ligand is released on the other face of the membrane after binding of protons to Glu14.
Keywords: ion-coupled transport/membrane proteins/multidrug resistance/multidrug transporter
Introduction
Multidrug transporters recognize a wide variety of substrates with high affinity and actively remove them from the cytoplasm. When the substrates are toxic to the cells, removal away from their cellular targets confers resistance against their harmful effects. This survival strategy is associated with multidrug resistance, a phenomenon that poses a serious problem in treatment of infectious diseases and resistant tumors (Nikaido, 1994; Gottesman et al., 1996). Multidrug transporters are ubiquitous and, based on primary amino acid sequence similarities, they have been classified into several families (Marger and Saier, 1993; Paulsen et al., 1996b).
The 110 amino acid multidrug transporter from Escherichia coli, EmrE, is a member of the family of MiniTexan or Smr drug transporters (Grinius and Goldberg, 1994; Paulsen et al., 1996a; Schuldiner et al., 1997). EmrE can transport acriflavine, ethidium bromide, tetraphenylphosphonium (TPP+), benzalkonium and several other drugs with relatively high affinities. EmrE is an H+/drug antiporter, utilizing the proton electrochemical gradient generated across the bacterial cytoplasmic membrane by exchanging two protons with one substrate molecule (Yerushalmi et al., 1995).
The EmrE multidrug transporter is unique in terms of its small size and hydrophobic nature (Yerushalmi et al., 1995). Hydropathic analysis of the EmrE sequence predicts four α–helical transmembrane segments. This model is supported experimentally by Fourier transform infrared studies that confirm the high α–helicity of the protein and by high-resolution heteronuclear NMR analysis of the protein structure (Arkin et al., 1996; Schwaiger et al., 1998). Negative dominance experiments suggest that EmrE forms a functional trimer (Yerushalmi et al., 1996). The membrane-embedded Glu14 is the only charged residue within the protein essential for activity (Paulsen et al., 1996a; Yerushalmi et al., 1996; Yerushalmi and Schuldiner, 2000). It is fully conserved in all members of the Mini Texan family, and even a conservative substitution by Asp at position 14 abolishes the ability of the transporter to confer resistance to the toxic chemicals (Paulsen et al., 1996a; Schuldiner et al., 1997). Interestingly, a variety of other multidrug transporters encode a negatively charged residue in their first transmembrane domain (Paulsen et al., 1996b). This includes a number of 12 and 14 transmembrane domain transporters. Additionally, in the E.coli multidrug transporter MdfA, such a glutamate residue (at position 26) was shown to be critical for transporter recognition of lipophilic cationic substrates (Edgar and Bibi, 1999). Edgar and Bibi (1999) propose that Glu26 of MdfA serves as a component of a drug-binding domain and probably interacts directly with the positive charge of cationic lipophilic substrates such as TPP+, ethidium bromide and benzalkonium. The staphylococcal transporter QacA contains at least one membrane-buried Asp residue that contributes significantly to its drug transport activity (Paulsen et al., 1996b; Edgar and Bibi, 1999). The transcriptional activator protein BmrR, of the multidrug resistance Bmr transporter from Bacillus subtilis, is also capable of binding lipophilic cations such as TPP+(Zheleznova et al., 1999). The structure of BmrR has been determined by X–ray crystallography in the presence and absence of TPP+. In the hydrophobic binding pocket, an electrostatic interaction between a buried glutamate residue (E134) and TPP+ is critical for substrate selectivity. In addition, the hydrophobic binding pocket in BmrR is exposed to the ligand after unfolding of an α–helix. Also in other transporters, negatively charged residues in the first transmembrane domain have been implicated in substrate recognition (Barker et al., 1999). E126 and D144 of the E.coli lac permease are required for substrate binding, providing further evidence that glutamate and aspartate residues are utilized for binding a vast array of dissimilar substrates by distantly related proteins (Sahin-Toth et al., 1999).
In this study, we demonstrate a role for Glu14 in both substrate and H+ binding and release from the EmrE transporter. We find that detergent-solubilized purified EmrE binds the toxic substrate TPP+ with high affinity. Binding is specific and is inhibited by other substrates of the transporter. A maximal binding of 1 mol of TPP+ has been observed per ∼3 mol of EmrE, supporting the proposal that EmrE functions as a trimer and that a single binding site is shared by the oligomer. Both the binding and release of TPP+ from EmrE are strongly influenced by pH in a way that suggests that protonation of residues within the transporter has a negative effect on its ability to bind to or retain previously bound TPP+. A study of the binding ability of mutant proteins with replacements at position 14 supports a model to explain the mechanism of action of EmrE in which two of the three Glu14s, in the postulated trimeric homooligomer, deprotonate upon ligand binding. After translocation of the ligand to the other face of the membrane, it is released only after binding of protons to Glu14. The cycle is terminated by return of the empty binding site to the opposite face of the membrane.
Results
Binding of [3H]TPP+ to purified EmrE–His
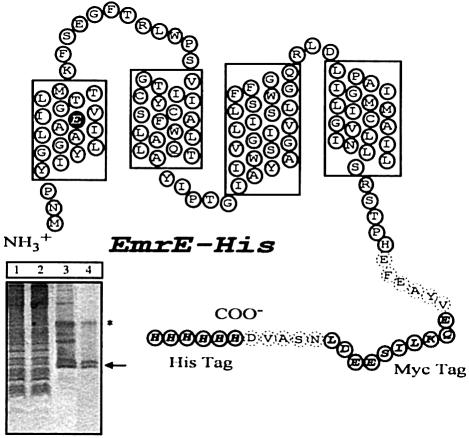
The ability of EmrE to convey drug resistance and transport various toxic substrates, in vivo and in vitro, has been clearly demonstrated (Schuldiner et al., 1997). There is little known, however, about the details of substrate binding or the specifics of how toxic compounds are translocated in exchange for H+ ions. In order to examine substrate binding to EmrE, we modified the transporter by the addition of a His6 tag (Figure 1). This tag is used for immobilization of the detergent-solubilized protein to a metal chelate adsorbent that allows for purification and also for a convenient assay. Escherichia coli expressing EmrE–His were able to grow in the presence of several toxic compounds including ethidium bromide, acriflavine and methyl viologen, verifying that the transporter is functional and capable of providing a drug-resistant phenotype similar to that of wild-type, untagged EmrE (data not shown). EmrE–His was also functional as determined by an in vitro drug transport assay, although the transport activity was at a reduced level compared with the wild-type EmrE (data not shown). Nearly homogeneous preparations of EmrE–His protein were obtained through a simple two-step purification protocol (see Materials and methods). We immobilized the detergent-solubilized transporter on Ni–NTA beads, and the immobilized EmrE–His transporter was examined subsequently for its ability specifically to bind labeled substrates.
Fig. 1. Model of EmrE–His with the four transmembrane regions predicted by hydropathy plots. The His and Myc tags are indicated by open ovals with one-letter amino acid codes shown in bold italicized text. Dashed ovals indicate residues that were incorporated into the transporter in the process of linking in the epitope tags. The Glu14 residue in the first transmembrane region is highlighted. The Myc tag is used here only as a linker to keep the His residues away from the membrane. Without the linker, the His-tagged transporter displays only residual activity. Inset, SDS–PAGE analysis of different stages of purification. Lane 1, total membranes; lane 2, detergent-solubilized extract after Ni–NTA purification, unbound fraction; lane 3, EmrE after Ni–NTA purification; lane 4, EmrE after size exclusion purification. The arrow indicates the monomeric form of EmrE; the asterisk the dimeric form. The apparent Mr of monomeric EmrE–His is 14 400 Da.
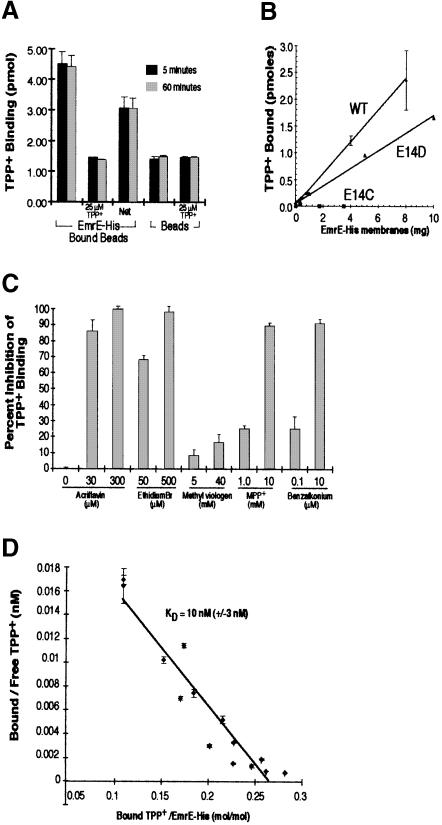
We chose to test TPP+ because of its high-affinity interaction with the transporter, demonstrated by its inhibition of methyl viologen transport (Yerushalmi et al., 1995). The results shown in Figure 2A demonstrate that purified EmrE–His bound to Ni–NTA beads specifically binds [3H]TPP+. [3H]TPP+ binding is dependent on the presence of EmrE–His and can be competed away by 25 μM unlabeled TPP+. The level of background binding reflects the presence of unbound [3H]TPP+ associated with the Ni–NTA beads rather than non-specific binding to EmrE–His, since the background levels of beads alone and EmrE–His-bound beads in the presence of unlabeled TPP+ are identical. Under these conditions, virtually all of the TPP+ binding occurs within the first 5 min of the binding reaction, with no increase seen with incubations up to 1 h. The technical limitations of the assay in its present form prevent accurate measurements of binding for time points of ⩽1 min, and therefore we were not able to determine the rate of TPP+ binding to EmrE–His. Figure 2B shows that binding is linear with the amount of protein used.
Fig. 2. Purified EmrE–His specifically binds [3H]TPP+. (A) Purified EmrE–His (4 μg) was bound to Ni–NTA beads and then incubated with 25 nM [3H]TPP+ in NH4–DM buffer. TPP+ binding reactions were incubated for 5 min (black bars) or 60 min (gray bars). Identical binding reactions were performed with beads that had not been bound with EmrE–His. Each TPP+ binding reaction was performed in the presence or absence of 25 μM cold TPP+. (B) Bacterial membranes expressing EmrE–His wild type, E14D or E14C were detergent solubilized and then incubated with Ni–NTA beads. Increasing amounts of solubilized membranes were bound to 20 μl of beads. The beads were then washed to remove proteins that did not bind. The bound beads were then incubated for 30 min with 25 nM [3H]TPP+ and the amount of bound TPP+ was determined as described above. (C) EmrE–beads were incubated with 50 nM [3H]TPP+ in the presence of various EmrE substrates. The concentration of inhibitor used is indicated below each bar. The values of TPP+ binding are plotted in terms of percentage inhibition, with binding in the absence of inhibitors representing zero inhibition. (D) EmrE–His transporter (0.35 μg) bound to Ni–NTA beads was incubated with TPP+ over a range of TPP+ concentrations (10–300 nM). Bound TPP+ was determined by measuring the EmrE–His-associated radioactivity after isolating the beads from the supernatant, and free TPP+ concentrations were measured by subtracting bound radioactivity from the total radioactivity present in each binding reaction. The KD was determined from the reciprocal of the slope obtained from the Scatchard plot. The KD for TPP+ under these conditions was 10 nM (±3 nM). In (A–D), all results are generated from the average of triplicate reactions and the error bars represent the average deviation.
Inhibition of TPP+ binding to EmrE–His
If the binding of TPP+ to the purified EmrE–His is specific, then other EmrE substrates should inhibit the binding of TPP+. Using known substrates, we tested whether they could compete with the TPP+ for access to the transporter-binding sites (Yerushalmi et al., 1995). We found that 500 μM ethidium bromide or 300 μM acriflavine included in a TPP+ binding assay reduced the levels of TPP+ binding to almost zero (Figure 2C). Two other EmrE substrates, 1–methyl-4–phenylpyridinium and benzalkonium, also completely inhibit TPP+ binding (at concentrations of 10 mM and 10 μM, respectively). This suggests that TPP+ binding to EmrE–His is specific and related to the drug efflux activity of the transporter. Surprisingly, methyl viologen, which is also a substrate for the EmrE transporter (Yerushalmi et al., 1995), inhibited TPP+ binding modestly and only at very high concentrations (>10 mM). These findings suggest that the detergent-solubilized protein displays drug-binding specificities very similar, albeit not identical, to those of the protein in the membrane.
Affinity of TPP+ binding to EmrE–His
The rapid binding of TPP+ to the EmrE–His transporter prevented us from determining kinetic constants using rate-based calculations. To circumvent this complication, we measured the equilibrium binding of TPP+ at several TPP+ concentrations. The results of such an experiment are shown in Figure 2D. The KD of TPP+ binding to purified EmrE–His measured by this method is 10 nM (±3 nM). This value is very close to the 8 nM IC50 value determined in experiments in which TPP+ was tested for its ability to inhibit [14C]methyl viologen transport mediated by purified wild-type EmrE (Yerushalmi et al., 1995). It is also practically identical to the KD determined for the wild-type EmrE (20 ± 15 nM) using Sephadex G–50 size exclusion chromatography (data not shown). The total number of TPP+-binding sites determined in these experiments is between 0.25 and 0.3 mol/mol (Figure 2D). This finding supports the suggestion that EmrE is a functional trimer (Yerushalmi et al., 1996). It also suggests that only one site per EmrE trimeric complex is occupied at any given time. Furthermore, it seems that the detergent-solubilized protein is likely to retain a structure similar to that which it assumes within the bacterial membrane.
pH dependence of TPP+ binding to EmrE–His
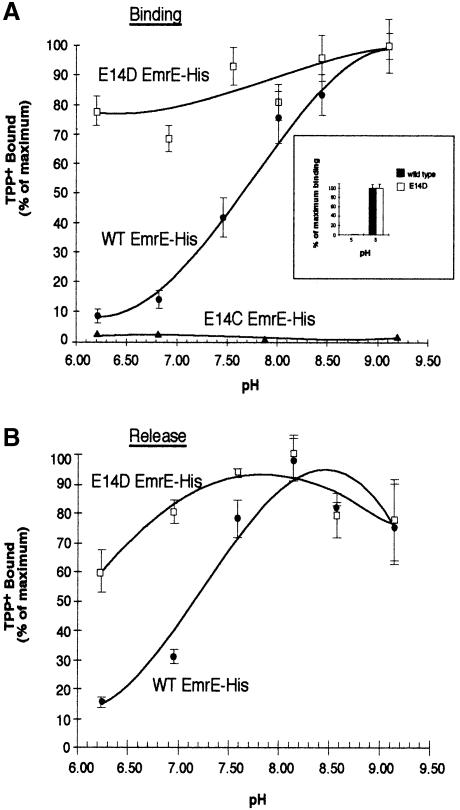
The EmrE multidrug antiporter uses the ΔμH+ across the membrane as a driving force to remove toxic substrates from within the bacteria (Schuldiner et al., 1997). This is achieved by exchange of two protons with one drug molecule in a process that involves coupled and coordinated binding and release of both substrates. Therefore, we determined the pH dependence of TPP+ binding to EmrE–His. To address this question, we performed TPP+ equilibrium binding reactions in buffers that covered a pH range from 6.2 to 9.2. We were unable to test a broader pH range because of the significant release of EmrE–His protein from the Ni–NTA beads at pH values <6.2 and >9.2 (data not shown). Figure 3A shows that TPP+ binding increased from low levels at pH 6.2, reaching near maximal binding above pH 8.5. These data are consistent with a substrate transport mechanism requiring release of protons from the transporter prior to ligand binding. Several lines of evidence predict that Glu14 is the only charged residue buried within the membrane (Yerushalmi et al., 1995; Arkin et al., 1996; Schwaiger et al., 1998; Steiner Mordoch et al., 1999). Glu14 may form part of a binding pocket and may interact directly with substrate molecules and/or protons, thereby playing a significant role in substrate translocation. To test this hypothesis, we tested the activity of mutants in which Glu14 was replaced by either aspartate or cysteine (E14D and E14C, respectively). Neither mutated EmrE transporter conveyed a resistant phenotype to bacteria grown in the presence of toxic substrates. E14D retains ∼12% of the ΔpH-driven transport activity, whereas the E14C mutant does not exhibit detectable transport activity (Yerushalmi et al., 1996; Yerushalmi and Schuldiner, 2000). In order to study the E14D and E14C mutant transporters further, we generated histidine epitope-tagged constructs. While E14D EmrE–His retains its ability specifically to bind TPP+, the E14C mutant was unable to bind TPP+ (Figure 2B). These results are consistent with the idea that a negative charge in a putative binding pocket is essential in order to bind substrate. The apparent importance of a charged residue at position 14 led us to investigate the effect of pH on substrate binding to the mutated transporters. Interestingly, substrate binding by E14D EmrE–His appeared to be affected only modestly by changes in pH (Figure 3A). Similar to the wild-type protein, the KD was 35 nM and the total number of binding sites was 0.3 per mole of EmrE. However, the pH dependence of binding of the E14D mutant was strikingly different: the binding of TPP+ at pH 6.2 was almost 80% of that measured at pH 9.0 for E14D EmrE–His. In contrast, wild-type EmrE–His exhibits only ∼10% of maximal binding at pH 6.2. E14C EmrE–His did not bind at any of the pH values tested. Aspartate lacks the methyl group present in glutamate and its charged carbonyl group would therefore almost certainly be positioned differently in a putative binding pocket compared with that of the wild-type glutamate carbonyl group. The pH-independent substrate binding of E14D EmrE–His is consistent with an alteration of the pKa of the carbonyl group at position 14 and this subsequently has a significant effect on the release of protons during the transport cycle.
Fig. 3. Effect of pH on the binding and release of TPP+ to EmrE–His wild type and the E14 replacement mutants. (A) EmrE–beads were incubated with 25 nM [3H]TPP+ for 1 h in solutions over a range of pH values. Values are graphed as a percentage of maximal TPP+ binding. Purified wild-type EmrE–His (•) and E14D EmrE–His mutant (□) or E14C EmrE–His membranes (▴) were assayed for TPP+ binding over a range of pH values. The inset graph shows TPP+ binding at pH 5 and 8. For this experiment, TPP+ bound to EmrE–His was separated from free TPP+ using size exclusion chromatography over a Sephadex G–50 column. After the TPP+ binding to EmrE–His at the desired pH, the binding reaction was run over the column and the bound [3H]TPP+ collected in scintillation vials by centrifugation and then counted. The black bars represent the data from wild-type EmrE–His and the open bars represent data from the E14D EmrE–His mutant construct. (B) EmrE–beads were incubated with 25 nM TPP+ for 30 min at 4°C. EmrE–beads bound to TPP+ at equilibrium levels were diluted 1:75 in 60 mM buffered solutions at the desired pH and incubated at 4°C for 2 h. This time was determined to be sufficient for binding reactions to reach equilibrium in experiments not shown here. Values are graphed as a percentage of maximal TPP+ binding. Wild-type EmrE–His (•) and the EmrE–His E14D mutant (□) were assayed. For (A) and (B), each point represents the average of triplicate binding reactions. The error bars represent the average deviation of the triplicate measurements.
Using a modified TPP+ binding assay that utilizes Sephadex G–50 size exclusion chromatography, we were able to demonstrate that E14D EmrE–His is unable to bind TPP+ at pH 5.0 (Figure 3A, inset).
pH dependence of TPP+ release from EmrE–His
To study further the interaction of H+, ligand and EmrE, we determined the pH dependence of the release of TPP+ from EmrE–His. TPP+ was bound at equilibrium to EmrE–His and then the release was measured over a range of pH values from 6.2 to 9.2. TPP+ release from EmrE–His was enhanced dramatically under acidic conditions and inhibited at basic pH values >8.0 (Figure 3B).
Again, in the E14D EmrE–His mutant, little effect of pH was seen on the equilibrium binding values after diluting the bound transporter into release buffer (Figure 3B). These results suggest that to allow for ligand release, re-protonation of the EmrE–His transporter occurs at the Glu14 residues. They are also consistent with a lower pK for the Asp residue at position 14.
The KD for TPP+ binding to EmrE–His varies with pH
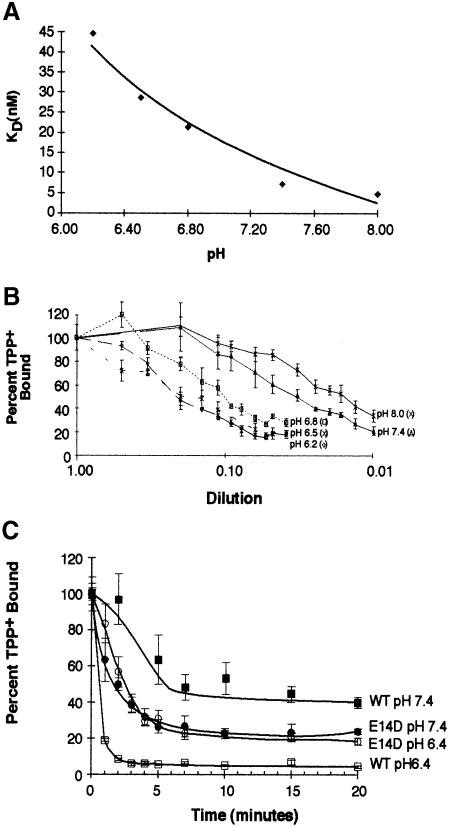
To analyze the effect of pH on the release reaction, we performed TPP+ release reactions at various dilutions and then determined the concentrations of bound and free TPP+ (Figure 4B). This information was used to generate Scatchard plots and to determine the KD of the transporter. Our data show that the KD for TPP+ increases to ∼45 nM as the pH for the release reactions decreases to pH 6.2 (Figure 4A). These results also suggest that the EmrE–His-bound TPP+ is released only after proton binding to the transporter.
Fig. 4. Effect of pH on the KD of TPP+ binding to EmrE–His and rate of release. EmrE–beads were incubated with 25 nM [3H]TPP+ for 30 min at 4°C. The EmrE–His bound to TPP+ at equilibrium was transferred into release buffer over a range of increasing dilutions and incubated at 4°C for 2 h. EmrE–His bound to TPP+ was separated from the binding solution by pelleting the beads and removing the supernatant. A fraction of the supernatant was counted to determine the concentration of free TPP+. EmrE–His was released from the beads by incubating with 150 mM imidazole buffer and the resulting supernatants were counted to measure the amount of bound TPP+ present. Using Scatchard analysis, we determined a KD for TPP+ binding at several pH values. (A) The resulting KDs were plotted against pH. (B) The percentage of bound TPP+ at each dilution. Each set of points represents the dilution-dependent release at a different pH. The values are plotted as a percentage of maximal TPP+ binding. Each point represents the average of triplicate binding reactions. The error bars represent the average deviation of the triplicate measurements. (C) EmrE–beads were incubated with 25 nM [3H]TPP+ for 30 min at 4°C. EmrE–His bound to TPP+ at equilibrium was diluted in 60 mM buffered solutions at pH 7.4 (▪, •) or pH 6.4 (□, ○) and incubated at 4°C for the times indicated. Values are plotted as a percentage of maximal TPP+ binding. Wild-type EmrE–His (squares) and the E14D EmrE–His (circles) were assayed. Each point represents the average of triplicate binding reactions. The error bars represent the average deviation of the triplicate measurements.
We examined the rate of release of bound TPP+ from EmrE–His at two pH values. Following a standard binding reaction in which TPP+ binding reached its equilibrium, we diluted the TPP+-bound EmrE–His into NH4–n–dodecyl-β–d–maltoside (DM) buffer at either pH 6.4 or 7.4. At various times, we removed the release buffer and determined the amount of TPP+ that remained bound to EmrE–His (Figure 4C). The rate of TPP+ release from EmrE–His was strikingly dependent on pH. While at pH 6.4 the bulk of the TPP is already released at the first time point, only 46% is released at pH 7.4 after 15 min. Full release at pH 7.4 is observed only after 14 h (data not shown). Again, the rate of TPP+ release was not significantly different in E14D EmrE–His under the acidic and neutral pH conditions. These results are consistent with our previous finding that, within the range tested, pH did not dramatically affect TPP+ binding to E14D EmrE–His.
Discussion
This work describes the binding of the high-affinity substrate, TPP+, to the purified, detergent-solubilized E.coli multidrug resistance transporter EmrE. The epitope-tagged EmrE–His transporter binds [3H]TPP+ with a KD of 10 nM. This value is in good agreement with the IC50 value of methyl viologen transport previously reported for TPP+ (Yerushalmi et al., 1995). Binding is inhibited by other substrates of EmrE with affinities close to their previously reported IC50 values, except for methyl viologen, which had only a modest inhibitory effect on TPP+ binding. That the binding activity is highly dependent on the protein conformation is supported by the fact that DM was the only detergent, among a dozen tested, which supports binding (data not shown). This experimental system provides a unique tool for studying the mechanism of action of the protein since it allows for analysis of binding independently from the rest of the steps in the catalytic cycle.
Previous studies that demonstrated dominant-negative inhibition by inactive mutants suggested that the transporter functions as a homooligomer, most likely a trimer (Yerushalmi et al., 1996). Our data indicate that purified EmrE–His binds between 0.25 and 0.3 mol of TPP+ per mol of transporter. These data suggest that a trimeric EmrE–His complex may form a single TPP+-binding site. A trimeric complex would comprise 12 transmembrane-spanning domains similar to larger multidrug transporters (Schuldiner et al., 1995; Gottesman et al., 1996).
The binding of TPP+ to EmrE–His is strongly pH dependent. The fact that EmrE is prevented from binding to TPP+ under acidic conditions suggests that release of one or more protons is necessary for substrate binding. Similarly, the effect of pH on the release of TPP+ from the transporter argues that protonation occurs and then allows for substrate release from its binding site. Kinetic studies were carried out on the release reaction because these allow for qualitative measurements of rates and also for determination of equilibrium values at low pH. pH affects both the rate of release and the affinity of the transporter for TPP+. Furthermore, the results suggest that both protons and ligand cannot bind simultaneously with high affinity.
The dependence on pH of both the binding and the release reactions is consistent with the presence of a residue with an apparent pK of 7.3–7.5. In EmrE there are five cationic residues and three carboxylates. None of the cationic residues are essential and they can be replaced by conservative residues with little loss of activity (Yerushalmi and Schuldiner, 2000). Among the carboxylates, two are found in hydrophilic loops, while a single negative charged residue, Glu14, is membrane embedded. The residues located in the hydrophilic loops can be replaced by Cys with practically no loss of activity, suggesting that they play no direct role in the transport cycle. Removal of both carboxylates leaves substantial residual activity, suggesting that these carboxylates have only an indirect effect on substrate recognition. In contrast, replacement of Glu14 even by Asp results in dramatic changes in the transporter activity.
The EmrE multidrug transporter specifically transports a variety of toxic substrates whose only obvious similarity is being cationic and aromatic (Schuldiner et al., 1997). It is not unreasonable to hypothesize from this fact that the substrate-binding site may be negatively charged. Here we show that replacement of Glu14 by an uncharged residue abolishes binding activity, whereas replacement by a negatively charged Asp creates a mutant protein that retains its ability to bind substrate. TPP+ binding to the E14D protein, however, displays a dramatically different pH profile, suggesting an apparent pK of <6. We suggest that this is a reflection of the different pK of the Glu14 carboxylate compared with that of the Asp14 carboxylate, most probably due to the different environments in which they reside. The high apparent pK displayed by the wild-type protein suggests that Glu14 is in a hydrophobic environment and/or interacting with other residues in the protein. On the other hand, the Asp14 residue has a pK closer to that of a free carboxylate.
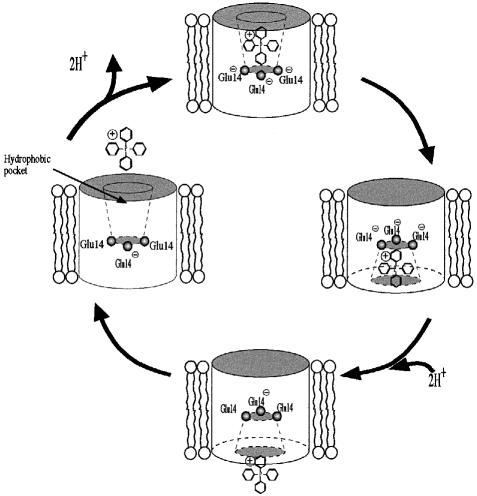
Figure 5 depicts our view of the alternative access model for EmrE. In this model, we postulate that TPP+ is bound in a hydrophobic pocket via an interaction with Glu14 (Steiner Mordoch et al., 1999). We assume that the Glu14 residue in each monomer participates in the binding, forming a charged cluster sharing one negative charge while the two others are neutralized by protons. The permanent negative charge in the binding site would also serve to enhance the interaction with a positively charged substrate. The proximity of the substrate to the cluster would influence it in such a way that it may induce release of the protons. At this stage, the binding site becomes modified so that it is accessible to the other face of the membrane. The interaction of the delocalized charge in the substrate with the three negative charges in the protein is likely to be a strong one in the hydrophobic environment present at the putative binding site. Such a stable complex can be dissociated efficiently when proton binding to the cluster occurs. The binding site relaxes back to the other face of the membrane so that a new cycle can start.
Fig. 5. Proposed mechanism of transport by EmrE. This schematic depicts a possible catalytic cycle for TPP+/H+ binding and release from the EmrE transporter. As the substrate approaches the hydrophobic binding pocket, two protons are released from the negatively charged glutamate triplet. The positively charged substrate is bound through electrostatic interactions with the negatively charged carboxylate groups extending from each glutamate residue. Following an unknown conformational transition, the opening to the binding pockets becomes accessible to the alternative face of the membrane, while being closed off from the opposite face. The subsequent movement of two protons towards the binding pocket catalyzes the release of the bound substrate. The transporter then relaxes, undergoing a conformational transition that converts the binding pocket accessibility back to the original membrane face.
The central role of acidic residues in membrane domains for H+ translocation has been postulated in a mechanism proposed for the lac permease. According to this model, substrate binding depends on the protonation state of Glu325, a carboxylic acid with an unusually high pK. Binding induces a series of conformational changes that bring about release of protons from Glu325. In the lac permease, substrate exchange can occur without H+ release because sugar is released prior to the H+. In EmrE, H+ binding is necessary for allowing release of the substrate. Unlike the case for the lac permease, we suggest that Glu14 is an essential part of the binding domain shared by substrates and protons. The binding domain cannot be occupied at the same time by both entities. This fact provides the molecular basis for the obligatory exchange catalyzed by EmrE.
Materials and methods
Plasmid construction
The plasmid encoding the EmrE–His transporter was constructed using standard methods of molecular cloning into the pT7–7 vector (Tabor and Richardson, 1985), which had been linearized with SmaI and NdeI. The protein sequence encoded by this EmrE-Myc/His plasmid is shown in Figure 1. The E14D and E14C mutant constructs were modified with the addition of the c-Myc and His tags similarly to that described above.
Purification of EmrE–His from E.coli
The wild-type and E14D histidine-tagged proteins were purified to near homogeneity using Ni–chelate chromatography as will be described elsewhere. Essentially, cells were grown and membranes were prepared as in Yerushalmi et al. (1995). The membranes were suspended in 15 mM Tris–HCl pH 7.5, 0.19 M NH4Cl and solubilized with 1% DM (Anatrace). The detergent-solubilized extract was purified using Ni–NTA beads (Qiagen). Following affinity purification, the protein was run in an Akta Purifier (Amersham Pharmacia Biotech) over a size exclusion column (Superdex 200) with the NH4–DM buffer (15 mM Tris–HCl pH 7.5, 0.15 M NaCl and 0.08% DM). The protein concentration was determined using the method described in Peterson (1977) and absorbance at 280 nm.
TPP+ binding assay
Ni–NTA beads were washed twice in distilled H2O, once in NH4–DM and suspended in at least 5 vols of NH4–DM. Purified EmrE–His protein was added (2.0 μg of purified protein per 100 μl of Ni–NTA beads) and incubated at 4°C for 40 min. The unbound material was discarded; the EmrE–His bound to beads (EmrE–beads) was washed and resuspended in the initial volume with NH4–DM. EmrE–beads (20 μl) were incubated at 4°C in a 200 μl suspension of NH4–DM containing 25 nM [3H]TPP+ (14.5 Ci/mmol) (Amersham Life Sciences). For negative control reactions, unlabeled TPP+ (25 μM) was added. The binding reactions were stopped by separating the beads from the supernatant by pulse centrifugation and then removing the supernatant. The bead fraction was then incubated for 10 min at room temperature with 450 μl of NH4–DM containing 150 mM imidazole in order to release the EmrE–His and [3H]TPP+ from the beads. After allowing the beads to settle out, the [3H]TPP+-associated radioactivity was measured by liquid scintillation. All binding reactions were performed in triplicate. TPP+ binding at pH values <6.2 was measured by size exclusion chromatography using Sephadex G–50 columns essentially as in Rudnick et al. (1990).
In experiments in which inhibition by other EmrE substrates was tested, [3H]TPP+ was present at 50 nM and the amount of bound Ni–NTA beads used was 100 μl.
TPP+ release assay
EmrE–His was bound to Ni–NTA beads as described above. The bound beads were then mixed in bulk with NH4–DM containing 25 nM [3H]TPP+ for 25 min at 4°C. A 20 μl aliquot of EmrE–beads was then transferred to tubes, 1.5 ml of NH4–DM buffer was added and the samples were allowed to incubate at 4°C for various times. Release reactions were stopped by separating beads from the supernatant fraction with a pulse spin and then removing the supernatant. EmrE–His and TPP+ were then released into the supernatant by incubating with imidazole buffer as described above. In the case of TPP+ release experiments performed at varying dilutions, an identical procedure was followed except that the release was from 20 μl of bound EmrE–beads into differing volumes of NH4–DM for 2 h at 4°C. In the case of release into buffers of varying pH, we prepared release solutions (140 mM KCl, 10 mM tricine, 5 mM MgCl2, 0.08% DM) buffered with 60 mM 2–(N–morpholino)ethane sulfonic acid (MES) (pH range 6.2–6.8), 60 mM MOPS (pH range 6.8–7.2) or 60 mM Tris–HCl (pH range 7.2–9.5) and adjusted to the final desired pH.
Acknowledgments
Acknowledgements
We thank H.Yerushalmi and S.Walter for providing the wild-type, E14C and E14D EmrE–His vectors, and H.Yerushalmi for her help in purifying the transporters. Dr M.Lebendiker provided invaluable assistance in the purification of the EmrE–His transporters. We also thank S.Steiner-Mordoch for her helpful suggestions and technical assistance, and R.Yelin for his help in preparing the manuscript. This work was supported by the Valazzi-Pikovsky Fellowship Fund (T.R.M.) and a grant from the Deutsche–Israeli Program (BMBF International Bureau at the DLR) and National Institutes of Health Grant NS16708NIH.
References
- Arkin I., Russ, W., Lebendiker, M. and Schuldiner, S. (1996) Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four helix bundle. Biochemistry, 35, 7233–7238. [DOI] [PubMed] [Google Scholar]
- Barker E.L., Moore, K.R., Rakhshan, F. and Blakely, R.D. (1999) Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J. Neurosci., 19, 4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. and Bibi, E. (1999) A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J., 18, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M.M., Pastan, I. and Ambudkar, S.V. (1996) P-glycoprotein and multidrug resistance. Curr. Opin. Genet. Dev., 6, 610–617. [DOI] [PubMed] [Google Scholar]
- Grinius L. and Goldberg, E. (1994) Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem., 269, 29998–30004. [PubMed] [Google Scholar]
- Marger M. and Saier, M. (1993) A major superfamily of transmembrane facilitators catalyzing uniport, symport and antiport. Trends Biochem. Sci., 18, 13–20. [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1994) Prevention of drug access to bacterial targets: permeability barrier and active efflux. Science, 264, 382–388. [DOI] [PubMed] [Google Scholar]
- Paulsen I.T., Skurray, R., Tam, R., Saier, M., Turner, R., Weiner, J., Goldberg, E. and Grinius, L. (1996a) The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol., 19, 1167–1175. [DOI] [PubMed] [Google Scholar]
- Paulsen I.T., Brown, M.H. and Skurray, R.A. (1996b) Proton-dependent multidrug efflux systems. Microbiol. Rev., 60, 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson G. (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem., 83, 346–356. [DOI] [PubMed] [Google Scholar]
- Rudnick G., Steiner-Mordoch, S.S., Fishkes, H., Stern-Bach, Y. and Schuldiner, S. (1990) Energetics of reserpine binding and occlusion by the chromaffin granule transporter. Biochemistry, 29, 603–608. [DOI] [PubMed] [Google Scholar]
- Sahin-Toth M., le Coutre, J., Kharabi, D., le Maire, G., Lee, J.C. and Kaback, H.R. (1999) Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli.Biochemistry, 38, 813–819. [DOI] [PubMed] [Google Scholar]
- Schuldiner S., Shirvan, A. and Linial, M. (1995) Vesicular neurotransmitter transporters: from bacteria to human. Physiol. Rev., 75, 369–392. [DOI] [PubMed] [Google Scholar]
- Schuldiner S., Lebendiker, M. and Yerushalmi, H. (1997) EmrE, the smallest ion coupled transporter, provides a unique paradigm for structure function studies. J. Exp. Biol., 200, 335–341. [DOI] [PubMed] [Google Scholar]
- Schwaiger M., Lebendiker, M., Yerushalmi, H., Coles, M., Groger, A., Schwarz, A., Schuldiner, S. and Kessler, H. (1998) NMR investigation of the multidrug transporter EmrE, an integral membrane protein. Eur. J. Biochem., 254, 610–619. [DOI] [PubMed] [Google Scholar]
- Steiner Mordoch S., Granot, D., Lebendiker, M. and Schuldiner, S. (1999) Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J. Biol. Chem., 274, 19480–19486. [DOI] [PubMed] [Google Scholar]
- Tabor S. and Richardson, C. (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA, 82, 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi H. and Schuldiner,S. (2000) An essential glutamyl residue in EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Lebendiker, M. and Schuldiner, S. (1995) EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem., 270, 6856–6863. [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Lebendiker, M. and Schuldiner, S. (1996) Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli.J. Biol. Chem., 271, 31044–31048. [DOI] [PubMed] [Google Scholar]
- Zheleznova E.E., Markham, P.N., Neyfakh, A.A. and Brennan, R.G. (1999) Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell, 96, 353–362. [DOI] [PubMed] [Google Scholar]