Abstract
Saccharomyces cerevisiae SCFMet30 ubiquitin–protein ligase controls cell cycle function and sulfur amino acid metabolism. We report here that the SCFMet30 complex mediates the transcriptional repression of the MET gene network by triggering degradation of the transcriptional activator Met4p when intracellular S–adenosylmethionine (AdoMet) increases. This AdoMet-induced Met4p degradation is dependent upon the 26S proteasome function. Unlike Met4p, the other components of the specific transcriptional activation complexes that are assembled upstream of the MET genes do not appear to be regulated at the protein level. We provide evidence that the interaction between Met4p and the F-box protein Met30p occurs irrespective of the level of intracellular AdoMet, suggesting that the timing of Met4p degradation is not controlled by its interaction with the SCFMet30 complex. We also demonstrate that Met30p is a short-lived protein, which localizes within the nucleus. Furthermore, transcription of the MET30 gene is regulated by intracellular AdoMet levels and is dependent upon the Met4p transcription activation function. Thus Met4p appears to control its own degradation by regulating the amount of assembled SCFMet30 ubiquitin ligase.
Keywords: protein degradation/SCF ubiquitin ligase/sulfur amino acid metabolism/transcriptional repression
Introduction
In response to environmental changes, cells commonly adapt by rapidly modifying their gene expression pattern. This requires the synthesis of new transcriptional factors as well as regulation of the activity of the pre-existing ones. Various molecular mechanisms underlying such controls have been described. These include covalent modifications, such as phosphorylation/dephosphorylation, which in turn can lead the modified factor to dissociate from DNA or to shuttle between the nucleus and the cytoplasm (Gorner et al., 1998; Komeili and O'Shea, 1999; Solow et al., 1999). More recently, it has been discovered that control of several transcriptional factors can arise through their proteolysis (Pahl and Baeuerle, 1996).
In eukaryotic cells, the major non-lysosomal pathway that targets proteins for their selective degradation is the ubiquitin–proteasome pathway. The proteasome is a large multisubunit complex protease that recognizes proteins modified by the addition of ubiquitin, a 76 amino acid polypeptide, to their lysine residues. The ubiquitylation of target proteins requires at least three classes of proteins, called E1, E2 and E3. E1 catalyzes the activation of ubiquitin to produce a thioester between itself and ubiquitin, which is then transferred to the ubiquitin-conjugating enzyme E2. Finally, the E3 ubiquitin ligase facilitates the recognition of the target by E2 or directly transfers the ubiquitin to the substrate (Hochstrasser, 1996). In addition, efficient polyubiquitylation was shown recently to need a new conjugation factor called E4 (Koegl et al., 1999). The required selectivity of the ubiquitylation complex is ensured by the E3 ubiquitin ligase, which interacts with both the E2 and the substrate (Hershko et al., 1983). Different classes of E3 ligases are now known, one of the largest and most versatile classes being the family of the SCF ubiquitin ligases. SCF ligases were first identified in the yeast Saccharomyces cerevisiae (Feldman et al., 1997; Skowyra et al., 1997; Patton et al., 1998a) and were shown to exist in virtually all groups of eukaryotic organisms, from fungi to mammals (for a review, see Koepp et al., 1999). SCF complexes are comprised of at least three common subunits, Skp1p, Cdc53p/cullin and the newly identified protein Hrt1p (Rbx1p, Roc1p). They also contain a modular receptor subunit that provides the substrate specificity and which is an F-box-containing protein (Patton et al., 1998b; Seol et al., 1999; Skowyra et al., 1999). The F-box domain is a degenerate motif of ∼40 amino acids in length that allows the protein that contains it to interact specifically with Skp1p (Bai et al., 1996). SCF complexes are tightly associated with a particular E2, Cdc34p, which was demonstrated to recognize an independent binding site within Cdc53p (Patton et al., 1998a). To date, although >15 F-box proteins have been identified within the yeast genome by sequence homology, only three SCF complexes have been described and characterized in yeast, namely the SCFCdc4, SCFGrr1 and SCFMet30 complexes. Each SCF complex has been demonstrated to target specific substrates for ubiquitin-mediated degradation: SCFCdc4 targets the CDK inhibitors Sic1p and Far1p; SCFGrr1 targets the G1 cyclins Cln1/Cln2; and SCFMet30 targets the CDK inhibitor Swe1p (for a review, see Koepp et al., 1999).
The F-box protein Met30p was identified originally as a factor implicated in the transcriptional regulation of the structural genes required for sulfur amino acid biosynthesis (Thomas et al., 1995). This metabolic pathway is comprised of ∼25 genes, most of which are strictly co–regulated: in response to an increase of intracellular S–adenosylmethionine (AdoMet), their transcription is turned off. Previous studies have revealed that at least five positive trans-acting factors are required for the overall transcriptional activation of the MET gene network. These factors include two leucine zipper factors Met4p and Met28p, two zinc finger-containing factors Met31p and Met32p, as well as Cbf1p, which is also a component of the yeast kinetochore machinery (Thomas and Surdin–Kerjan, 1997). Both in vivo studies and in vitro reconstitution experiments have demonstrated that these five different factors cooperate by forming large multisubunit complexes, which assemble on the 5′ upstream regions of the MET genes. Depending on the gene, different combinations of these factors are used to form multiprotein complexes that recognize specific DNA target sequences. For example, the Cbf1–Met4–Met28 complex assembles on the TCACGTG element present upstream of the MET16 gene, while the Met4–Met28–Met31 and Met4–Met28–Met32 complexes are both capable of binding the core motif AAACTGTG present upstream of the MET3 and MET28 genes (Kuras et al., 1997; Blaiseau and Thomas, 1998). It is noteworthy that within each of these multiprotein complexes transcriptional activation is dependent upon only one activation domain found within the Met4p subunit. The MET regulatory system thus appears to exemplify how one particular transcriptional activator can be tethered to multiple target sequences by several DNA-binding factor combinations.
New insights into the regulatory mechanisms underlying the AdoMet-induced repression of the MET network were gained when Met30p was shown to associate with Skp1p and Cdc53p in an F-box-dependent manner, thus leading to the identification of the SCFMet30 complex (Patton et al., 1998a). This complex may also contain the Hrt1p protein since this factor recently was shown, along with Cdc34p, Cdc53p and Skp1p, to be required for the repression of MET25 gene expression when the intracellular level of AdoMet is high (Seol et al., 1999). Here we present evidence that Met4p is targeted for degradation by the SCFMet30 complex, and provide a novel model that links SCF-dependent proteolysis to a transcriptional feedback mechanism required to regulate the essential methionine biosynthesis pathway.
Results
SCFMet30 controls the overall sulfate assimilation pathway
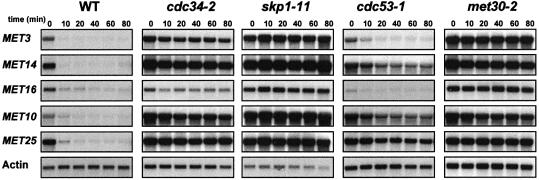
Structural genes of the MET network ultimately are regulated by AdoMet levels, although they are transcriptionally activated by different molecular mechanisms. Therefore, it seemed important to determine whether SCFMet30 triggers the repression of most or only a subset of MET genes when intracellular AdoMet increases, as originally described for the MET25 gene (Patton et al., 1998a). In all the following repression experiments, the negative regulation of the MET gene network was triggered by growing the cells in the presence of 1 mM l-methionine. As already demonstrated, repression of the MET genes actually results from the rapid conversion of methionine into AdoMet (Thomas et al., 1988; Thomas and Surdin–Kerjan, 1997). Repression kinetics of five MET genes, each specific for one step of the sulfate assimilation pathway, were monitored by Northern analysis in cdc34–2, cdc53-1, met30-2 and skp1-11 SCF mutant strains. Cells were grown in non-repressive growth conditions (0.2 mM dl-homocysteine) for eight generations at 28°C, shifted to 37°C for 2 h, transferred to a medium containing a repressive concentration of methionine (1 mM l-methionine), and RNA samples extracted at the time intervals indicated. As shown in Figure 1, in all but two instances the repression of the MET genes is impeded in the mutant cells, thus confirming the involvement of the SCFMet30 complex in the overall regulation of the MET network. The only two exceptions were observed in cells carrying the cdc53-1 mutation, the presence of which does not affect the regulation of the MET3 and MET16 genes, perhaps suggesting that some mechanistic variations might underlie the SCFMet30 operation.
Fig. 1. Regulation of the sulfate assimilation pathway in SCFMet30 mutants. The strains were grown for eight generations in B medium with 0.2 mM dl-homocysteine as sulfur source, shifted to 37°C for 2 h and a repressing amount of l-methionine (1 mM) was then added. Total RNA was extracted at the indicated times after l-methionine addition, and expression of MET genes was determined by Northern blot analysis. The actin probe was used as a control of the amount of RNA loaded.
Met4p destabilization induced by the increase of intracellular AdoMet
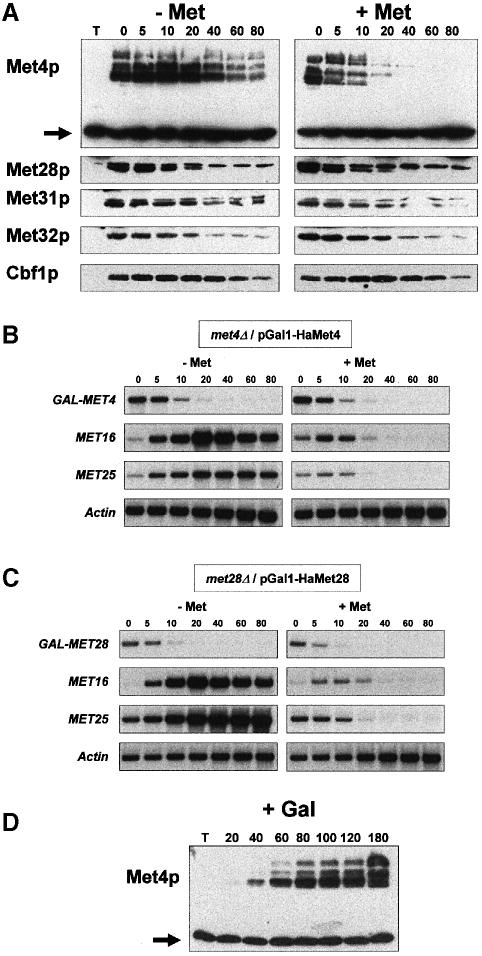
The requirement for SCFMet30 in MET gene regulation suggested that proteolysis might be central to the control of the MET pathway. To understand how the SCFMet30 complex triggers the repression of the MET genes when intracellular AdoMet increases, we attempted to determine whether one or several subunits of the complexes tethering Met4p to DNA are destabilized specifically upon the increase in intracellular AdoMet. We chose to analyze the stability of Met4p and its associated factors by promoter shut-off experiments, 35S pulse–chase experiments being clearly incompatible with studies of the sulfate assimilation pathway. Each factor was thus expressed as a hemagglutinin (HA)-tagged derivative under the control of the GAL1 promoter in cells carrying a deletion mutation inactivating the corresponding chromosomal gene. This allowed us to verify that each tagged derivative was functional, being able to complement the methionine auxotrophy phenotype associated with the deletion mutation as well as to monitor by Northern blot analyses the transcription level of both the fusion genes and the structural MET genes (see below). Expression of the tagged derivatives was induced by growing the cells for 90 min in a non-repressive, galactose-containing medium. Cultures were then divided, and transferred to fresh glucose medium with or without 1 mM l-methionine, and protein samples were extracted at regular intervals and analyzed by immunoblotting with anti-HA antibodies. As shown in Figure 2A, in non-repressive growth conditions the half-lives of the different factors (Cbf1p, Met4p, Met28p, Met31p and Met32p) are equivalent, ranging from ∼20 to 40 min. In striking contrast, repressive concentrations of extracellular methionine, while not affecting the stability of Cbf1p, Met28p, Met31p and Met32p, severely decreased the stability of Met4p, whose half-life was <10 min in these growth conditions. Thus high extracellular methionine levels, which result in an increase in intracellular AdoMet, specifically induce the rapid proteolysis of Met4p. Total RNAs corresponding to the above-described experiments were analyzed by Northern blotting. These experiments demonstrated first that the kinetics of the glucose-induced repression of the HA-Met4 mRNA expressed from the GAL1 promoter were the same in the presence or absence of high extracellular methionine levels (Figure 2B). Secondly, they revealed that the tagged proteins were not overexpressed: after a 90 min galactose induction, Met4p as well as Met28p levels were just high enough for activating the transcription of two of their target genes, MET16 and MET25 (Figure 2B and C, left panels). Further, the promoter shut-off experiments appeared to be physiologically relevant, the AdoMet-mediated repression of the structural MET genes being measured when either MET4 or MET28 was expressed from the GAL1 promoter (Figure 2B and C, right panels).
Fig. 2. Met4p is destabilized specifically in repressive growth conditions. (A) Cells carrying a plasmid coding for the indicated HA–tagged proteins under the regulation of the GAL1 promoter were grown in raffinose and expression was induced by resuspending the cells in fresh galactose-containing medium (2% galactose) for 90 min. Cultures were then divided in two, filtered, transferred to glucose-containing medium in the presence or absence of 1 mM l–methionine, and samples taken at the times indicated and immunoblotted with anti-HA antibodies. The non-specific band revealed by anti-HA antibodies (indicated by an arrow in the case of Met4p stability determination) was used as a control of the amount of loaded extracts in each experiment (T = extract of cells expressing no HA-tagged protein). (B and C) Total RNA was extracted from cells expressing either the HA-Met4 or the HA-Met28 fusion proteins grown as in (A) and analyzed with MET4, MET16, MET25 and MET28 probes. (D) Induction of Met4p modification was followed by first growing the cells in raffinose-containing medium. Cells were then transferred to galactose-containing medium (2% galactose) to induce GAL1–HA-MET4 expression, and samples were taken at the times indicated.
We also observed that Met4p is revealed by Western blot analyses as multiple bands of low electrophoretic mobility in both non-repressive and repressive growth conditions. The calculated molecular weight of the faster migrating HA-Met4 form is ∼95 kDa, while those of the upper bands are ∼120 and 140 kDa, respectively. It is noteworthy that these bands have an apparent molecular weight higher than the predicted molecular weight of the HA-Met4 derivative, which is ∼81 kDa. Therefore, we concluded that the multiple bands correspond to Met4p modifications and are not the result of degradation events. In addition, we followed by immunoblotting the appearance of the different forms of the tagged Met4 derivative after its induction from the GAL1 promoter. Modifications of Met4p are visible as soon as the protein is synthesized, arguing that these modifications are constitutive (Figure 2D).
Met4p degradation is triggered by the SCFMet30 complex
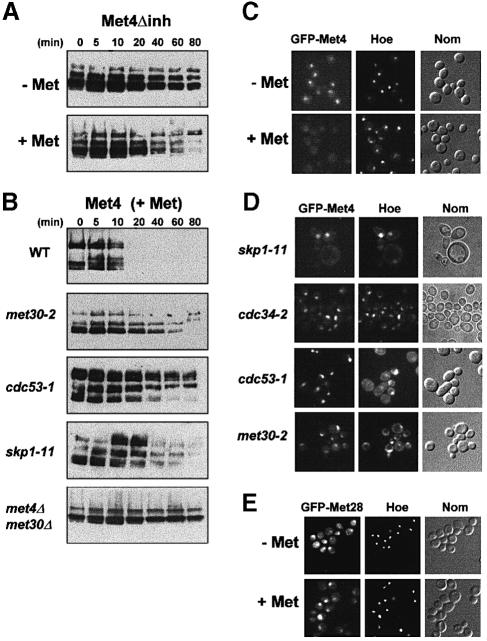
The rapid degradation of Met4p observed in cells grown in the presence of high levels of methionine strongly suggested that Met4p might be the target of the SCFMet30 complex. To substantiate this possibility further, we first examined the stability of a Met4p derivative (Met4Δinh), which is unable to respond to AdoMet levels and is defective in binding Met30p (Kuras and Thomas, 1995; Thomas et al., 1995). Promoter shut-off experiments (Figure 3A) demonstrated that an HA-tagged Met4Δinh derivative is degraded less rapidly than the wild-type HA–Met4 protein when the cells are grown in repressive growth conditions (compare Figures 3A and 2A). We then monitored the stability of the wild-type Met4p in the presence of mutations impairing SCFMet30 function by expressing a tagged Met4p from the GAL1 promoter in skp1-11, cdc53-1 and met30-2 mutant cells. The cells were grown in non-repressive raffinose medium, filtered and transferred in galactose-based medium to induce GAL1–HA-MET4 expression. They were shifted to non-permissive temperature 30 min after, and the growth in galactose was continued for another 60 min. Then the cells were transferred to fresh glucose medium containing 1 mM l–methionine and samples were taken at regular intervals. As shown in Figure 3B, in the presence of methionine, Met4p appears to be stabilized in the skp1-11, cdc53-1 and met30-2 mutant cells as compared with wild-type cells grown in the same conditions. We also took advantage of the fact that a met4 null mutation suppresses the lethality associated with a met30 disruption mutation (E.E.Patton, C.Peyraud, A.Rouillon, Y.Surdin-Kerjan, M.Tyers and D.Thomas, manuscript in preparation) and found that in the absence of Met30p and under repressive conditions, Met4p stability was dramatically increased. Furthermore, Met4p appears to be less modified in the absence of Met30p than in its presence, suggesting that the SCFMet30 complex might be required for some of the modifications observed.
Fig. 3. Met4p degradation depends on the SCFMet30 complex. (A) The stability of HA-Met4Δinh expressed in wild-type cells grown in repressive and non-repressive conditions was analyzed as described in Figure 2A. (B) The stability of HA-Met4p was studied in SCFMet30 mutants: wild-type and mutant cells were grown in raffinose at 28°C for eight generations, filtered and resuspended in galactose medium for 30 min at 28°C. Cells were then shifted to 37°C and incubation in galactose medium was continued for another 60 min. Cells were then transferred to fresh glucose medium containing 1 mM l-methionine and proteins were extracted at the indicated times and processed for immunoblotting with anti-HA antibodies. Equal loading was determined by the presence of the non-specific band revealed by the anti-HA antibodies (not shown). (C and D) GFP–Met4 localization was monitored in wild-type cells and in SCFMet30 mutants. Cells expressing the GFP–Met4 fusion protein from the GAL1 promoter were grown in raffinose at 28°C, filtered and resuspended in galactose medium for 60 min at 28°C. Cells were then shifted to 37°C and incubation in galactose medium was continued for another 60 min. In repressive growth conditions (+Met), 1 mM l-methionine was added 15 min before observation. Nuclei were visualized by adding the dye Hoescht 33342 (Hoe) to the culture 20 min before observation (Nom = Nomarski interferential observation). (E) Localization of the GFP–Met28 derivative was monitored in cells expressing the fusion protein from the MET28 promoter and grown in the presence (+Met) or absence (–Met) of a repressing amount of l-methionine (1 mM).
Cellular localization of a green fluorescent protein (GFP)–Met4 fusion protein in living cells corroborated the above results. A GFP–MET4 fusion gene placed downstream of a GAL1 promoter region was integrated into the chromosome of wild-type and SCF mutant strains at the URA3 locus. In a wild-type background, GFP–Met4 fusion protein localizes within the nucleus when the cells are grown in non-repressive growth conditions, but is not observed when 1 mM l-methionine is added to the medium (Figure 3C; immunoblotting confirmed that this corresponds to a rapid degradation of the GFP–Met4 fusion protein, not shown). In contrast, when expressed in SCFMet30 mutant cells, the GFP–Met4 fusion protein remained visible in the nucleus after 1 mM l-methionine was added to the medium (Figure 3D). As a control, we monitored the localization of GFP–Met28 fusion protein, which is visualized within the nucleus of cells grown in the absence or presence of methionine (Figure 3E). These results are consistent with the Met4p degradation data presented above, and together provide strong evidence that the SCFMet30 complex is responsible for the degradation of Met4p in response to increased levels of intracellular AdoMet.
Degradation of Met4p involves the proteasome
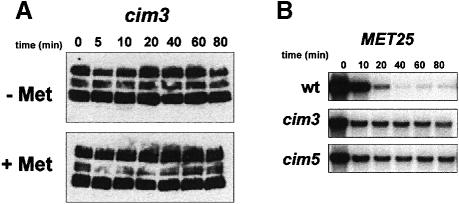
While SCFMet30-dependent degradation of Met4p strongly suggested that Met4p was targeted for ubiquitin-mediated degradation, we wanted to test directly whether Met4p was modified by ubiquitin in vivo. However, as with other short-lived proteins, ubiquitin conjugates proved difficult to detect in vivo (data not shown). We therefore tested if Met4p stability was dependent on the 26S proteasome, by using a cim3 mutation that inhibits proteasome function (Ghislain et al., 1993). As shown in Figure 4A, the HA–Met4 derivative is dramatically stabilized in cim3 cells grown in the presence of 1 mM l-methionine, as compared with wild-type cells.
Fig. 4. Met4p is stabilized in the presence of a cim3 mutation. (A) The stability of HA-Met4p expressed in cim3-1 mutant cells grown in repressive and non-repressive conditions was analyzed as described in Figure 3B. (B) Repression of the MET25 gene was monitored in cim3-1 and cim5-1 mutant cells. Cells were grown and RNA extracted as described in Figure 1. The wild-type (wt) strain used in this assay was the YPH499 strain, which was congenic to both mutant strains.
To confirm this result, Northern analyses were carried out with both cim3 and cim5 mutant cells and compared with their congenic parent strains. Total RNA was extracted after the cells were grown at restrictive temperature for 2 h and then shifted from a repressive growth condition to a non-repressive growth condition. Repression kinetics of the MET25 gene were assayed in the different strains. As shown in Figure 4B, repression of the MET25 gene indeed appears to be impaired by the presence of either the cim3 or cim5 mutation (Figure 4B), consistent with the idea that degradation of Met4p occurs via the 26S proteasome.
Met4p–Met30p interactions are constitutive
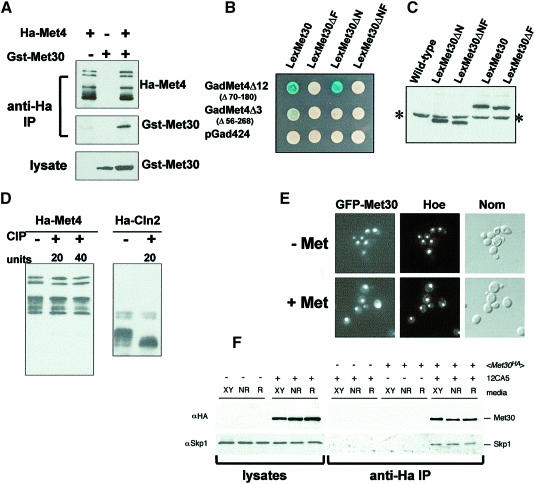
Most of the biochemical data acquired to date on the SCF complexes have demonstrated that F-box proteins interact with their substrate in a phosphorylation-dependent manner (Feldman et al., 1997; Skowyra et al., 1997; Winston et al., 1999). Substrate degradation thus appears to be controlled by its interaction with its cognate SCF complex. Since in the present case the degradation of the substrate is induced specifically by adding large amounts of methionine to the medium, we first addressed this point by determining whether an interaction between Met4p and Met30p could be measured in cells growing in the absence of methionine. HA-Met4 and GST–Met30 derivatives, expressed from the GAL1 promoter, were expressed in cells for 2 h in the absence of methionine. Immunoprecipitation of HA-Met4, followed by immunoblotting with anti-GST antibodies, revealed a strong interaction between Met4p and Met30p in cells growing in the absence of methionine (Figure 5A). These results were corroborated by two-hybrid analysis. As reported previously, a LexA–Met30 fusion protein comprising the entire Met30p interacts with a Gal4–Met4 fusion protein providing that the latter contains the inhibitory domain (Figure 5B). Moreover, the Gal4–Met4–LexA–Met30 interaction appears to be stronger when the LexA derivative protein lacks the first 158 N–terminal residues of Met30p. In all cases, the interactions are seen on plates containing ammonium sulfate as the sole sulfur source, a growth condition in which Met4p activity is required. This confirms the results of the co-immunoprecipitation assays and shows that Met30p can bind Met4p when the intracellular concentration of AdoMet is low. Unexpectedly, we found that LexA–Met30 derivatives lacking the F-box (amino acids 187–202) failed to interact with a Gal4–Met4 fusion protein. Western control assays showed that all the LexA–Met30 derivatives were expressed at equivalent levels (Figure 5C). Together, these results suggest that other SCFMet30 interactions might be critical for Met4p binding.
Fig. 5. Met4p–Met30p interactions. (A) Co-immunoprecipitation analysis of the Met4p–Met30p interaction. HA-Met4p and GST–Met30p were expressed, alone or together, from the GAL1 promoter by growing the cells for 2 h in the presence of 2% galactose. In non-repressive growth conditions, anti-HA immunoprecipitates were probed with either anti-HA or anti-GST antibodies, while anti-GST antibodies revealed the presence of the GST–Met30 fusion protein in the lysates. (B) Two-hybrid analysis of the interaction established between Gal4–Met4 and LexA–Met30 fusion proteins. β-galactosidase activities were revealed on X-gal plates containing ammonium sulfate as the sulfur source. (C) Western assays of LexA–Met30 derivatives. Proteins were extracted from cells expressing the various LexA–Met30 derivatives and processed for immunoblotting with anti-LexA antibodies. The asterisk indicates the non-specific band revealed by the anti-LexA antibodies. (D) Met4p is not modified by incubation in the presence of alkaline phosphatase. Immunoprecipitates of HA-Met4 were incubated in the presence of 20 or 40 U of alkaline phosphatase (CIP) at 37°C for 15 min. Incubations were loaded onto a gel and probed with anti-HA antibodies. As a control, an immunoprecipitate of an HA-Cln2 fusion protein expressed from the GAL1 promoter was incubated in the presence of 20 U of CIP at 37°C for 15 min. (E) GFP–Met30 localization. The GFP–Met30 fusion protein was expressed from the GAL1 promoter and its localization in living cells was monitored as in Figure 3C in the presence or absence of 1 mM l-methionine. (F) Skp1p–Met30p interaction is not affected by methionine concentration. Cells expressing HA-Met30 protein from the ADH1 promoter were grown in rich (XY), non-repressive (NR) or repressive (R) media conditions. Western blots of lysates and anti-HA immunoprecipitates were probed with anti-HA antibodies and anti-Skp1 antibodies. ‘+’ and ‘–’ denote the presence or absence of the protein or antibody.
In a next step, we tried to determine whether the multiple bands of Met4p might correspond to phosphorylation events. Immunoprecipitations of HA-Met4 were carried out with cells growing in the absence of methionine or with cells grown in the presence of 1 mM l-methionine for 5 min. Immunoprecipitates were incubated for 15 min in the presence of alkaline phosphatase and analyzed by immunoblotting. With both repressive and non-repressive concentrations of methionine, most, if not all, of the Met4p forms were resistant to the phosphatase treatment (Figure 5D shows the results obtained with cells growing in a low methionine concentration; the same results were obtained with cells grown in the presence of high extracellular methionine concentrations for 5 min; data not shown). As a control, alkaline phosphatase was incubated with an immunoprecipitate of an HA-tagged Cln2p that was expressed from the GAL1 promoter. As expected, the majority of slow migrating forms of Cln2p were converted to fast migrating forms, as previously reported (Lanker et al., 1996). These experiments strongly suggest that the majority of Met4p modifications that are visible by gel electrophoresis do not correspond to phosphorylation events.
To verify the above results, we next looked at the cellular localization of a GFP–Met30 fusion protein expressed transiently from the GAL1 promoter. Microscopic observations showed that the GFP–Met30 protein is located within the nucleus of cells grown in the absence as well as in the presence of a high extracellular methionine concentration (Figure 5E). Taken together, all these results are consistent with the possibility that, in contrast to what was observed for the substrates of the SCFCdc4 and SCFGrr1 complexes for instance, the timing of Met4p degradation is not controlled at the level of its interaction with the SCFMet30 complex.
Finally, we tested the possibility that the Skp1p–Met30p interaction might be regulated by the presence or absence of high extracellular methionine concentrations, in a way similar to the Skp1p–Grr1p interaction, which has been reported to be stimulated by the presence of glucose in the growth medium (Li and Johnston, 1997). Immunoprecipitation of HA-Met30 followed by immunoblotting with polyclonal antibodies directed against Skp1p revealed that the specific Skp1p–Met30p interaction is neither increased nor decreased by the presence of high extracellular methionine concentrations (Figure 5F).
Met30p is itself a short-lived protein
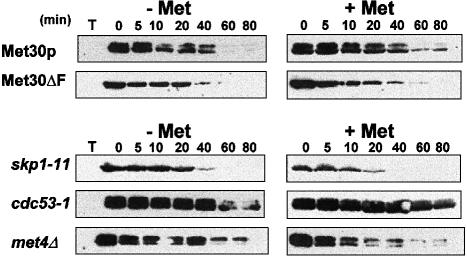
Recent studies have revealed that the F-box subunits of SCF complexes are themselves unstable proteins, and that control of such an instability might provide a general mechanism to regulate SCF function (Zhou and Howley, 1998; Mathias et al., 1999). We thus attempted to determine whether Met30p itself might constitute a target for the regulatory mechanisms that allow the cells to respond to variations in sulfur availability. Met30p stability was examined in repressive and non-repressive conditions by expressing the HA-Met30 derivative under the control of a GAL1 promoter. After the culture was divided and transferred to fresh glucose medium with or without 1 mM l-methionine, protein samples were extracted at regular intervals and analyzed by immunoblot assays. These experiments demonstrated that Met30p is indeed a short-lived protein (half-life <20 min, Figure 6), similar to the other F-box proteins, as demonstrated for Cdc4p and Grr1p (Zhou and Howley, 1998; Mathias et al., 1999). However, in contrast to Met4p, the half-life of the HA–Met30 derivative remained the same whatever the growth medium was, suggesting that AdoMet levels do not regulate Met30p stability.
Fig. 6. Characterization of Met30p stability. The HA-tagged full-length Met30 and Met30ΔF proteins were expressed from a GAL1 promoter by growing the cells for 2 h in 2% galactose. Glucose (2%) and cycloheximide (50 μg/ml) were added and the cells were allowed to grow for the indicated times in the presence or absence of 1 mM l–methionine. Upper panel: the stability of both HA-Met30 and HA–Met30ΔF was monitored in W303-1A wild-type cells. Lower panel: the stability of HA-Met30 was monitored in the SCF mutants skp1-11 and cdc53-1 by shifting the cells to 37°C before the addition of galactose as well as in met4Δ mutant cells.
We noticed that Met30p was revealed as a doublet in these experiments carried out with cells disrupted by a fast trichloroacetic acid (TCA)-based extraction method (see Materials and methods). These Met30p forms were not modified by alkaline phosphatase (data not shown). The instability of Cdc4p was postulated to result from its interaction with Skp1p as Cdc4p destabilization was shown to be dependent on its F-box and as the full-length Cdc4p was stabilized in skp1-11 mutant strains (Zhou and Howley, 1998). Our promoter shut-off experiments revealed that it is not the case for Met30p. Indeed, expressed in wild-type cells, an HA-tagged Met30ΔF derivative is slightly less stable than the wild-type HA-Met30 fusion protein. Likewise, in the presence of the skp1-11 mutation, the full-length HA-Met30 is slightly less stable than in wild-type cells, especially when the cells are grown in the presence of methionine (Figure 6). It is important, moreover, to note that in both cases the HA-Met30 derivative is revealed as a single band. This strongly suggests that Met30p should interact with Skp1p in order to be modified. Together with the results of the two-hybrid assays, this also suggests that these modifications might be involved in the recognition of Met4p. Finally, the stability of Met30p was monitored in the presence of a cdc53-1 mutation. In contrast to what was observed with the skp1–11 mutation, the HA-Met30 derivative was stabilized in the presence of the cdc53-1 mutation, suggesting that Met30p degradation might indeed involve the ubiquitin-dependent pathway.
Met30p synthesis is controlled by Met4p
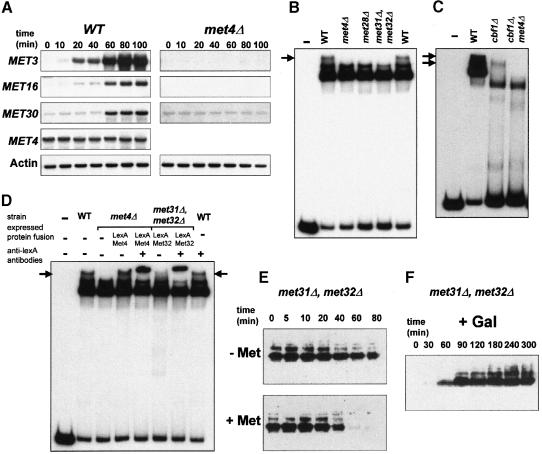
To understand further how Met4p stability is regulated in response to modifications of intracellular AdoMet levels, we next addressed whether MET30 gene expression might be regulated. Such a possibility was indeed suggested by the presence of both a TCACGTG motif and an AAACTGTG-related sequence (at positions –177 bp and –166 bp, respectively, numbered from the start codon) within the 5′ upstream region of the MET30 gene that therefore displays an organization resembling that of several structural genes from the sulfate assimilation pathway. To test formally if the MET30 gene is under the control of AdoMet and Met4p, we monitored MET30 gene expression by Northern assays with RNA extracted from wild-type cells after they were shifted from repressive to non-repressive growth conditions. As shown in Figure 7A, the transcription of the MET30 gene appears to be activated in the absence of methionine. Northern analyses performed with met4Δ cells showed that this activation of MET30 transcription requires Met4p (Figure 7A) as well as Met31p and Met32p (data not shown). It must be noted, however, that in the presence of a large amount of methionine or in the absence of Met4p, a low but reproducibly detectable level of MET30 transcription is measured. One might imagine that such basal (i.e. Met4p independent) transcription is required to ensure Met30p function in cellular mechanisms other than sulfur metabolism, such as Swe1p degradation (Kaiser et al., 1998).
Fig. 7. MET30 transcription is regulated by intracellular AdoMet and Met4p. (A) Derepression kinetics of MET3, MET4, MET16 and MET30 transcription were monitored in wild-type and met4Δ cells. Cells were grown in 100 ml of B medium in the presence of a repressing amount (1 mM) of l-methionine as sulfur source. When the cells reached a density of ∼107 cells/ml, they were harvested by filtration and washed with 100 ml of B medium. The cells were then suspended in 100 ml of B medium without methionine and shaken at 28°C. Total RNA was extracted at the times indicated. The actin probe was used as a control of the amounts of RNA loaded. (B) Gel retardation assays were performed with a MET30 radiolabeled probe corresponding to nucleotides –255 to –61. The binding reactions were performed with 40 μg extracts from strains W303-1A (wild–type), CD106 (met4::TRP1), CD130-7D (met28::LEU2) and CD179 (met31::LEU2, met32::TRP1). (C) The binding reactions were performed with 40 μg extracts from strains W303-1A (wild-type) and 60 μg extracts from strains CC857-2A (cbf1::TRP1) and CC653-3C (met4::TRP1, cbf1::TRP1). (D) The binding reactions were performed with extracts from strains W303-1A (wild-type), CD106 (met4::TRP1) and CD179 (met31::LEU2, met32::TRP1), and from strains CD106 and CD179 expressing, respectively, a LexA–Met4 or a LexA–Met32 fusion protein. All the reactions were carried out in the presence of 40 μg of proteins. Where indicated, 1 μl of a polyclonal antibody raised against LexA used at a 1/10 dilution was added. Arrows indicate the Met4-containing high molecular weight complexes. (E) The stability of HA-Met4 was assayed in the met31Δ, met32Δ mutant cells grown in the absence (–Met) or presence (+ Met) of 1 mM l-methionine. Experiments were carried out as described in Figure 2A. (F) Induction of Met4p in met31Δ, met32Δ mutant cells was followed by first growing the cells in raffinose-containing medium. Cells were then transferred to galactose-containing medium (2% galactose) to induce GAL1–HA-MET4 expression and samples were taken at the times indicated.
Next, mobility shift assays were performed with various cellular extracts and a MET30 probe that corresponds to nucleotides –255 to –61 and therefore contains the two sequence motifs TCACGTG and AAACTGTG. Mobility shift assays performed with wild-type cell extracts show three prominent complexes of low mobility (Figure 7B). Strikingly, the complex of lower mobility is not formed with extracts of met4Δ, met28Δ or the double met31Δ, met32Δ mutant cells. It thus seemed possible that, as already shown for the MET3 and MET28 genes (Blaiseau and Thomas, 1998), a multisubunit complex involving Met4p, Met28p and Met31p/Met32p could be assembled on the 5′ upstream region of the MET30 gene. To confirm this hypothesis, we next performed a mobility shift assay with extracts of cbf1Δ or cbf1Δ, met4Δ double mutant cells. As shown in Figure 7C, disruption of cbf1 results in the absence of the massive complex of higher electrophoretic mobility, suggesting that it does correspond to the binding of Cbf1p alone to the MET30 probe. In addition, a complex of low mobility is seen specifically in cbf1Δ cells but not in cbf1Δ, met4Δ cells, indicating that it contains Met4p. To demonstrate further that Met4p is indeed tethered to the 5′ upstream region of the MET30 gene, met4Δ and met31Δ, met32Δ cells were transformed using a plasmid expressing a LexA–Met4 and a LexA–Met32 fusion protein, respectively. Mobility shift assays were performed with extracts of the transformed cells. As shown in Figure 7D, in met4Δ cells, the expression of a LexA–Met4 derivative restores the formation of a complex of low electrophoretic mobility, which moreover is supershifted by the addition of LexA antibodies to the binding reaction, therefore proving that Met4p is a component of the complex. Similar results were obtained with the met31Δ, met32Δ cells expressing a LexA–Met32 fusion protein, proving that Met32p is also a component of the complex. Taken together, all these results demonstrate that Met4p is tethered to the 5′ upstream region of the MET30 gene through a high molecular weight complex containing Met28p, Met31p and Met32p.
To substantiate further the possibility that the control of Met4p stability was indeed a feedback-regulated mechanism, we next analyzed the stability of a tagged Met4 derivative in cells that do not express Met31p and Met32p. In such cells, the recruitment of Met4p to DNA is impeded and therefore the transcription of the MET30 gene is reduced. Immunoblotting assays demonstrate that, as compared with wild-type cells, Met4p stability is increased in met31Δ, met32Δ cells in both non-repressive and repressive growth conditions (Figure 7E, compare with Figure 2A). We thus conclude that variations of Met30p synthesis affect the regulation of Met4p stability. In addition, it must be noted that Met4p appears to be less modified in the met31Δ, met32Δ mutant cells than in wild-type cells, a result resembling that obtained with the double met4Δ, met30Δ mutant strain (Figure 3B). To confirm this point, accumulation of the tagged Met4 derivative was followed after the induction of the GAL1 promoter. Even after 5 h of galactose induction, the Met4p form of 140 kDa apparent molecular weight is not seen in the met31Δ, met32Δ cells. Together, these results reinforce the possibility that some of the modifications of Met4p require the synthesis of sufficient amounts of Met30p.
Discussion
The present report provides evidence that the SCFMet30 ubiquitin ligase complex mediates the transcriptional repression of the MET gene network by triggering the elimination of the Met4p activator when the intracellular concentration of AdoMet increases. AdoMet-induced proteolysis is a very effective mechanism, the half-life of Met4p being <10 min under repressive growth conditions. In the presence of mutations impairing the SCFMet30 complex, Met4p is stabilized, as seen by immunoblotting experiments and GFP localization studies, and the transcription of the MET genes becomes constitutive. The identification of Met4p as the substrate of the SCFMet30 complex supports previous studies that have identified Met30p as a factor regulating the Met4p activation function (Thomas et al., 1995). Met4p stabilization in a proteasome mutant strongly indicates that proteolysis of Met4p involves the ubiquitin–proteasome pathway.
The experiments reported here suggest that among the different transcription factors that control MET gene expression, Met4p constitutes the only target of the SCFMet30 complex. Unlike Met4p, the stability of either Cbf1p, Met28p, Met31p or Met32p is not altered in the presence of high extracellular methionine concentrations. This provides an immediate insight into the rules that govern the regulatory mechanisms operating in the MET gene network. Met4p is indeed the only factor endowed with transcription activation function, while Cbf1p, Met28p, Met31p and Met32p act by promoting the recruitment of Met4p to the DNA. Thus, inducing the degradation of the only transcriptionally competent subunit appears to constitute a simple and efficient mechanism to control the activity of the transcriptional activation complexes assembled upstream of the MET genes.
Contrary to what has been observed for the interactions between the other F-box proteins and their substrates, we were unable to detect any evidence that the recognition of Met4p by Met30p was regulated by the level of intracellular AdoMet. Both co-immunoprecipitation assays and two-hybrid analysis strongly argue for the fact that the Met4p–Met30p interaction is established when the intracellular AdoMet concentration is low without resulting in Met4p degradation. Moreover, phosphatase assays failed to alter the multiple Met4p isoforms, in either low or high methionine concentrations. Therefore, as SCFMet30 degradation of Met4 appears not to be controlled at the level of the Met4p–Met30p interaction, the exact mechanism of switching Met4p degradation on or off remains to be deciphered. At least two working hypotheses could be postulated. In the first model, an increase in the intracellular AdoMet concentration is needed to activate the SCFMet30 activity or assembly. SCFGrr1 complex assembly has been reported to be stimulated by glucose, a requirement for the induction of the HXT glucose transporter genes (Li and Johnston, 1997). However, in the present case, our results indicate that the interaction between Skp1p and Met30p is not modified by the presence of high extracellular methionine concentrations. As a second model, we can imagine that when the intracellular AdoMet concentration is low, the SCFMet30 complex interacts with Met4p but fails to ubiquitylate it because the ubiquitylation sites on Met4p are masked as a result of its assembly into multiprotein activation complexes. Similar protective effects of protein–protein interactions against ubiquitin-dependent degradation were reported for the mating type transcription factors a1/α2, as well as for the p58 component of the CBF3 subunit of the yeast kinetochore (Kaplan et al., 1997; Johnson et al., 1998; Russell et al., 1999). In such a model, an increase in AdoMet might lead to the dissociation or alteration of the Met4p-containing complexes, thereby allowing Met4 to be ubiquitylated and degraded. Accordingly, several protein–protein interactions needed for the assembly of the Cbf1–Met4–Met28 complex seem to be regulated in response to variations in the level of intracellular AdoMet (A.Rouillon and D.Thomas, unpublished results). The existence of another regulatory level of Met4p activity might also account for the fact that, in the presence of the cdc53-1 or cim3-1 mutations, the transcription of some of the MET genes was repressed although Met4p is stabilized.
The SCFMet30 complex displays several additional distinctive features. Like the other F-box proteins Cdc4p and Grr1p, Met30p is an unstable protein. However, unlike Cdc4p (Zhou and Howley, 1998), Met30p was not stabilized by the presence of a skp1-11 mutation or by the deletion of its F-box. Furthermore, we found by two-hybrid assay that deletion of the F-box abolishes the interaction between Met30p and its substrate Met4p. Moreover, our experiments reveal that both Met4p and Met30p undergo covalent modifications, which vary depending upon the different interactions in which each protein is engaged. Although the nature of these modifications is unknown, it is tempting to postulate that they could in some way regulate either the substrate recognition or the activity of the SCFMet30 complex. Other subunits of the SCF complexes are already known to undergo post-translational modifications, e.g. Cdc53p, which is modified by the attachment of Rub1p, a ubiquitin-like protein (Liakopoulos et al., 1998); Rub1p conjugation to Cdc53p was postulated further to be required for optimal assembly or function of the SCFCdc4 complex (Lammer et al., 1998). Likewise, in Dictyostelium discoideum, Skp1p was demonstrated to be modified by the addition of a linear pentasaccharide to a proline residue (Teng-umnuay et al., 1998).
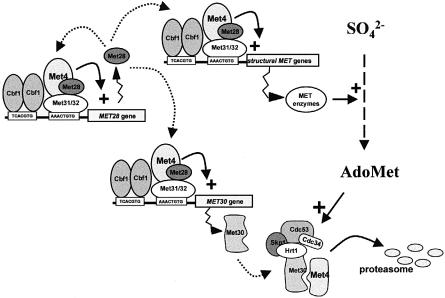
A novel important finding of this present study is that the amount of SCFMet30 formed is regulated by its own substrate. The 5′ upstream region of the MET30 gene resembles the structure of the MET genes, comprising both the TCACGTG and AAACTGTG motifs. Northern blot assays demonstrated that MET30 transcription is repressed when the intracellular level of AdoMet is high, and that MET30 transcription is dependent on Met4p, Met31p and Met32p. Mobility shift assays, moreover, allowed us to demonstrate that Met4p is recruited to the 5′ upstream region of the MET30 gene through the assembly of high molecular weight complexes identical or closely related to the Met4–Met28–Met31 and Met4–Met28–Met32 complexes, which were shown to tether Met4p upstream of the MET3 and MET28 genes (Blaiseau and Thomas, 1998). By regulating the transcription of the MET30 gene, Met4p thus appears to control its own fate directly. Accordingly, in the presence of the met31Δ, met32Δ mutations, which lower MET30 gene expression, Met4p is stabilized. This constitutes the first report of an SCF ubiquitin ligase complex whose substrate regulates its own degradation by controlling the synthesis of the F-box recognition subunit. Given its efficiency, such a regulatory mechanism is expected to be widely encountered in the SCF field and, for instance, degradation of the oncoprotein E2F1 by the SCFSkp2 complex has been postulated to be regulated on such a level (Harper and Elledge, 1999).
This feedback-regulated degradation of Met4p by the SCFMet30 complex actually constitutes the second regulatory loop shown to date within the MET gene network. Met4p previously has been demonstrated to regulate the transcription of the MET28 regulatory gene whose product stimulates Met4p tethering to DNA (Kuras et al., 1997). Here we report that another regulatory loop is established by showing that the expression of a second protein, Met30p, capable of modifying the biochemical properties of Met4p, is under the control of Met4p (see Figure 8 for a simplified scheme recapitulating these regulatory connections). These regulatory loops function in opposite ways. The Met4p–Met28p loop is expected to increase dynamically the response of the system when the intracellular AdoMet concentration is low, while the Met4p–Met30p loop is expected to control high detrimental accumulation of AdoMet. The fact that the transcription of the MET4 gene is constitutive (Kuras et al., 1997; this study) seems to allow the system to escape the repressed state.
Fig. 8. Relationships between the trans-acting factors regulating the MET network and how AdoMet-induced degradation of the Met4p activator is triggered by the SCFMet30 complex.
Finally, it must be asked why regulation of a basic metabolic pathway expected to be required throughout the yeast cell life relies on protein degradation, which appears to constitute an expensive regulatory mechanism and is therefore seen in time-dependent systems such as cell cycle control or long-term adaptations such as stress responses. The fact that the MET cluster recently was shown by micro-array analyses to be regulated in a cell cycle-dependent fashion may provide a first explanation of such a result (Spellman et al., 1998; Tavazoie et al., 1999). Moreover, unpublished results from our laboratories demonstrate another unexpected link between sulfur amino acid metabolism regulation and cell cycle progression: the Met30p-mediated regulation of Met4p stability is indeed shown to be essential for G1/S transition (E.E.Patton, C.Peyraud, A.Rouillon, Y.Surdin-Kerjan, M.Tyers and D.Thomas, manuscript in preparation).
Materials and methods
Yeast strains and media
The S.cerevisiae strains used in this work are listed in Table I. Standard yeast media and B sulfur-less medium were prepared as described by Cherest and Surdin-Kerjan (1992). Saccharomyces cerevisiae was transformed after acetate chloride treatment as described by Gietz et al. (1992). Genetic crosses, sporulation, dissection and scoring of nutritional markers were as described by Sherman et al. (1979).
Table I. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| C301 | MATα, his3, leu2, trp1, met4::TRP1, ura3::GAL1-GFPMET4::URA3 | this study |
| C302 | MATα, his3, leu2, trp1, met30-2, ura3::GAL1-GFPMET4::URA3 | this study |
| C312 | MATα, his3, leu2, trp1, ura3::GAL1-GFPMET4::URA3 | this study |
| C319 | MATα, his3, leu2, trp1, cdc34-2, ura3::GAL1-GFPMET4::URA3 | this study |
| C323 | MATα, his3, leu2, trp1, cdc53-1, ura3::GAL1-GFPMET4::URA3 | this study |
| C327 | MATα, his3, leu2, trp1, skp1-11, ura3::GAL1-GFPMET4::URA3 | this study |
| CC640-6C | MATα, his3, leu2, trp1, met30-2 | this study |
| CC653-3C | MATa, ade2, his3, leu2, trp1,ura3, met4::TRP1, cbf1::TRP1 | this study |
| CC807-1C | MATa, ade2, his3, leu2, trp1,ura3, met4::TRP1, met30::LEU2 | Patton et al. (1999) |
| CC849-8A | MATα, ade2, his3, leu2, trp1,ura3, met4::TRP1 | this study |
| CC857-2A | MATa, ade2, his3, leu2, trp1,ura3, cbf1::TRP1 | this study |
| CD130-7D | MATα, ade2, his3, leu2, trp1,ura3, met28::LEU2 | H.Chérest |
| CD168 | MATα, ade2, his3, leu2, trp1, ura3, met31::LEU2, met32::HIS3 | Blaiseau and Thomas (1998) |
| CD179 | MATα, ade2, his3, leu2, trp1,ura3, met31::LEU2 met32::TRP1 | Blaiseau and Thomas (1998) |
| CMY763 | MATα, leu2, ura3, cim3 | Ghislain et al. (1993) |
| CMY765 | MATα, leu2, ura3, cim5 | Ghislain et al. (1993) |
| MT1166 | MATa, ade2, his3, leu2, trp1, ura3, skp1-11 | Patton et al. (1998) |
| MT670 | MATa, ade2, his3, leu2, trp1, ura3, cdc34-2 | Patton et al. (1998) |
| MT871 | MATa, ade2, his3, leu2, trp1, ura3, cdc53-1 | Patton et al. (1998) |
| YPH499 | MATa, ade2, his3, leu2, lys2, trp1, ura3 | Ghislain et al. (1993) |
| W303-1A | MATa, ade2, his3, leu2, trp1, ura3 | R.Rothstein |
Plasmid constructions
Escherichia coli strain DH10B was used to propagate the plasmids. Standard procedures were used for recombinant DNA manipulation (Table II). To express HA-tagged proteins from the GAL1 promoter, we first cloned the GAL1 promoter region from the pJN1 plasmid (Nehlin and Ronne, 1990) into the pRS316 and pRS306 vectors (Sikorski and Hieter, 1989). Next a cassette encoding three copies of the HA epitope was inserted into the resulting pGal316 and pGal306 plasmids. The resulting plasmids, pGal316Flu and pGal306Flu, were then digested by appropriate restriction enzymes to clone in-frame various DNA fragments encoding the different proteins that will be expressed as HA-tagged derivatives. To express the GST–Met30 fusion in yeast cells, the Met30 open reading frame was cloned into the pEG(KG) vector (Mitchell et al., 1993). To express a GFP–Met4 fusion protein from the GAL1 promoter, a GFP-encoding gene (containing the two mutations S65G and S72A) was amplified by PCR from the yGFP3 plasmid (Cormack et al., 1996) and used to replace the HA casssette present in the pGal306FluMet4 plasmid. This plasmid was then cleaved with StuI within the URA3 gene and used to transform wild-type and SCF mutant strains. Stable uracil prototroph transformants were selected and correct integration events were verified by Southern blot analysis. To construct the GFP–Met28 and GFP–Met30 fusion proteins, the same GFP-encoding gene was amplified by PCR with appropriate oligonucleotides and introduced by the gap repair method into the p314Met28 and pGal316FluMet30 plasmids. In both cases, the GFP moiety was fused in-phase at the C–terminal end of the proteins. The sequences of the oligonucleotides and the details of cloning strategies used are available upon request.
Table II. Plasmids used in this study.
| Plasmids | Relevant characteristics | Source |
|---|---|---|
| pGal306Flu | pGal1-HA(x3), URA3 | this study |
| pGal316Flu | pGal1-HA(x3), URA3, CEN | this study |
| pGal316FluMet4 | pGal1-HA-MET415–666, URA3, CEN | this study |
| pFL39FluMet4 | pGal1-HA-MET415–666, TRP1, CEN | this study |
| pGal316FluMet28 | pGal1-HA-MET281–166, URA3, CEN | this study |
| pGal316FluMet30 | pGal1-HA-MET307–640, URA3, CEN | this study |
| pGal316FluMet30ΔF | pGal1-HA-Met30Δ187–202, URA3, CEN | this study |
| pGal316FluCbf1 | pGal1-HA-CBF18–352, URA3, CEN | this study |
| pGal316FluMet32 | pGal1-HA-MET321–192, URA3, CEN | this study |
| pFL39FluMet31 | pGal1-HA-MET311–177, URA3, CEN | this study |
| pGal316GFPMet4 | pGal1-GFP–MET415–666, URA3, CEN | this study |
| p314Met28GFP | MET28-GFP, TRP1, CEN | this study |
| pGal316FluMet30GFP | pGal1-HA-MET307–640-GFP, URA3, CEN | this study |
| pEG(KT)Met30 | pGal1-GST-MET307–640, URA3, CEN | this study |
| pLexMet30 | pADH1-LEXA-MET307–640, HIS3, CEN | Thomas et al. (1995) |
| pLexMet30ΔF | pADH1-LEXA-MET30Δ187–202, HIS3, CEN | this study |
| pLexMet30ΔN | pADH1-LEXA-MET30158–640, HIS3, CEN | this study |
| pLexMet30ΔNF | pADH1-LEXA-MET30[158–187]–[202–640], HIS3, CEN | this study |
| pLexMet32 | pADH1-LEXA-MET321–191, HIS3, CEN | Blaiseau and Thomas (1998) |
| pLexMet4-1 | pADH1-LEXA-MET415–666, HIS3, CEN | Kuras and Thomas (1995) |
| pGadMet4Δ12 | pADH1-GAL4-MET4Δ79-180, LEU2, CEN | Thomas et al. (1995) |
| pGadMet4Δ3 | pADH1-GAL4-MET4Δ56-268, LEU2, CEN | Blaiseau and Thomas (1998) |
| pMT634 | pGAL1-HA-CLN2, LEU2, CEN | Willems et al. (1996) |
Northern blot analyses
Northern blotting was performed as described by Thomas (1980), with total cellular RNA extracted from yeast by the hot phenol method as described by Schmitt et al. (1990) and oligolabeled probes (Hodgson and Fisk, 1987).
Microscopy
Localization of proteins tagged with GFP was monitored with viable cells on a Nikon eclipse fluorescence microscope using an Omega XF116 filter and all the images were collected with a CCD camera using identical settings (Fluograb, Graftek, France). For the GFP–Met4 and GFP–Met30 fusion proteins, the cells were grown overnight at 28°C in raffinose-based medium to an OD650 nm of 0.6–0.8. Galactose was added at a final concentration of 4% and the cells grown for either 2 h at 28°C, or 1 h at 28°C and 1 h at 37°C for the temperature-sensitive strains. A 2 ml aliquot of cells was harvested by a brief centrifugation, resuspended in 100 μl of galactose-containing medium and 2 μl were placed on a microscope slide for observation. When required, l-methionine (1 mM) and the dye Hoechst 33342 (Sigma; 1 μg/ml) were added to the culture 15 and 20 min, respectively, prior to the observations.
Protein extraction and immunoblotting
The half-lives of Met4p and its associated factors were determined by the use of the promoter shut-off method. Various strains harboring plasmids allowing the expression of the HA-tagged proteins from the GAL1 promoter were grown at 28°C to early log phase in raffinose medium (2% raffinose). Cells were filtered and resuspended in fresh medium containing 2% galactose to induce the transcription of the gene of interest. At 30 min after galactose induction, temperature-sensitive strains were shifted to 37°C. The cells were filtered 90 min after the addition of galactose, resuspended in glucose medium to turn off the GAL1 promoter, in the presence or absence of 1 mM l-methionine, and aliquots were collected at regular intervals. In the case of the HA-Met30 fusion protein, cycloheximide (50 μg/ml) was added to the glucose-containing medium.
For the immunoblotting experiments, total cellular extracts were prepared as follows: 2 ml of cells were harvested by centrifugation, resuspended in 500 μl of 20% cold TCA and disrupted by vortexing three times for 15 s with an equal volume of 0.45–0.50 mm glass beads. The supernatant was collected and the beads washed twice with 400 μl of 5% TCA. Total supernatant was spun for 15 min at 4°C. The pellet was resuspended in 100 μl of loading buffer and boiled for 5 min. Then 20 μg of each extract were loaded onto SDS–(8–12%) polyacrylamide gels (Miniprotean II system, Bio-Rad). Proteins were transferred to a nitrocellulose membrane (Optitran, Schleicher & Schuell) by a semi-dry blotting device (Bio-Rad). Anti-HA antibodies (12CA5, Boehringer Mannheim) and peroxidase-conjugated anti-mouse antibodies (Sigma) were used at a 1:1000 dilution. Immunodetection was carried out using Supersignal (Pierce) chemiluminescent substrate.
Co-immunoprecipitations and phosphatase assays
In vivo interaction between HA-Met4 and GST–Met30 fusion proteins was assayed by co-immunoprecipitation experiments using the following protocol: after an overnight culture at 28°C in raffinose medium to early log phase, cells were filtered and resuspended in galactose medium for 2 h to induce transcription from the GAL1 promoter. Total proteins were then extracted with glass beads in buffer containing 0.3% deoxycholate, 50 mM NaCl, 50 mM Tris–HCl pH 8.0, 5 mM EDTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF) and complete-Mini antiprotease inhibitors (Boehringer Mannheim) as specified by the manufacturer. Anti-HA antibodies were pre-adsorbed on protein A–Sepharose (Pharmacia). Then 1 mg of each extract was added and gently mixed for 90 min at 4°C. The beads were washed four times with 100 mM Tris–HCl pH 8.0. Immunoprecipitated proteins were resuspended by vortexing in either SDS gel loading buffer for the immunoblots or in 100 mM Tris–HCl pH 8.0 for the phosphatase assays. Assays of Met30–Skp1 interactions were performed according to Willems et al. (1996).
For the phosphatase assays, immunoprecipitation of HA-Met4 or HA-Cln2 was performed as described above except that in addition the extraction buffer contained 1% Triton and 0.2% SDS. Immunoprecipitates were incubated for 15 min at 37°C in the presence of 20 or 40 U of alkaline phosphatase (Boehringer Mannheim). SDS loading buffer (4×) was added to stop the reaction, and samples were processed for immunoblotting.
Electrophoretic mobility shift assays
The cellular extracts were prepared as described (Blaiseau and Thomas, 1998). The binding reaction mixtures (20 μl volume) contained 25 mM HEPES pH 7.6, 60 mM KCl, 7.5% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 5 mM MgCl2 and 0.8 μg of poly(dI–dC)–poly(dI–dC). The amount of protein used in each binding reaction is indicated in the figure legends. Probes were labeled by filling in the ends with the Klenow fragment and using [α-32P]dATP (800 Ci/mmol; Amersham). Approximately 40 000 c.p.m. of probe (∼0.5 ng) were used in each binding mixture. Samples were incubated for 30 min in ice before being loaded onto a 5% polyacrylamide gel in 0.25× TBE (22 mM Tris pH 8.3, 22 mM boric acid, 0.6 mM EDTA) and electrophoresed at 15 V/cm at 7°C. Gels were pre-electrophoresed for 1 h at 7.5 V/cm at 7°C. Gels were run for 6 h, dried and autoradiographed for 15 h with an intensifying screen.
Acknowledgments
Acknowledgements
We are grateful to Carl Mann for helpful discussions and for providing us with mutant strains. A.R. is supported by a thesis fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur. E.P. is a recipient of a National Sciences and Engineering Research Council of Canada studentship. M.T. is supported by the National Cancer Institute of Canada and the Medical Research Council of Canada. D.T. is supported by the Centre National de la Recherche Scientifique and the Association de la Recherche sur le Cancer.
References
- Bai C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W. and Elledge, S.J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Blaiseau P.L. and Thomas, D. (1998) Multiple transcription activation complexes tether the yeast activator Met4 to DNA. EMBO J., 17, 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chérest H. and Surdin-Kerjan, Y. (1992) Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics, 130, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia, R.H. and Falkow, S. (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Feldman R.M.R., Correll, C.C., Kaplan, K.B. and Deshaies, R.J. (1997) A complex of Cdc4p, Skp1p and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell, 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy, A. and Mann, C. (1993) S.cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature, 366, 358–362. [DOI] [PubMed] [Google Scholar]
- Gietz D., St. Jean, A., Woods, R.A. and Schiestl, R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag, E., Martinez-Pastor, M.T., Estruch, F., Ammerer, G., Hamilton, B., Ruis, H. and Schuller, C. (1998) Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev., 12, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W. and Elledge,S.J. (1999) Skipping into the E2F1-destruction pathway. Nature Cell Biol., 1, E5–E7. [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller, H., Elias, S. and Ciechanover, A. (1983) Components of ubiquitin–protein ligase system. Resolution, affinity purification and role in protein breakdown. J. Biol. Chem., 258, 8206–8214. [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hodgson C.P. and Fisk, R.Z. (1987) Hybridization probe size control: optimized ‘oligolabelling’. Nucleic Acids Res., 15, 6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R., Swanson, R., Rakhilina, L. and Hochstrasser, M. (1998) Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin–proteasome pathway. Cell, 94, 217–227. [DOI] [PubMed] [Google Scholar]
- Kaiser P., Sia, R.A., Bardes, E.G., Lew, D.J. and Reed, S.I. (1998) Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev., 12, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K.B., Hyman, A.A. and Sorger, P.K. (1997) Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell, 91, 491–500. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U. and Jentsch, S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Koepp D.M., Harper, J.W. and Elledge, S.J. (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell, 97, 431–434. [DOI] [PubMed] [Google Scholar]
- Komeili A. and O'Shea, E.K. (1999) Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science, 284, 977–980. [DOI] [PubMed] [Google Scholar]
- Kuras L., Barbey, R. and Thomas, D. (1997) Assembly of a bZIP/bHLH transcription activation complex: formation of the yeast Cbf1–Met4–Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J., 16, 2441–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L. and Thomas, D. (1995) Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol. Cell. Biol., 15, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M. and Estelle, M. (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev., 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker S., Valdivieso, M.H. and Wittenberg, C. (1996) Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science, 271, 1597–1601. [DOI] [PubMed] [Google Scholar]
- Li F.N. and Johnston, M. (1997) Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J., 16, 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Doenges, G., Matuschewski, K. and Jentsch, S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J., 17, 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias N., Johnson, S., Byers, B. and Goebl, M. (1999) The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol. Cell. Biol., 19, 1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.A., Marshall, T.K. and Deschenes, R.J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- Nehlin J. and Ronne, H. (1990) Yeast Mig1 repressor is related to the mammalian early growth response and Wilm's tumor finger protein. EMBO J., 9, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl H.L. and Baeuerle, P.A. (1996) Control of gene expression by proteolysis. Curr. Opin. Cell Biol., 8, 340–347. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Willems, A.R., Sa, D., Kuras, L., Thomas, D., Craig, K.L. and Tyers, M. (1998a) Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev., 12, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E.E., Willems, A.R. and Tyers, M. (1998b) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet., 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Peyraud, C., Rouillon, A., Surdin-Kerjan, Y., Tyers, M. and Thomas, D. (1999) manuscript in preparation. [DOI] [PMC free article] [PubMed]
- Russell I.D., Grancell, A.S. and Sorger, P.K. (1999) The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol., 145, 933–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M., Brown, T. and Trumpower, B. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol J.H. et al. (1999) Cdc53/cullin and the essential hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme cdc34. Genes Dev., 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink, G.R. and Hicks, J.B. (1979) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sikorski R.S. and Hieter, P. (1989) A system of shuttle vectors and yeast strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J. and Harper, J.W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Koepp, D.M., Kamura, T., Conrad, M.N., Conaway, R.C., Conaway, J.W., Elledge, S.J. and Harper, J.W. (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science, 284, 662–665. [DOI] [PubMed] [Google Scholar]
- Solow S.P., Lezina, L. and Lieberman, P.M. (1999) Phosphorylation of TFIIA stimulates TATA binding protein–TATA interaction and contributes to maximal transcription and viability in yeast. Mol. Cell. Biol., 19, 2846–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D. and Futcher, B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes, J.D., Campbell, M.J., Cho, R.J. and Church, G.M. (1999) Systematic determination of genetic network architecture. Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay P. Morris, H.R., Dell, A., Panico, M., Paxton, T. and West, C.M. (1998) The cytoplasmic F-box binding protein SKP1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem., 273, 18242–18249. [DOI] [PubMed] [Google Scholar]
- Thomas D., and Surdin-Kerjan, Y. (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 61, 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Rothstein, R., Rosenberg, N. and Surdin-Kerjan, Y. (1988) SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol. Cell. Biol., 8, 5132–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Kuras, L., Barbey, R., Cherest, H., Blaiseau, P.L. and Surdin-Kerjan, Y. (1995) Met30, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol. Cell. Biol., 15, 6526–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.S. (1980) Hybridization of denatured DNA and small DNA fragments transferred to nitrocellulose. Proc. Natl Acad. Sci. USA, 77, 5201–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A.R., Lanker, S., Patton, E.E., Craig, K.L., Nason, T.F., Mathias, N., Kobayashi, R., Wittenberg, C. and Tyers, M. (1996) Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell, 86, 453–463. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack, P., Beer-Romero, P., Chu, C.Y., Elledge, S.J. and Harper, J.W. (1999) The SCFβ–TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev., 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. and Howley, P.M. (1998) Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell, 2, 571–580. [DOI] [PubMed] [Google Scholar]